Birth Weight and Early Postnatal Outcomes: Association with the Cord Blood Lipidome
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Population, Study Design & Ethics
2.2. Clinical, Endocrine–Metabolic, and Body Composition Assessments
2.3. Lipidomics Fingerprinting by UHPLC-ESI-QTOF MS
2.4. Data Processing
2.5. Lipidomic Data Normalization and Analysis
2.6. Lipid Annotation
2.7. Statistical Analysis
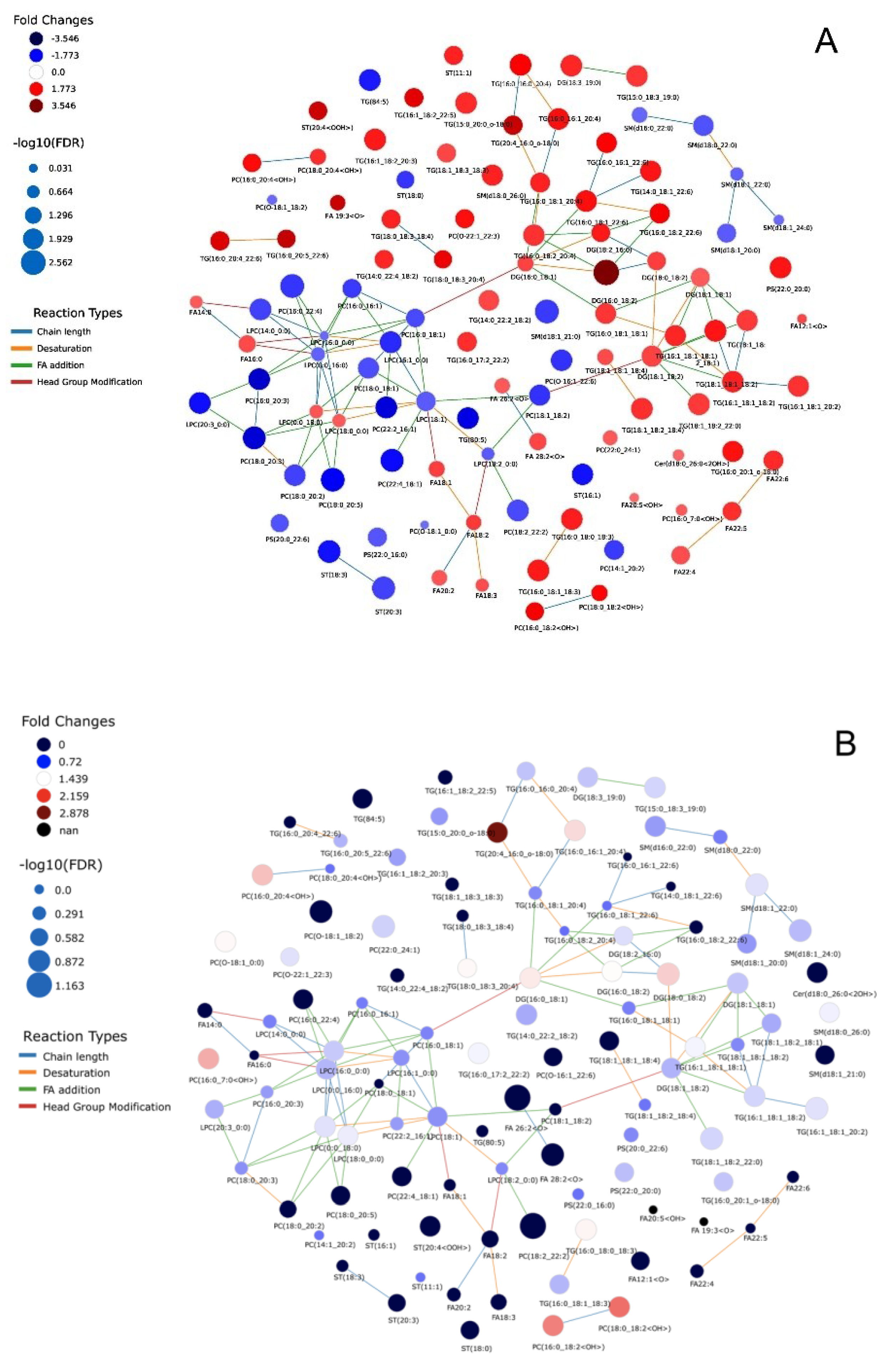
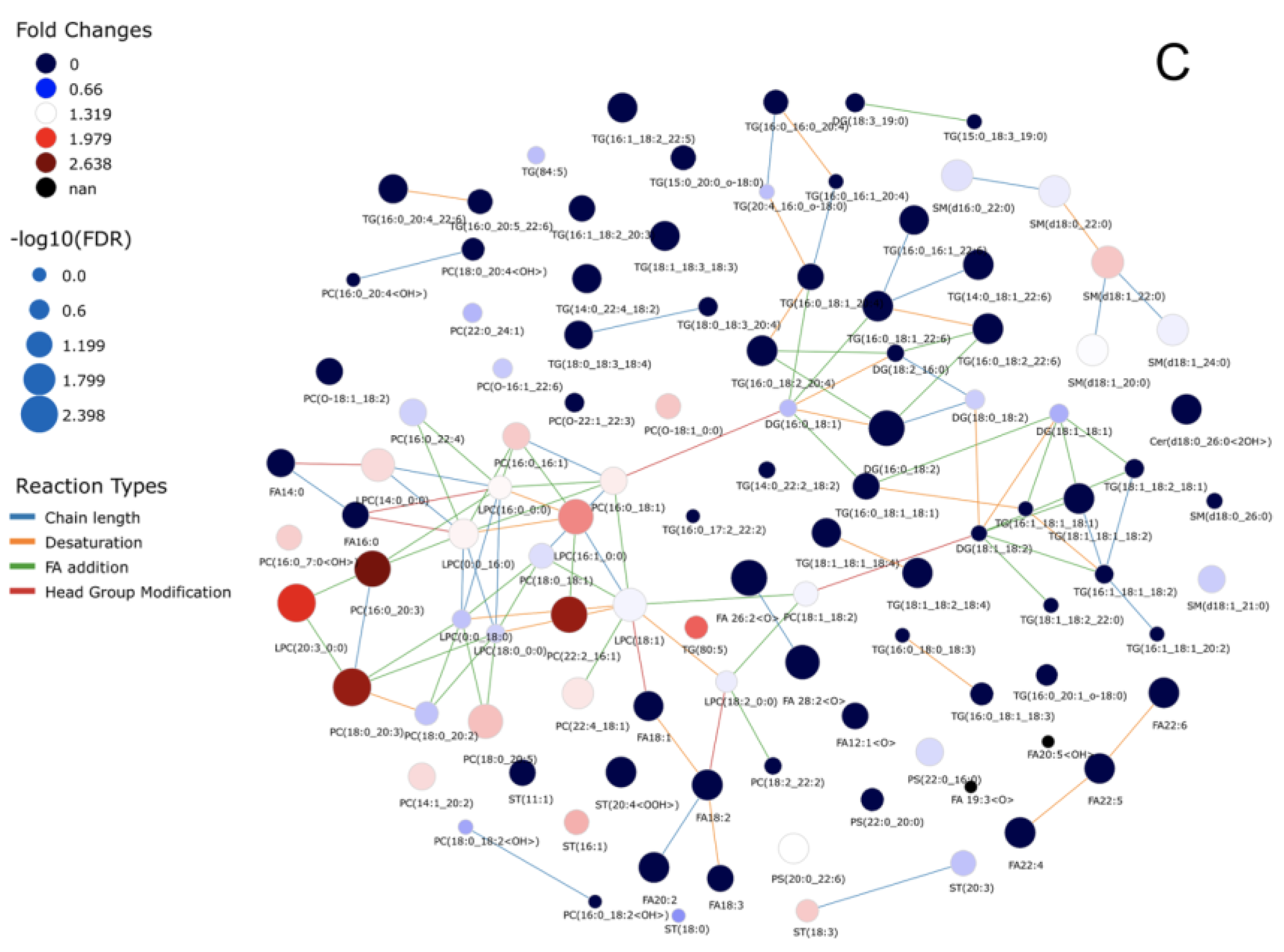
3. Results
3.1. Anthropometric, Endocrine–Metabolic, and Body Composition Variables
3.2. Non-Targeted Lipidomics-Based Analysis
3.3. Lipid Species Modulated in SGA, AGA, and LGA Infants
3.4. Correlations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clayton, P.E.; Cianfarani, S.; Czernichow, P.; Johannsson, G.; Rapaport, R.; Rogol, A. Management of the Child Born Small for Gestational Age through to Adulthood: A Consensus Statement of the International Societies of Pediatric Endocrinology and the Growth Hormone Research Society. J. Clin. Endocrinol. Metab. 2007, 92, 804–810. [Google Scholar] [CrossRef] [PubMed]
- de Zegher, F.; Díaz, M.; Lopez-Bermejo, A.; Ibáñez, L. Recognition of a Sequence: More Growth before Birth, Longer Telomeres at Birth, More Lean Mass after Birth. Pediatr. Obes. 2017, 12, 274–279. [Google Scholar] [CrossRef] [PubMed]
- de Zegher, F.; Malpique, R.; Garcia-Beltran, C.; Ibáñez, L. Towards a Simple Marker of Hepato-Visceral Adiposity and Insulin Resistance: The Z-Score Change from Weight-at-Birth to BMI-in-Childhood. Pediatr. Obes. 2019, 14, e12533. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Díaz, M.; Bassols, J.; Aragonés, G.; López-Bermejo, A.; Zegher, F.; Ibáñez, L. The Sequence of Prenatal Growth Restraint and Post-natal Catch-up Growth Leads to a Thicker Intima-media and More Pre-peritoneal and Hepatic Fat by Age 3–6 Years. Pediatr. Obes. 2016, 11, 251–257. [Google Scholar] [CrossRef]
- Sebastiani, G.; García-Beltran, C.; Pie, S.; Guerra, A.; López-Bermejo, A.; de Toledo, J.S.; de Zegher, F.; Rosés, F.; Ibáñez, L. The Sequence of Prenatal Growth Restraint and Postnatal Catch-up Growth: Normal Heart but Thicker Intima-Media and More Pre-Peritoneal Fat in Late Infancy. Pediatr. Obes. 2019, 14, e12476. [Google Scholar] [CrossRef]
- Xie, C.; Wang, Y.; Li, X.; Wen, X. Childhood Growth Trajectories of Etiological Subgroups of Large for Gestational Age Newborns. J. Pediatr. 2016, 170, 60–66.e1–5. [Google Scholar] [CrossRef]
- Ibáñez, L.; Ong, K.; Dunger, D.B.; de Zegher, F. Early Development of Adiposity and Insulin Resistance after Catch-Up Weight Gain in Small-for-Gestational-Age Children. J. Clin. Endocrinol. Metab. 2006, 91, 2153–2158. [Google Scholar] [CrossRef]
