Protective Effects and Mechanism of a Novel Probiotic Strain Ligilactobacillus salivarius YL20 against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cell Culture
2.2. Intestinal Organoid Culture
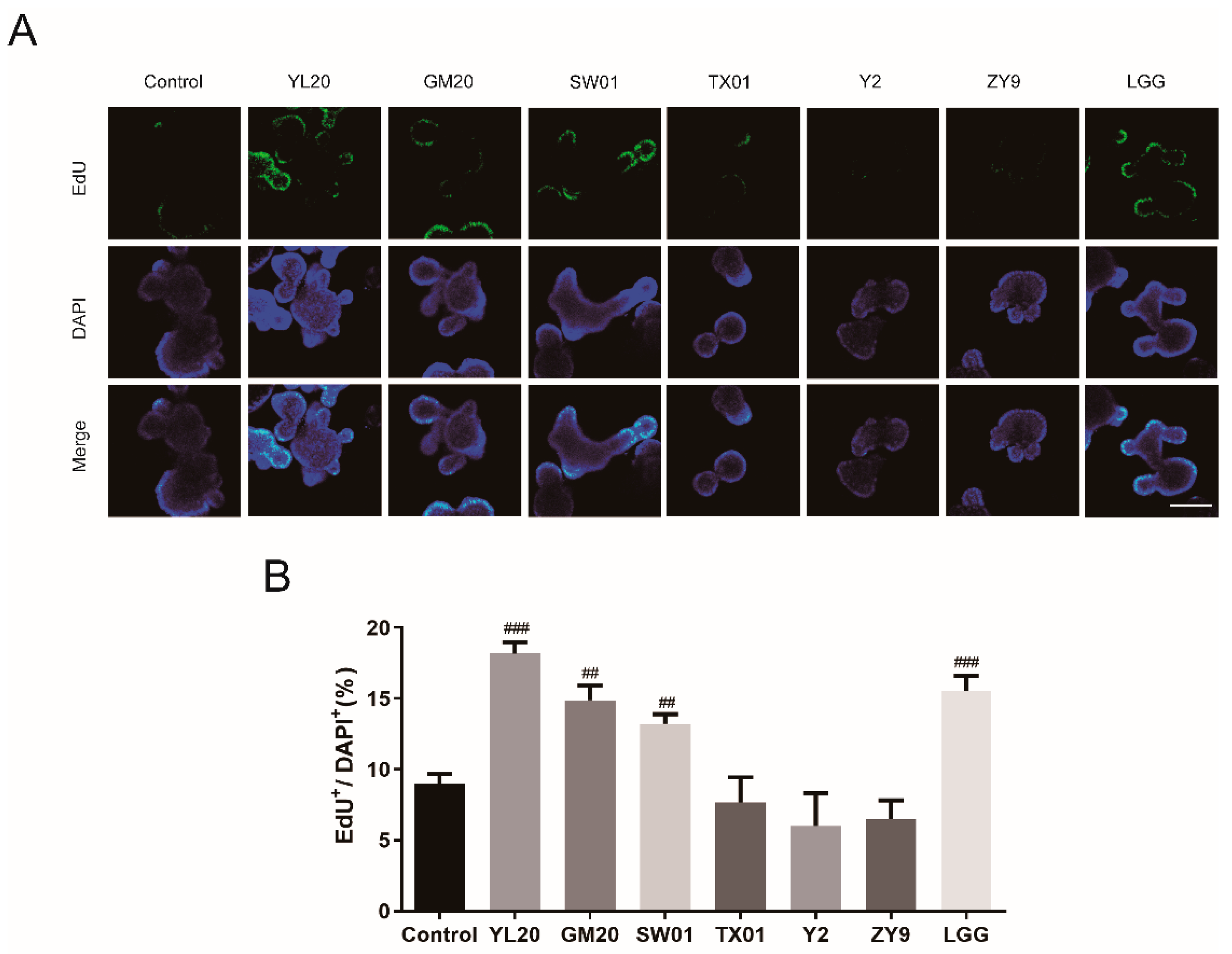
2.3. EdU Staining
2.4. Quantitative Real-Time PCR (qRT-PCR)
2.5. Enzyme Linked Immunosorbent Assay (ELISA)
2.6. Antagonistic Activity of Probiotics against C. sakazakii Using Agar Well Diffusion Method
2.7. Sensitivity to Antibiotics
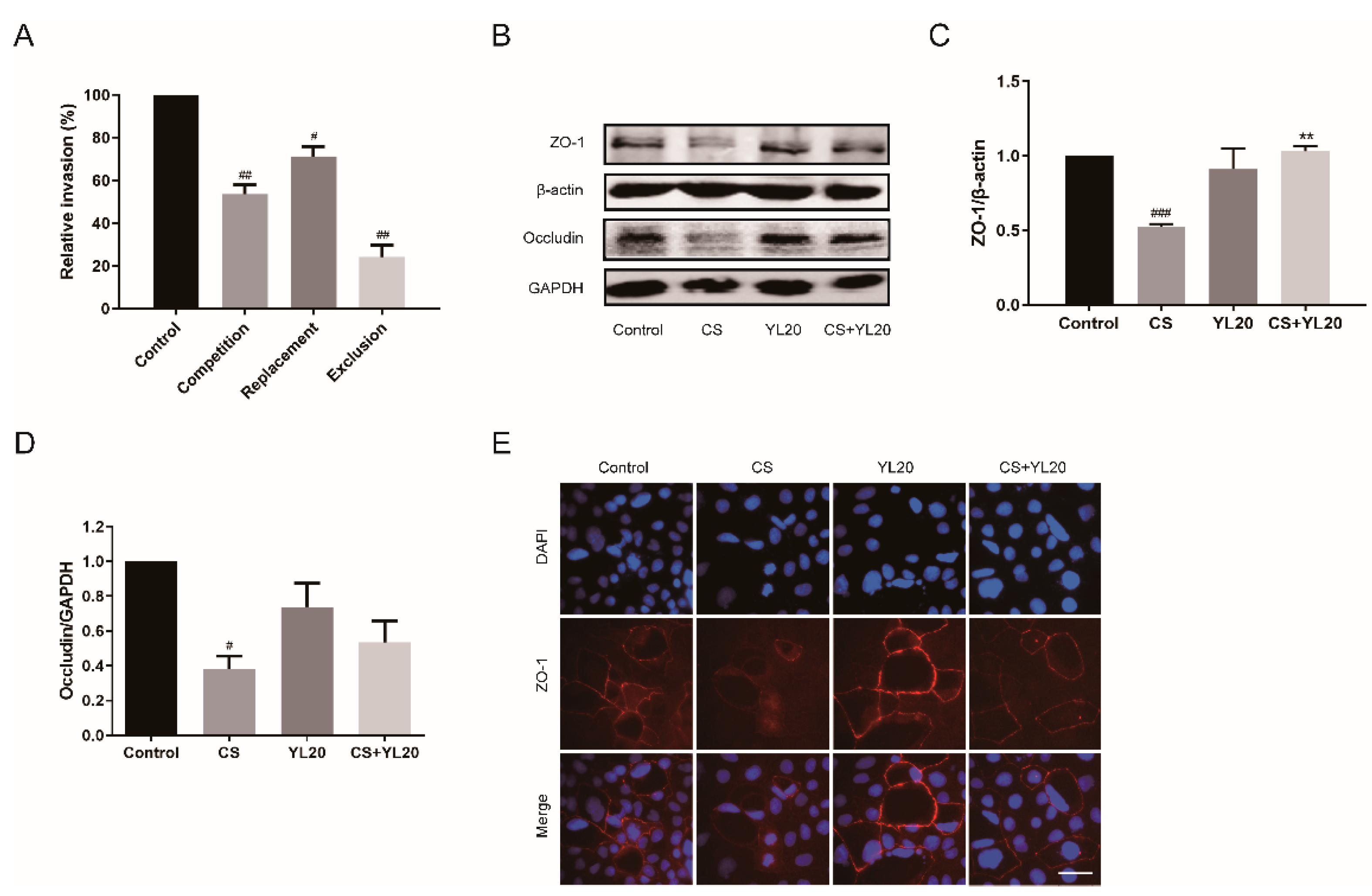
2.8. Invasion Assay
2.9. Western Blotting Analysis
2.10. Immunofluorescence Analysis
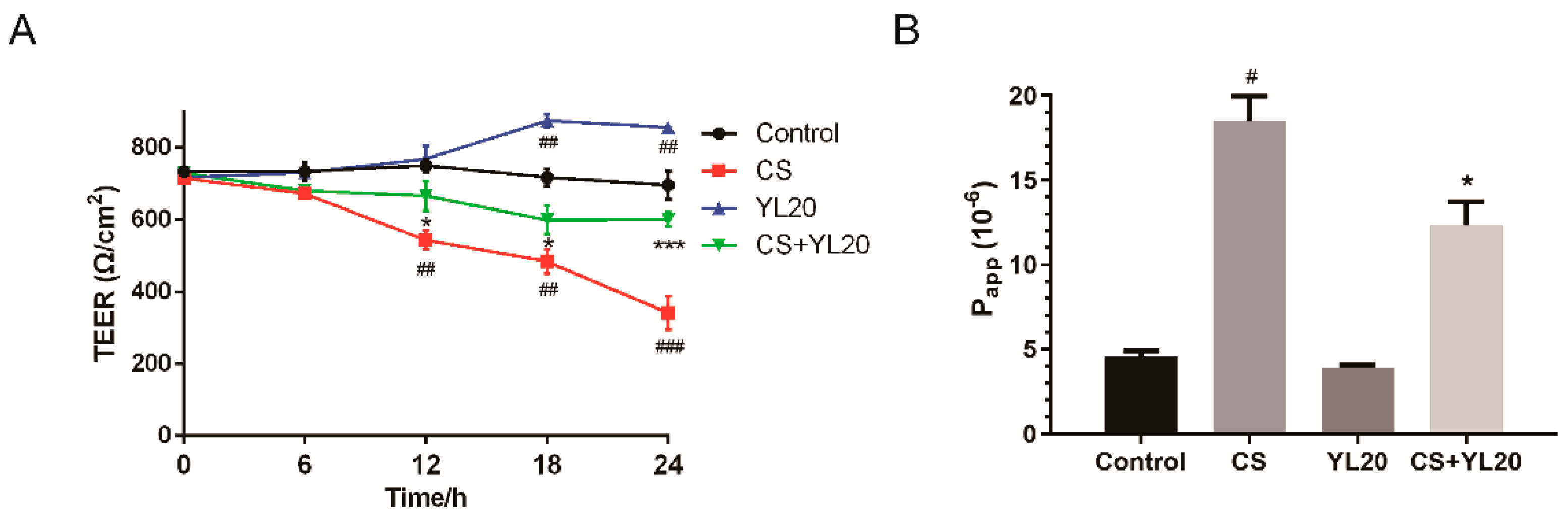
2.11. Measurement of Transepithelial Electrical Resistance (TEER) and Permeability across Caco-2 Monolayers
2.12. Animal Experiment
2.13. H&E Staining, PAS Staining, and Immunohistochemical Staining
2.14. Quantification of the Predominant Bacterial Groups in the Intestinal Contents
2.15. Statistical Analysis
3. Results
3.1. Effects of Lactic Acid Bacteria on Intestinal Organoid Proliferation
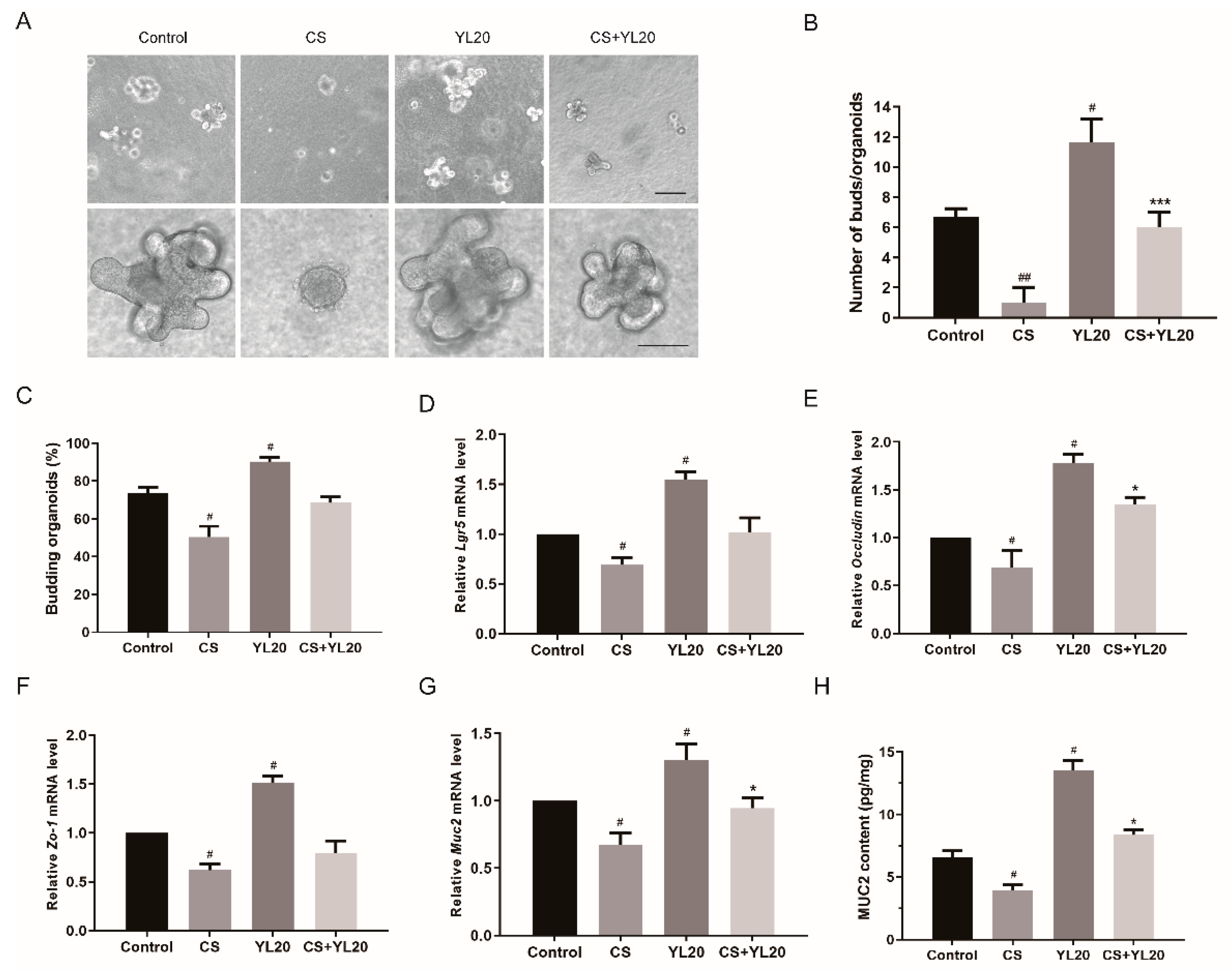
3.2. L. salivarius YL20 Attenuates C. sakazakii-Induced Damage of Intestinal Organoids by Promoting ISC-Mediated Epithelial Cell Proliferation and Enhancing Intestinal Barrier Function
3.3. L. salivarius YL20 Inhibits C. sakazakii Adherence to Intestinal Cells and Enhances the Expression of TJ Proteins in Intestinal Epithelial Cells
3.4. L. salivarius YL20 Reverses TEER Decrease and Corresponding Membrane Permeability Increase across C. sakazakii-Infected Caco-2 Monolayers
3.5. YL20 Administration Attenuates C. sakazakii-Induced Clinical Symptoms and Intestinal Epithelial Damage in Newborn Mice
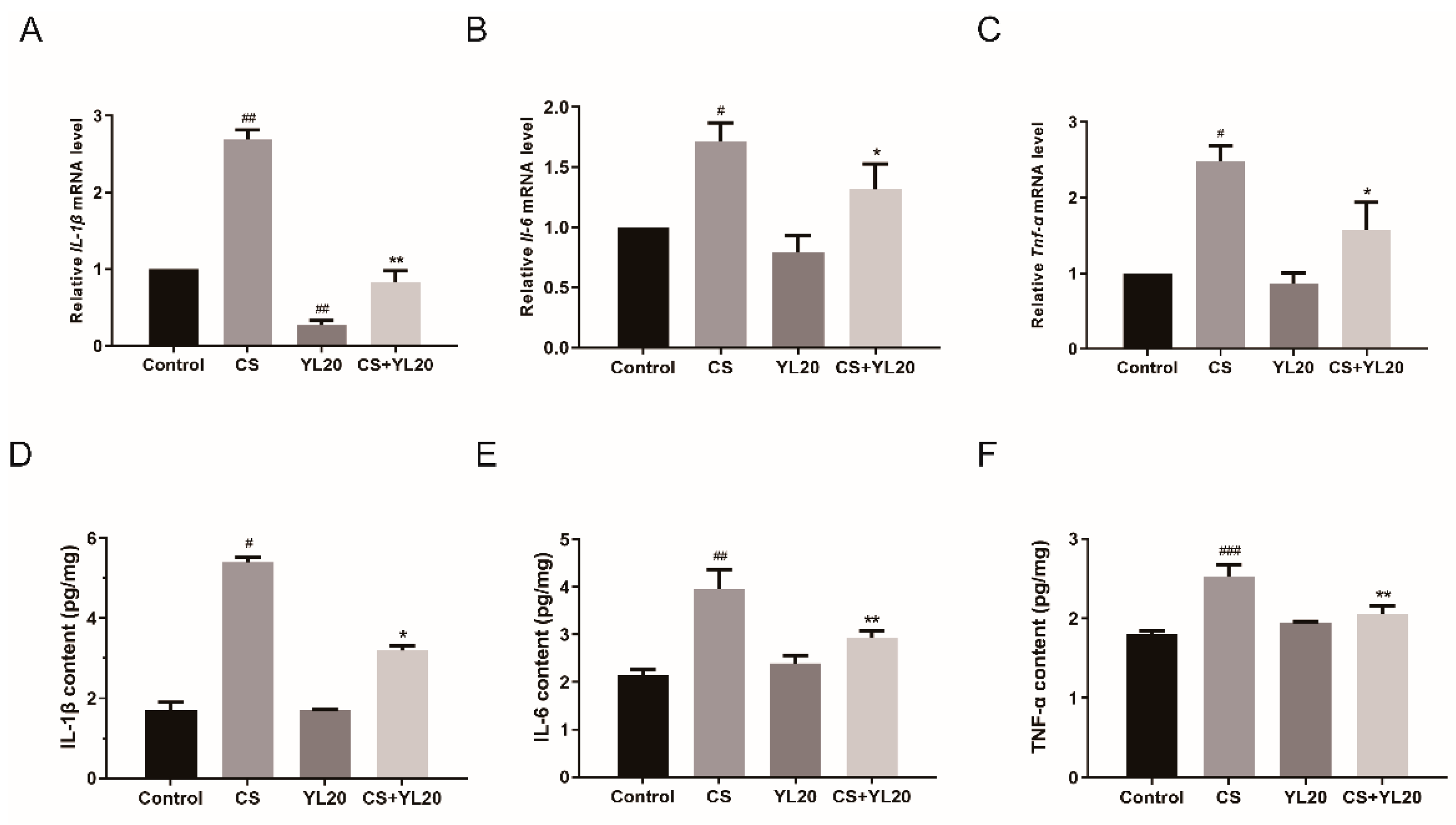
3.6. YL20 Administration Inhibits C. sakazakii-Induced Rise of Inflammatory Factors in the Intestinal Tract of Newborn Mice
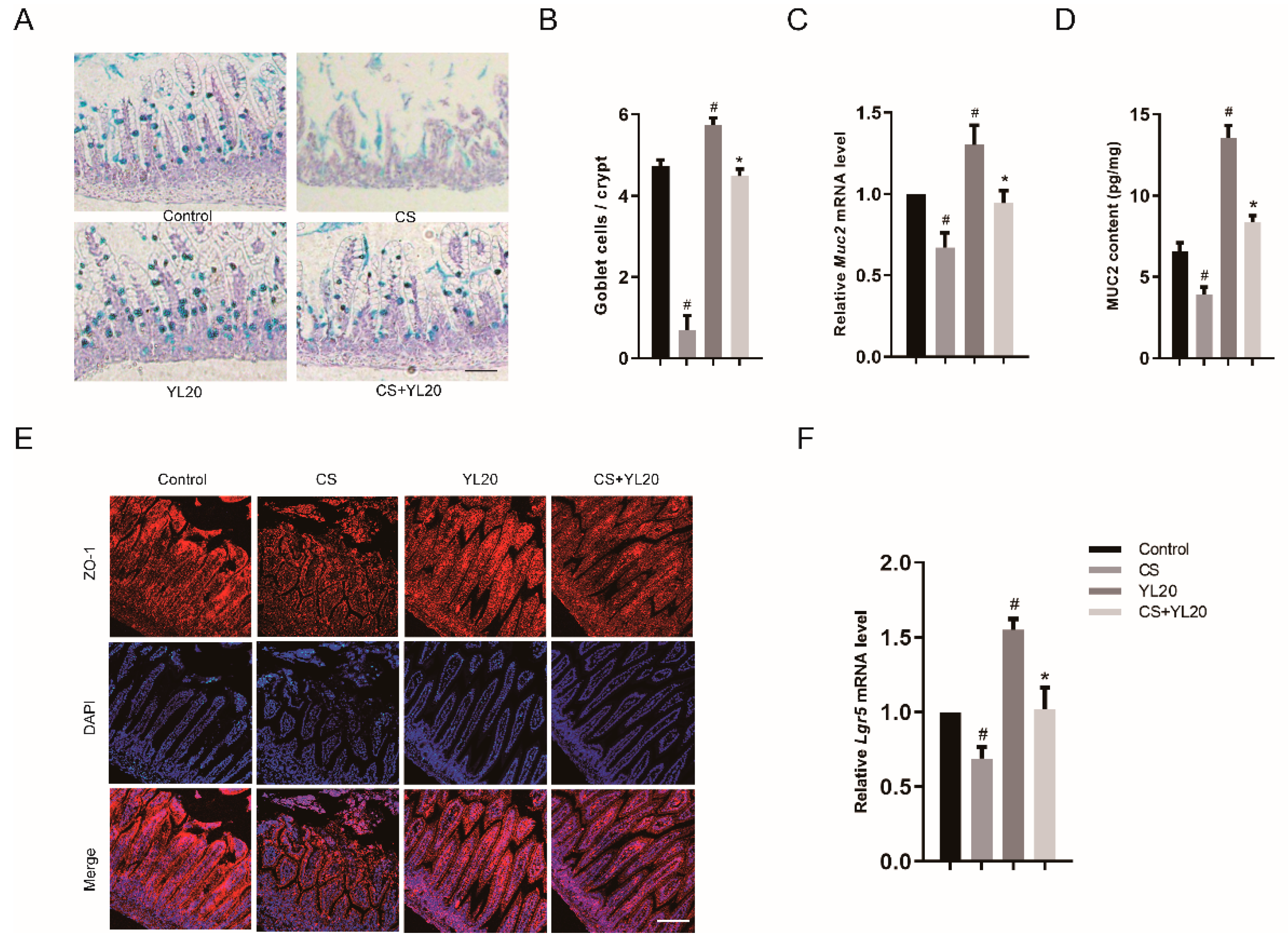
3.7. YL20 Administration Improves Intestinal Barrier Integrity and Inhibits C. sakazakii-Induced Intestinal Barrier Damage
3.8. YL20 Administration Improves Intestinal Microbiota Composition