- Palatianou, M.; Simos, Y.; Andronikou, S.; Kiortsis, D. Long-Term Metabolic Effects of High Birth Weight: A Critical Review of the Literature. Horm. Metab. Res. 2014, 46, 911–920. [Google Scholar] [CrossRef]
- Nobili, V.; Alisi, A.; Panera, N.; Agostoni, C. Low Birth Weight and Catch-up-Growth Associated with Metabolic Syndrome: A Ten Year Systematic Review. Pediatr. Endocrinol. Rev. 2008, 6, 241–247. [Google Scholar]
- Maguolo, A.; Olivieri, F.; Zusi, C.; Miraglia Del Giudice, E.; Morandi, A.; Maffeis, C. The Risk of Metabolic Derangements Is Higher in Children and Adolescents with Overweight or Obesity Born Small for Gestational Age. Nutr. Metab. Cardiovasc. Dis. 2021, 31, 1903–1910. [Google Scholar] [CrossRef]
- Ibáñez, L.; Lopez-Bermejo, A.; Diaz, M.; Marcos, M.V.; de Zegher, F. Early Metformin Therapy to Delay Menarche and Augment Height in Girls with Precocious Pubarche. Fertil Steril 2011, 95, 727–730. [Google Scholar] [CrossRef] [Green Version]
- Ibáñez, L.; de Zegher, F. Polycystic Ovary Syndrome in Adolescent Girls. Pediatr. Obes. 2020, 15, e12586. [Google Scholar] [CrossRef]
- de Zegher, F.; Pérez-Cruz, M.; Sebastiani, G.; Díaz, M.; López-Bermejo, A.; Ibáñez, L. Large for Gestational Age Newborns from Mothers Without Diabetes Mellitus Tend to Become Tall and Lean Toddlers. J. Pediatr. 2016, 178, 278–280. [Google Scholar] [CrossRef]
- Meikle, T.G.; Huynh, K.; Giles, C.; Meikle, P.J. Clinical Lipidomics: Realizing the Potential of Lipid Profiling. J. Lipid Res. 2021, 62, 100127. [Google Scholar] [CrossRef]
- LaBarre, J.L.; Puttabyatappa, M.; Song, P.X.K.; Goodrich, J.M.; Zhou, L.; Rajendiran, T.M.; Soni, T.; Domino, S.E.; Treadwell, M.C.; Dolinoy, D.C.; et al. Maternal Lipid Levels across Pregnancy Impact the Umbilical Cord Blood Lipidome and Infant Birth Weight. Sci. Rep. 2020, 10, 14209. [Google Scholar] [CrossRef]
- Patel, N.; Hellmuth, C.; Uhl, O.; Godfrey, K.; Briley, A.; Welsh, P.; Pasupathy, D.; Seed, P.T.; Koletzko, B.; Poston, L.; et al. Cord Metabolic Profiles in Obese Pregnant Women: Insights Into Offspring Growth and Body Composition. J. Clin. Endocrinol. Metab. 2018, 103, 346–355. [Google Scholar] [CrossRef]
- Robinson, O.; Keski-Rahkonen, P.; Chatzi, L.; Kogevinas, M.; Nawrot, T.; Pizzi, C.; Plusquin, M.; Richiardi, L.; Robinot, N.; Sunyer, J.; et al. Cord Blood Metabolic Signatures of Birth Weight: A Population-Based Study. J. Proteome Res. 2018, 17, 1235–1247. [Google Scholar] [CrossRef]
- Perng, W.; Rifas-Shiman, S.L.; McCulloch, S.; Chatzi, L.; Mantzoros, C.; Hivert, M.-F.; Oken, E. Associations of Cord Blood Metabolites with Perinatal Characteristics, Newborn Anthropometry and Cord Blood Hormones in Project Viva. Metabolism 2017, 76, 11–22. [Google Scholar] [CrossRef]
- Thompson, M.; Ulu, A.; Yuil-Valdes, A.G.; Mukherjee, M.; Thoene, M.; van Ormer, M.; Slotkowski, R.; Lyden, E.; Anderson Berry, A.; Hanson, C.K.; et al. Omega-6 and Omega-3 Fatty Acid-Derived Oxylipins from the Lipoxygenase Pathway in Maternal and Umbilical Cord Plasma at Delivery and Their Relationship with Infant Growth. Int. J. Mol. Sci. 2022, 23, 708. [Google Scholar] [CrossRef]
- Wang, Y.; Yutuc, E.; Griffiths, W.J. Standardizing and Increasing the Utility of Lipidomics: A Look to the next Decade. Expert Rev. Proteom. 2020, 17, 699–717. [Google Scholar] [CrossRef]
- Ismail, I.T.; Showalter, M.R.; Fiehn, O. Inborn Errors of Metabolism in the Era of Untargeted Metabolomics and Lipidomics. Metabolites 2019, 9, 242. [Google Scholar] [CrossRef] [Green Version]
- De Zegher, F.; Sebastiani, G.; Diaz, M.; Sánchez-Infantes, D.; Lopez-Bermejo, A.; Ibáñez, L. Body Composition and Circulating High-Molecular-Weight Adiponectin and IGF-I in Infants Born Small for Gestational Age. Diabetes 2012, 61, 1969–1973. [Google Scholar] [CrossRef]
- de Zegher, F.; Sebastiani, G.; Diaz, M.; Gómez-Roig, M.D.; López-Bermejo, A.; Ibáñez, L. Breast-Feeding vs Formula-Feeding for Infants Born Small-for-Gestational-Age: Divergent Effects on Fat Mass and on Circulating IGF-I and High-Molecular-Weight Adiponectin in Late Infancy. J. Clin. Endocrinol. Metab. 2013, 98, 1242–1247. [Google Scholar] [CrossRef]
- Ferrández-Longas, A.; Baguer, L.; Labarta, J.; Labena, C.; Mayayo, E.; Puga, B.; Rueda, C.; Ruiz-Echarri, M. Longitudinal Growth Study of Normal Spanish Children from Birth to Adulthood. Anthropometric, Pubertal, Radiological and Intellectual Data. Pediatr. Endocrinol. Rev. (PER) 2005, 2, 425–642. [Google Scholar]
- Díaz, M.; García, C.; Sebastiani, G.; de Zegher, F.; López-Bermejo, A.; Ibáñez, L. Placental and Cord Blood Methylation of Genes Involved in Energy Homeostasis: Association With Fetal Growth and Neonatal Body Composition. Diabetes 2017, 66, 779–784. [Google Scholar] [CrossRef]
- Pellegrino, R.M.; di Veroli, A.; Valeri, A.; Goracci, L.; Cruciani, G. LC/MS Lipid Profiling from Human Serum: A New Method for Global Lipid Extraction. Anal. Bioanal. Chem. 2014, 406, 7937–7948. [Google Scholar] [CrossRef]
- Sarkar, D.; Lim, S.H.; Sinclair, E.; Jafri, K.; Walton-Doyle, C.; Milne, J.; Vissers, H.; Trivedi, D.; Richardson, K.; Silverdale, M.; et al. Paper Spray Ionisation Ion Mobility Mass Spectrometry of Sebum Classifies Biomarker Classes for the Diagnosis of Parkinson’s Disease. ChemRxiv 2021. [Google Scholar] [CrossRef]