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samuels, N.; van de Graaf, R.; Been, J.V.; de Jonge, R.C.; Hanff, L.M.; Wijnen, R.M.; Kornelisse, R.F.; Reiss, I.K.; Vermeulen, M.J. Necrotising enterocolitis and mortality in preterm infants after introduction of probiotics: A quasi-experimental study. Sci. Rep. 2016, 6, 31643. [Google Scholar] [CrossRef] [PubMed]
- Battersby, C.; Santhalingam, T.; Costeloe, K.; Modi, N. Incidence of neonatal necrotising enterocolitis in high-income countries: A systematic review. Arch. Dis. Child. Fetal Neonatal Ed. 2018, 103, F182–F189. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.J.; Podd, B.; Ford, H.R.; Camerini, V. Evidence vs. experience in neonatal practices in necrotizing enterocolitis. J. Perinatol. 2008, 28, S9–S13. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, P.W.; Nasr, T.R.; Stoll, B.J. Necrotizing enterocolitis: Recent scientific advances in pathophysiology and prevention. Semin. Perinatol. 2008, 32, 70–82. [Google Scholar] [CrossRef]
- Fan, H.; Chen, Z.; Lin, R.; Liu, Y.; Wu, X.; Puthiyakunnon, S.; Wang, Y.; Zhu, B.; Zhang, Q.; Bai, Y.; et al. Bacteroides fragilis Strain ZY-312 Defense against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and in a Neonatal Rat Model. mSystems 2019, 4, e00305-19. [Google Scholar] [CrossRef]
- Nanthakumar, N.N.; Fusunyan, R.D.; Sanderson, I.; Walker, W.A. Inflammation in the developing human intestine: A possible pathophysiologic contribution to necrotizing enterocolitis. Proc. Natl. Acad. Sci. USA 2000, 97, 6043–6048. [Google Scholar] [CrossRef]
- Henry, M.; Fouladkhah, A. Outbreak History, Biofilm Formation, and Preventive Measures for Control of Cronobacter sakazakii in Infant Formula and Infant Care Settings. Microorganisms 2019, 7, 77. [Google Scholar] [CrossRef]
- Bowen, A.B.; Braden, C.R. Invasive Enterobacter sakazakii disease in infants. Emerg. Infect. Dis. 2006, 12, 1185–1189. [Google Scholar] [CrossRef]
- Ke, A.; Parreira, V.R.; Goodridge, L.; Farber, J.M. Current and Future Perspectives on the Role of Probiotics, Prebiotics, and Synbiotics in Controlling Pathogenic Cronobacter Spp. in Infants. Front. Microbiol. 2021, 12, 755083. [Google Scholar] [CrossRef]
- Holý, O.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Hochel, I.; Parra-Flores, J.; Petrželová, J.; Fačevicová, K.; Forsythe, S.; Alsonosi, A. Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microb. Pathog. 2019, 127, 250–256. [Google Scholar] [CrossRef]