- Gonzalez-Riano, C.; Gradillas, A.; Barbas, C. Exploiting the Formation of Adducts in Mobile Phases with Ammonium Fluoride for the Enhancement of Annotation in Liquid Chromatography-High Resolution Mass Spectrometry Based Lipidomics. J. Chromatogr. Open 2021, 1, 100018. [Google Scholar] [CrossRef]
- Gil-de-la-Fuente, A.; Godzien, J.; Saugar, S.; Garcia-Carmona, R.; Badran, H.; Wishart, D.S.; Barbas, C.; Otero, A. CEU Mass Mediator 3.0: A Metabolite Annotation Tool. J. Proteome Res. 2019, 18, 797–802. [Google Scholar] [CrossRef]
- Gil de la Fuente, A.; Godzien, J.; Fernández López, M.; Rupérez, F.J.; Barbas, C.; Otero, A. Knowledge-Based Metabolite Annotation Tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A Metabolite Mass Spectral Database. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. MS-DIAL 4: Accelerating Lipidomics Using an MS/MS, CCS and Retention Time Atlas. BioRxiv 2020, 1159–1163. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS Structure Database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed]
- Koelmel, J.P.; Li, X.; Stow, S.M.; Sartain, M.J.; Murali, A.; Kemperman, R.; Tsugawa, H.; Takahashi, M.; Vasiliou, V.; Bowden, J.A.; et al. Lipid Annotator: Towards Accurate Annotation in Non-Targeted Liquid Chromatography High-Resolution Tandem Mass Spectrometry (LC-HRMS/MS) Lipidomics Using A Rapid and User-Friendly Software. Metabolites 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Pieke, E.N.; Granby, K.; Trier, X.; Smedsgaard, J. A Framework to Estimate Concentrations of Potentially Unknown Substances by Semi-Quantification in Liquid Chromatography Electrospray Ionization Mass Spectrometry. Anal. Chim. Acta 2017, 975, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics. a Review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS Classification, Nomenclature and Shorthand Notation for MS-Derived Lipid Structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Köhler, N.; Rose, T.D.; Falk, L.; Pauling, J.K. Investigating Global Lipidome Alterations with the Lipid Network Explorer. Metabolites 2021, 11, 488. [Google Scholar] [CrossRef]
- Weber, D.; Stuetz, W.; Bernhard, W.; Franz, A.; Raith, M.; Grune, T.; Breusing, N. Oxidative Stress Markers and Micronutrients in Maternal and Cord Blood in Relation to Neonatal Outcome. Eur. J. Clin. Nutr. 2014, 68, 215–222. [Google Scholar] [CrossRef]
- Gveric-Ahmetasevic, S.; Sunjic, S.B.; Skala, H.; Andrisic, L.; Stroser, M.; Zarkovic, K.; Skrablin, S.; Tatzber, F.; Cipak, A.; Jaganjac, M.; et al. Oxidative Stress in Small-for-Gestational Age (SGA) Term Newborns and Their Mothers. Free. Radic. Res. 2009, 43, 376–384. [Google Scholar] [CrossRef]
- Szczuko, M.; Kikut, J.; Komorniak, N.; Bilicki, J.; Celewicz, Z.; Ziętek, M. The Role of Arachidonic and Linoleic Acid Derivatives in Pathological Pregnancies and the Human Reproduction Process. Int. J. Mol. Sci. 2020, 21, 9628. [Google Scholar] [CrossRef]
- Reilly, K.B.; Srinivasan, S.; Hatley, M.E.; Patricia, M.K.; Lannigan, J.; Bolick, D.T.; Vandenhoff, G.; Pei, H.; Natarajan, R.; Nadler, J.L.; et al. 12/15-Lipoxygenase Activity Mediates Inflammatory Monocyte/Endothelial Interactions and Atherosclerosis in Vivo. J. Biol. Chem. 2004, 279, 9440–9450. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.S. Prostacyclin: A Major Prostaglandin in the Regulation of Adipose Tissue Development. J. Cell Physiol. 2019, 234, 3254–3262. [Google Scholar] [CrossRef]
- Nazarewicz, R.R.; Zenebe, W.J.; Parihar, A.; Parihar, M.S.; Vaccaro, M.; Rink, C.; Sen, C.K.; Ghafourifar, P. 12(S)-Hydroperoxyeicosatetraenoic Acid (12-HETE) Increases Mitochondrial Nitric Oxide by Increasing Intramitochondrial Calcium. Arch. Biochem. Biophys. 2007, 468, 114–120. [Google Scholar] [CrossRef]
- Gilani, A.; Agostinucci, K.; Hossain, S.; Pascale, J.V.; Garcia, V.; Adebesin, A.M.; Falck, J.R.; Schwartzman, M.L. 20-HETE Interferes with Insulin Signaling and Contributes to Obesity-Driven Insulin Resistance. Prostaglandins Other Lipid Mediat. 2021, 152, 106485. [Google Scholar] [CrossRef]
- van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The Critical Role of Phosphatidylcholine and Phosphatidylethanolamine Metabolism in Health and Disease. Biochim. Biophys. Acta (BBA)—Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Law, S.-H.; Chan, M.-L.; Marathe, G.K.; Parveen, F.; Chen, C.-H.; Ke, L.-Y. An Updated Review of Lysophosphatidylcholine Metabolism in Human Diseases. Int. J. Mol. Sci. 2019, 20, 1149. [Google Scholar] [CrossRef]
- Lu, Y.-P.; Reichetzeder, C.; Prehn, C.; Yin, L.-H.; Yun, C.; Zeng, S.; Chu, C.; Adamski, J.; Hocher, B. Cord Blood Lysophosphatidylcholine 16: 1 Is Positively Associated with Birth Weight. Cell. Physiol. Biochem. 2018, 45, 614–624. [Google Scholar] [CrossRef]
- Hellmuth, C.; Uhl, O.; Standl, M.; Demmelmair, H.; Heinrich, J.; Koletzko, B.; Thiering, E. Cord Blood Metabolome Is Highly Associated with Birth Weight, but Less Predictive for Later Weight Development. Obes. Facts 2017, 10, 85–100. [Google Scholar] [CrossRef]
- Takatera, A.; Takeuchi, A.; Saiki, K.; Morioka, I.; Yokoyama, N.; Matsuo, M. Blood Lysophosphatidylcholine (LPC) Levels and Characteristic Molecular Species in Neonates: Prolonged Low Blood LPC Levels in Very Low Birth Weight Infants. Pediatr. Res. 2007, 62, 477–482. [Google Scholar] [CrossRef]