- Seghesio, E.; De Geyter, C.; Vandenplas, Y. Probiotics in the Prevention and Treatment of Necrotizing Enterocolitis. Pediatr. Gastroenterol. Hepatol. Nutr. 2021, 24, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.C.; Hsu, C.H.; Chen, H.L.; Chung, M.Y.; Hsu, J.F.; Lien, R.I.; Tsao, L.Y.; Chen, C.H.; Su, B.H. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: A multicenter, randomized, controlled trial. Pediatrics 2008, 122, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Braga, T.D.; da Silva, G.A.; de Lira, P.I.; de Carvalho Lima, M. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: A double-blind, randomized, controlled trial. Am. J. Clin. Nutr. 2011, 93, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Gurien, L.A.; Stallings-Archer, K.; Smith, S.D. Probiotic Lactococcus lactis decreases incidence and severity of necrotizing enterocolitis in a preterm animal model. J. Neonatal. Perinatal. Med. 2018, 11, 65–69. [Google Scholar] [CrossRef]
- Weng, M.; Ganguli, K.; Zhu, W.; Shi, H.N.; Walker, W.A. Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G779–G787. [Google Scholar] [CrossRef]
- Williams, J.M.; Duckworth, C.A.; Burkitt, M.D.; Watson, A.J.; Campbell, B.J.; Pritchard, D.M. Epithelial cell shedding and barrier function: A matter of life and death at the small intestinal villus tip. Vet. Pathol. 2015, 52, 445–455. [Google Scholar] [CrossRef]
- Hollister, E.B.; Gao, C.; Versalovic, J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology 2014, 146, 1449–1458. [Google Scholar] [CrossRef]
- Sun, J. Intestinal organoid as an in vitro model in studying host-microbial interactions. Front. Biol. 2017, 12, 94–102. [Google Scholar] [CrossRef]
- Lee, S.B.; Han, S.H.; Park, S. Long-Term Culture of Intestinal Organoids. Methods Mol. Biol. 2018, 1817, 123–135. [Google Scholar]
- Hou, Q.; Ye, L.; Liu, H.; Huang, L.; Yang, Q.; Turner, J.R.; Yu, Q. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018, 25, 1657–1670. [Google Scholar] [CrossRef]