- Lerin, C.; Goldfine, A.B.; Boes, T.; Liu, M.; Kasif, S.; Dreyfuss, J.M.; de Sousa-Coelho, A.L.; Daher, G.; Manoli, I.; Sysol, J.R.; et al. Defects in Muscle Branched-Chain Amino Acid Oxidation Contribute to Impaired Lipid Metabolism. Mol. Metab. 2016, 5, 926–936. [Google Scholar] [CrossRef]
- McCann, M.R.; George De la Rosa, M.V.; Rosania, G.R.; Stringer, K.A. L-Carnitine and Acylcarnitines: Mitochondrial Biomarkers for Precision Medicine. Metabolites 2021, 11, 51. [Google Scholar] [CrossRef]
- Beken, S.; Abali, S.; Yildirim Saral, N.; Guner, B.; Dinc, T.; Albayrak, E.; Ersoy, M.; Kilercik, M.; Halici, M.; Bulbul, E.; et al. Early Postnatal Metabolic Profile in Neonates With Different Birth Weight Status: A Pilot Study. Front. Pediatr. 2021, 9, 646860. [Google Scholar] [CrossRef]
- Marston, N.A.; Giugliano, R.P.; Im, K.; Silverman, M.G.; O’Donoghue, M.L.; Wiviott, S.D.; Ference, B.A.; Sabatine, M.S. Association Between Triglyceride Lowering and Reduction of Cardiovascular Risk Across Multiple Lipid-Lowering Therapeutic Classes. Circulation 2019, 140, 1308–1317. [Google Scholar] [CrossRef]
- Kelishadi, R.; Badiee, Z.; Adeli, K. Cord Blood Lipid Profile and Associated Factors: Baseline Data of a Birth Cohort Study. Paediatr. Perinat. Epidemiol. 2007, 21, 518–524. [Google Scholar] [CrossRef]
- Kaser, S.; Ebenbichler, C.F.; Wolf, H.J.; Sandhofer, A.; Stanzl, U.; Ritsch, A.; Patsch, J.R. Lipoprotein Profile and Cholesteryl Ester Transfer Protein in Neonates. Metabolism 2001, 50, 723–728. [Google Scholar] [CrossRef]
- Merzouk, H.; Meghelli-Bouchenak, M.; El-Korso, N.; Belleville, J.; Prost, J. Low Birth Weight at Term Impairs Cord Serum Lipoprotein Compositions and Concentrations. Eur. J. Pediatr. 1998, 157, 321–326. [Google Scholar] [CrossRef]

| LGA vs. AGA | SGA vs. AGA | LGA vs. SGA | |||||||||||||
| Name | m/z | RT (min) | Formula | CV (%) | p-Value | p-Value BH | FC | p-Value | VIP | FC | p-Value | VIP | FC | p-Value | VIP |
| Fatty Acyls | |||||||||||||||
| 1(3)-Glyceryl-6-keto-PGF1α/ 2-glyceryl-6-keto-PGF1α | 443.2649 | 0.84 | C23H40O8 | 2.8 | 4.9 × 10−6 | 1.4 × 10−4 | −LGA | - | 4.40 | 5.41 | 0.20 | 1.73 | −LGA | - | 4.07 |
| 11-HEDE | 301.1990 | 0.89 | C20H36O3 | 8.6 | 0.0051 | 0.051 | 0.35 | 0.77 | 0.61 | 2.99 | 0.045 * | 2.04 | 0.12 | 0.0029 * | 1.75 |
| 12,20-DiHETE | 355.2276 | 2.52 | C20H32O4 | 3.9 | 6.9 × 10−6 | 1.4 × 10−4 | −LGA | - | 4.61 | 1.59 | 0.83 | 1.25 | −LGA | - | 3.97 |
| 12-HETE | 339.2329 | 3.21 | C20H32O3 | 1.1 | 0.0017 | 0.018 | 0.01 | 0.011 | 2.47 | 0.87 | 0.97 | 1.36 | 0.01 | 0.0043 * | 2.50 |
| 19-Hydroxy-PGE2 | 429.2497 | 0.82 | C20H34O6 | 5.4 | 2.2 × 10−7 | 1.4 × 10−5 | −LGA | - | 3.41 | 22.20 | 0.0025 * | 3.45 | −LGA | - | 3.84 |