- Deng, K.; Chen, T.; Wu, Q.; Xin, H.; Wei, Q.; Hu, P.; Wang, X.; Wang, X.; Wei, H.; Shah, N.P. In vitro and in vivo examination of anticolonization of pathogens by Lactobacillus paracasei FJ861111.1. J. Dairy Sci. 2015, 98, 6759–6766. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Tao, X.; Wan, C.; Li, S.; Xu, H.; Xu, F.; Shah, N.P.; Wei, H. In vitro probiotic characteristics of Lactobacillus plantarum ZDY 2013 and its modulatory effect on gut microbiota of mice. J. Dairy Sci. 2015, 98, 5850–5861. [Google Scholar] [CrossRef] [PubMed]
- Béduneau, A.; Tempesta, C.; Fimbel, S.; Pellequer, Y.; Jannin, V.; Demarne, F.; Lamprecht, A. A tunable Caco-2/HT29-MTX co-culture model mimicking variable permeabilities of the human intestine obtained by an original seeding procedure. Eur. J. Pharm. Biopharm. 2014, 87, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Bian, G.; Su, Y.; Zhu, W. Comparison of faecal microbial community of lantang, bama, erhualian, meishan, xiaomeishan, duroc, landrace, and yorkshire sows. Asian-Australas. J. Anim. Sci. 2014, 27, 898–906. [Google Scholar] [CrossRef]
- Kang, M.; Mischel, R.A.; Bhave, S.; Komla, E.; Cho, A.; Huang, C.; Dewey, W.L.; Akbarali, H.I. The effect of gut microbiome on tolerance to morphine mediated antinociception in mice. Sci. Rep. 2017, 7, 42658. [Google Scholar] [CrossRef] [PubMed]
- Steed, H.; Macfarlane, G.T.; Blackett, K.L.; Macfarlane, S.; Miller, M.H.; Bahrami, B.; Dillon, J.F. Bacterial translocation in cirrhosis is not caused by an abnormal small bowel gut microbiota. FEMS Immunol. Med. Microbiol. 2011, 63, 346–354. [Google Scholar] [CrossRef]
- Bartosch, S.; Fite, A.; Macfarlane, G.T.; McMurdo, M.E. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 2004, 70, 3575–3581. [Google Scholar] [CrossRef]
- Rinttilä, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef]
- Wise, M.G.; Siragusa, G.R. Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl. Environ. Microbiol. 2005, 71, 3911–3916. [Google Scholar] [CrossRef]
- Giri, C.P.; Shima, K.; Tall, B.D.; Curtis, S.; Sathyamoorthy, V.; Hanisch, B.; Kim, K.S.; Kopecko, D.J. Cronobacter spp. (previously Enterobacter sakazakii) invade and translocate across both cultured human intestinal epithelial cells and human brain microvascular endothelial cells. Microb. Pathog. 2012, 52, 140–147. [Google Scholar] [CrossRef]