| 3-Hexenyl 3-hydroxybutanoate; WE 10:1; O | 245.1397 | 0.78 | C10H18O3 | 8.1 | 1.8 × 10−7 | 1.4 × 10−5 | - | - | - | +SGA | - | 4.80 | −LGA | - | 3.39 |
| 5S-HpEPE | 353.2122 | 1.71 | C20H30O4 | 2.8 | 6.9 × 10−6 | 1.4 × 10−4 | −LGA | - | 4.76 | 0.99 | 0.67 | 0.22 | −LGA | - | 4.15 |
| 6-Hydroxypentadecanedioic acid | 287.1866 | 0.84 | C15H28O5 | 6.1 | 5.5 × 10−6 | 1.4 × 10−4 | −LGA | - | 3.93 | 4.83 | 0.28 | 1.73 | −LGA | - | 3.66 |
| 9-HODE | 297.2421 | 2.88 | C18H32O3 | 7.1 | 0.0033 | 0.021 | 3.55 | 0.045 | 1.56 | 1.98 | 0.013 * | 0.96 | 1.79 | 0.62 | 1.11 |
| Ascorbyl palmitate | 473.2750 | 0.85 | C22H38O7 | 4.1 | 1.8 × 10−7 | 1.4 × 10−5 | - | - | - | +SGA | - | 5.49 | −LGA | - | 3.80 |
| Eicosadienoic acid | 307.264 | 3.84 | C20H36O2 | 2.4 | 0.053 | 0.23 | 0.91 | 0.37 | 0.16 | 1.18 | 0.14 | 0.67 | 0.77 | 0.022 * | 0.56 |
| FA 26:2; O | 467.3739 | 2.72 | C26H48O3 | 2.3 | 5.2 × 10−4 | 0.0069 | 0.69 | 0.048 * | 0.90 | 1.17 | 0.13 | 0.55 | 0.58 | 0.0030 * | 0.90 |
| FA 28:2; O | 495.4052 | 3.78 | C28H52O3 | 4.2 | 0.0012 | 0.014 | 0.70 | 0.048 * | 0.82 | 1.28 | 0.093 | 0.80 | 0.55 | 0.0071 * | 0.97 |
| FAHFA(18:1-(10-O-16:0) | 595.4939 | 3.80 | C34H64O4 | 1.4 | 0.0014 | 0.016 | 0.71 | 0.048 * | 0.75 | 1.33 | 0.082 | 0.83 | 0.54 | 0.0077 * | 0.94 |
| FAHFA(30:1) | 539.4310 | 2.95 | C30H56O4 | 2.6 | 1.1 × 10−4 | 0.0017 | 0.65 | 0.037 * | 0.92 | 1.36 | 0.041 | 0.92 | 0.47 | 0.00038 * | 1.10 |
| Hexacosanedioic acid; FA26:1;O2 | 425.3636 | 3.52 | C26H50O4 | 2.4 | 9.4 × 10−5 | 0.0016 | 0.67 | 0.037 * | 0.96 | 1.36 | 0.021 | 0.91 | 0.49 | 0.0027 * | 1.07 |
| Methyl-FA 18:3;2OOH | 415.2363 | 0.81 | C19H32O6 | 5.1 | 1.3 × 10−6 | 6.7 × 10−5 | −LGA | - | 3.74 | 8.42 | 0.045 * | 2.52 | −LGA | - | 3.82 |
| Decanoylcarnitine | 316.2482 | 0.92 | C17H33NO4 | 7.9 | 0.045 | 0.067 | 1.30 | 0.027 | 0.38 | 1.50 | 0.033 | 1.12 | 0.87 | 0.36 | 0.15 |
| Decenoylcarnitine | 314.2326 | 0.86 | C17H31NO4 | 8.6 | 0.034 | 0.061 | 1.31 | 0.039 | 0.38 | 1.49 | 0.045 * | 1.12 | 0.88 | 0.58 | 0.09 |
| Oleoylcarnitine | 426.3576 | 2.43 | C25H47NO4 | 5.5 | 0.0091 | 0.043 | 1.46 | 0.0035 | 0.65 | 1.15 | 0.22 | 0.49 | 1.27 | 0.038 * | 0.40 |
| Palmitoylcarnitine | 400.3420 | 2.25 | C23H45NO4 | 5.4 | 0.0017 | 0.018 | 1.46 | 0.029 * | 0.64 | 1.18 | 0.14 | 0.48 | 1.24 | 0.029 | 0.38 |
| α-Linolenic acid | 277.217 | 2.55 | C18H30O2 | 2.4 | 0.080 | 0.26 | 0.86 | 0.34 | 0.28 | 1.25 | 0.16 | 0.67 | 0.68 | 0.045 | 0.64 |
| Docosatetraenoic acid | 331.264 | 3.65 | C22H36O2 | 1.4 | 0.013 | 0.094 | 0.91 | 0.18 | 0.30 | 1.18 | 0.093 | 0.65 | 0.76 | 0.0194 * | 0.56 |
| Docosahexaenoic acid; DHA | 327.2329 | 2.77 | C22H32O2 | 2.1 | 0.0057 | 0.054 | 0.97 | 0.83 | 0.14 | 1.55 | 0.045 * | 1.07 | 0.63 | 0.020 * | 0.75 |