- Li, A.; Jiang, X.; Wang, Y.; Zhang, L.; Zhang, H.; Mehmood, K.; Li, Z.; Waqas, M.; Li, J. The impact of Bacillus subtilis 18 isolated from Tibetan yaks on growth performance and gut microbial community in mice. Microb. Pathog. 2019, 128, 153–161. [Google Scholar] [CrossRef]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef]
- Yan, F.; Liu, L.; Cao, H.; Moore, D.J.; Washington, M.K.; Wang, B.; Peek, R.M.; Acra, S.A.; Polk, D.B. Neonatal colonization of mice with LGG promotes intestinal development and decreases susceptibility to colitis in adulthood. Mucosal Immunol. 2017, 10, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Aoki-Yoshida, A.; Saito, S.; Fukiya, S.; Aoki, R.; Takayama, Y.; Suzuki, C.; Sonoyama, K. Lactobacillus rhamnosus GG increases Toll-like receptor 3 gene expression in murine small intestine ex vivo and in vivo. Benef. Microbes 2016, 7, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, T.Y.; Kim, Y.; Lee, S.H.; Kim, S.; Kang, S.W.; Yang, J.Y.; Baek, I.J.; Sung, Y.H.; Park, Y.Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef] [PubMed]
- Sittipo, P.; Pham, H.Q.; Park, C.E.; Kang, G.U.; Zhi, Y.; Ji, H.J.; Jang, A.; Seo, H.S.; Shin, J.H.; Lee, Y.K. Irradiation-Induced Intestinal Damage Is Recovered by the Indigenous Gut Bacteria Lactobacillus acidophilus. Front. Cell. Infect. Microbiol. 2020, 10, 415. [Google Scholar] [CrossRef]
- Mack, D.R.; Michail, S.; Wei, S.; McDougall, L.; Hollingsworth, M.A. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 1999, 276, G941–G950. [Google Scholar]
- Mack, D.R.; Ahrne, S.; Hyde, L.; Wei, S.; Hollingsworth, M.A. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 2003, 52, 827–833. [Google Scholar] [CrossRef]
- Lutgendorff, F.; Akkermans, L.M.; Söderholm, J.D. The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr. Mol. Med. 2008, 8, 282–298. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Ahl, D.; Liu, H.; Schreiber, O.; Roos, S.; Phillipson, M.; Holm, L. Lactobacillus reuteri increases mucus thickness and ameliorates dextran sulphate sodium-induced colitis in mice. Acta Physiol. (Oxf.) 2016, 217, 300–310. [Google Scholar] [CrossRef] [PubMed]
- González-Mariscal, L.; Betanzos, A.; Nava, P.; Jaramillo, B.E. Tight junction proteins. Prog. Biophys. Mol. Biol. 2003, 81, 1–44. [Google Scholar] [CrossRef]
- Qiao, J.; Sun, Z.; Liang, D.; Li, H. Lactobacillus salivarius alleviates inflammation via NF-κB signaling in ETEC K88-induced IPEC-J2 cells. J. Anim. Sci. Biotechnol. 2020, 11, 76. [Google Scholar] [CrossRef]
- Mange, J.P.; Stephan, R.; Borel, N.; Wild, P.; Kim, K.S.; Pospischil, A.; Lehner, A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 2006, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Campana, R.; van Hemert, S.; Baffone, W. Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 2017, 9, 12. [Google Scholar] [CrossRef]
- Collado, M.C.; Isolauri, E.; Salminen, S. Specific probiotic strains and their combinations counteract adhesion of Enterobacter sakazakii to intestinal mucus. FEMS Microbiol. Lett. 2008, 285, 58–64. [Google Scholar] [CrossRef]
- Jariwala, R.; Mandal, H.; Bagchi, T. Indigenous lactobacilli strains of food and human sources reverse enteropathogenic E. coli O26:H11-induced damage in intestinal epithelial cell lines: Effect on redistribution of tight junction proteins. Microbiology (Reading) 2017, 163, 1263–1272. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Rocha-Ramírez, L.M.; Ochoa, S.A.; González-Pedrajo, B.; Espinosa, N.; Eslava, C.; Hernández-Chiñas, U.; Mendoza-Hernández, G.; Rodríguez-Leviz, A.; Valencia-Mayoral, P.; et al. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 2012, 7, e52091. [Google Scholar] [CrossRef]
- Jin, T.; Guan, N.; Du, Y.; Zhang, X.; Li, J.; Xia, X. Cronobacter sakazakii ATCC 29544 Translocated Human Brain Microvascular Endothelial Cells via Endocytosis, Apoptosis Induction, and Disruption of Tight Junction. Front. Microbiol. 2021, 12, 675020. [Google Scholar] [CrossRef]
- Ke, A.; Parreira, V.R.; Farber, J.M.; Goodridge, L. Inhibition of Cronobacter sakazakii in an infant simulator of the human intestinal microbial ecosystem using a potential synbiotic. Front. Microbiol. 2022, 13, 947624. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Yang, I.; Corwin, E.J.; Brennan, P.A.; Jordan, S.; Murphy, J.R.; Dunlop, A. The infant microbiome: Implications for infant health and neurocognitive development. Nurs. Res. 2016, 65, 76. [Google Scholar] [CrossRef] [PubMed]
- Cukrowska, B.; Bierła, J.B.; Zakrzewska, M.; Klukowski, M.; Maciorkowska, E. The relationship between the infant gut microbiota and allergy. The role of Bifidobacterium breve and prebiotic oligosaccharides in the activation of anti-allergic mechanisms in early life. Nutrients 2020, 12, 946. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequence |
|---|---|
| Tnf-α | GCTCTGTGAAGGGAATGGGTGTT GTCCAGGTCACTGTCCCAGCATC |
| Il-6 | ACTTCCATCCAGTTGCCTTCTTG TGTTGGGAGTGGTATCCTCTGTG |
| Il-1β | CAGAGTTCCCCAACTGGTACATC GGGAAGGCATTAGAAACAGTCC |
| Gapdh | TGCCCAGAACATCATCCCT TCCTCAGTGTAGCCCAAG |
| Muc2 | GACGGCGATGTCTACCGATT TCCTTGTAGGAGTCTCGGCA |
| Lgr5 | CCTACTCGAAGACTTACCCAGT GCATTGGGGTGAATGATAGCA |
| Zo-1 | GTTGGTACGGTGCCCTGAAAGA GCTGACAGGTAGGACAGACGAT |
| Occludin | TGGCAAGCGATCATACCCAGAG CTGCCTGAAGTCATCCACACTC |
| Target Organism | Oligonucleotide Sequence (5′–3′) | Reference |
|---|---|---|
| Total bacterial | F′-CGGTGAATACGTTCYCGG R′-GGWTACCTTGTTACGACTT | [24] |
| Bacteroidetes | F′-GGTGTCGGCTTAAGTGCCAT R′-CGGAYGTAAGGGCCGTGC | [25] |
| Lactobacillus | F′-AGCAGTAGGGAATCTTCCA R′-CACCGCTACACATGGAG | [26] |
| Bifidobacterium | F′-AGGGTTCGATTCTGGCTCAG R′-CATCCGGCATTACCACCC | [26] |
| Enterobacteriaceae | F′-CATTGACGTTACCCGCAGAAGAAG R′-CTCTACGAGACTCAAGCTTGC | [27] |
| Enterococcus | F′-CCCTTATTGTTAGTTGCCATCAT R′-ACTCGTTGTACTTCCCATTGT | [28] |
| Clostridium perfringens | F′-CGCATAACGTTGAAAGATGG R′-CCTTGGTAGGCCGTTACCC | [29] |
| Bacterial Strain | Diameter (mm) |
|---|---|
| Lacticaseibacillus salivarius YL20 | 12.5 a ± 0.3 |
| Lactobacillus gasseri GM20 | 12.1 a ± 0.8 |
| Lacticaseibacillus rhamnosus SW01 | 11.1 a ± 0.7 |
| Lacticaseibacillus rhamnosus GG | 11.9 a ± 0.7 |
| Antibiotic | Content | Inhibition Zone Diameter (mm) | Sensitivity |
|---|---|---|---|
| Penicillin | 10 IU | 7.1 ± 0.2 | R |
| Streptomycin | 10 μg | 10.3 ± 0.3 | R |
| Gentamicin | 10 μg | 17.2 ± 0.6 | S |
| kanamycin | 30 μg | 19.2 ± 0.8 | S |
| Tetracycline | 30 μg | 35.5 ± 0.5 | S |
| Chloramphenicol | 30 μg | 26.3 ± 0.8 | S |
| Ciprofloxacin | 5 μg | 27.9 ± 0.9 | S |
| Rifampicin | 5 μg | 32.5 ± 0.7 | S |
| Cephalothi | 30 μg | 30.2 ± 0.8 | S |
| Cefotaxime Sodium | 30 μg | 34.8 ± 0.6 | S |
| Ceftazidime | 30 μg | 22.5 ± 0.7 | S |
| Erythromycin | 15 μg | 24.0 ± 0.7 | S |
| Vancomycin | 30 μg | 15.8 ± 0.8 | S |
| Cotrimoxazole | 1.25 μg | 20.3 ± 0.8 | S |
| Imipenem | 10 μg | 36.0 ± 0.8 | S |
| Target Organism | Control | CS | YL20 | CS + YL20 |
|---|---|---|---|---|
| Total bacterial | 42.45 ± 0.33 a | 42.16 ± 0.20 a | 42.39 ± 0.38 a | 41.97 ± 0.19 a |
| Bacteroidetes | 4.98 ± 0.32 a | 3.95 ± 0.13 b | 4.22 ± 0.17 ab | 4.01 ± 0.14 ab |
| Lactobacillus | 7.17 ± 0.12 b | 5.87 ± 0.07 c | 8.32 ± 0.05 a | 7.33 ± 0.04 b |
| Bifidobacterium | 5.68 ± 0.12 a | 4.32 ± 0.13 b | 5.35 ± 0.25 ab | 5.13 ± 0.19 ab |
| Enterobacteriaceae | 5.20 ± 0.04 b | 6.30 ± 0.08 a | 4.28 ± 0.08 c | 5.33 ± 0.05 b |
| Enterococcus | 5.53 ± 0.11 b | 6.69 ± 0.23 a | 4.93 ± 0.15 c | 5.81 ± 0.32 ab |
| Clostridium perfringens | 7.10 ± 0.16 a | 7.31 ± 0.20 a | 6.93 ± 0.28 a | 7.45 ± 0.13 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Geng, M.; Zhu, C.; Huang, L.; Zhang, Y.; Zhang, T.; Zhao, C.; Zhang, T.; Du, X.; Wang, N. Protective Effects and Mechanism of a Novel Probiotic Strain Ligilactobacillus salivarius YL20 against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and In Vivo. Nutrients 2022, 14, 3827. https://doi.org/10.3390/nu14183827
Wang W, Geng M, Zhu C, Huang L, Zhang Y, Zhang T, Zhao C, Zhang T, Du X, Wang N. Protective Effects and Mechanism of a Novel Probiotic Strain Ligilactobacillus salivarius YL20 against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and In Vivo. Nutrients. 2022; 14(18):3827. https://doi.org/10.3390/nu14183827
Chicago/Turabian StyleWang, Weiming, Meng Geng, Caixia Zhu, Lei Huang, Yue Zhang, Tengxun Zhang, Chongjie Zhao, Tongcun Zhang, Xinjun Du, and Nan Wang. 2022. "Protective Effects and Mechanism of a Novel Probiotic Strain Ligilactobacillus salivarius YL20 against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and In Vivo" Nutrients 14, no. 18: 3827. https://doi.org/10.3390/nu14183827
APA StyleWang, W., Geng, M., Zhu, C., Huang, L., Zhang, Y., Zhang, T., Zhao, C., Zhang, T., Du, X., & Wang, N. (2022). Protective Effects and Mechanism of a Novel Probiotic Strain Ligilactobacillus salivarius YL20 against Cronobacter sakazakii-Induced Necrotizing Enterocolitis In Vitro and In Vivo. Nutrients, 14(18), 3827. https://doi.org/10.3390/nu14183827