| Docosapentaenoic acid; DPA | 329.249 | 3.16 | C22H34O2 | 5.8 | 0.030 | 0.16 | 1.03 | 0.76 | 0.13 | 1.56 | 0.019 | 0.99 | 0.66 | 0.026 * | 0.61 |
| Eicosadienoic acid | 307.264 | 3.84 | C20H36O2 | 2.4 | 0.053 | 0.23 | 0.91 | 0.37 | 0.16 | 1.18 | 0.14 | 0.67 | 0.77 | 0.022 * | 0.56 |
| Hexacosanedioic acid | 425.364 | 3.52 | C26H50O4 | 2.4 | 9.4 × 10−5 | 0.0016 | 0.67 | 0.037 * | 0.96 | 1.36 | 0.021 | 0.91 | 0.49 | 0.0027 * | 1.07 |
| Linoleic acid | 279.233 | 3.05 | C18H32O2 | 1.5 | 0.037 | 0.19 | 0.80 | 0.37 | 0.32 | 1.40 | 0.081 | 0.87 | 0.57 | 0.022 * | 0.81 |
| Myristic acid | 227.202 | 2.56 | C14H28O2 | 3.2 | 0.13 | 0.36 | 0.91 | 0.45 | 0.16 | 1.22 | 0.21 | 0.68 | 0.75 | 0.054 * | 0.57 |
| Oleic acid | 281.249 | 3.63 | C18H34O2 | 1.1 | 0.058 | 0.23 | 0.95 | 1.00 | 0.05 | 1.41 | 0.070 | 0.94 | 0.68 | 0.032 * | 0.59 |
| Palmitic acid | 255.233 | 3.46 | C16H32O2 | 2.4 | 0.11 | 0.30 | 0.98 | 0.85 | 0.10 | 1.26 | 0.074 | 0.74 | 0.78 | 0.045 | 0.48 |
| Glycerolipids | |||||||||||||||
| DG 16:0/18:2/0:0 | 610.5393 | 11.34 | C37H68O5 | 6.0 | 1.5 × 10−5 | 0.0015 | 1.45 | 0.053 | 0.70 | 3.55 | 0.011 * | 3.46 | 0.41 | 0.0083 * | 1.33 |
| TG 14:0/18:3/22:3 | 917.6996 | 13.57 | C57H98O6 | 6.7 | 0.023 | 0.057 | 0.86 | 0.18 | 0.27 | 1.80 | 0.061 | 1.46 | 0.48 | 0.055 * | 1.00 |
| TG 14:0/22:2/18:2 | 921.7310 | 13.94 | C57H102O6 | 4.8 | 0.024 | 0.057 | 1.20 | 0.078 | 0.31 | 1.29 | 0.021 * | 0.71 | 0.93 | 0.62 | 0.29 |
| TG 15:0/18:3/19:0 | 895.7153 | 13.94 | C55H100O6 | 6.0 | 0.016 | 0.049 | 1.32 | 0.056 | 0.48 | 1.41 | 0.020 * | 0.93 | 0.94 | 0.65 | 0.40 |
| TG 15:0/20:0/O-18:0 | 901.8020 | 13.55 | C57H98O6 | 14.7 | 0.017 | 0.049 | 1.15 | 0.21 | 0.27 | 1.48 | 0.019 * | 1.05 | 0.78 | 0.068 | 0.42 |
| TG 16:0/16:0/20:4 | 872.7700 | 13.55 | C55H98O6 | 4.3 | 0.017 | 0.049 | 1.27 | 0.21 | 0.42 | 1.80 | 0.019 * | 1.54 | 0.70 | 0.095 | 0.66 |
| TG 16:0/18:0/18:3 | 879.7411 | 13.94 | C55H100O6 | 3.8 | 0.014 | 0.047 | 1.47 | 0.054 | 0.59 | 1.56 | 0.020 * | 1.18 | 0.94 | 0.59 | 0.50 |
| TG 16:0/18:1/18:1 | 876.8000 | 13.56 | C55H102O6 | 8.8 | 0.030 | 0.059 | 1.08 | 0.48 | 0.19 | 1.40 | 0.026 * | 0.95 | 0.77 | 0.052 | 0.45 |
| TG 16:0/18:1/18:3 | 877.7253 | 13.55 | C55H98O6 | 5.1 | 0.014 | 0.047 | 1.23 | 0.14 | 0.36 | 1.58 | 0.019 * | 1.22 | 0.78 | 0.15 | 0.52 |
| TG 16:0/18:1/20:4 | 898.7850 | 13.56 | C57H100O6 | 16.5 | 0.055 | 0.073 | 1.10 | 0.47 | 0.25 | 1.53 | 0.035 * | 1.21 | 0.72 | 0.052 | 0.56 |
| TG 16:0/18:1/22:6 | 922.7880 | 13.56 | C59H100O6 | 7.3 | 0.021 | 0.055 | 1.02 | 0.87 | 0.19 | 1.69 | 0.020 * | 1.38 | 0.60 | 0.055 * | 0.78 |
| TG 16:0/18:2/20:4 | 896.7700 | 13.24 | C57H98O6 | 7.5 | 0.027 | 0.058 | 1.01 | 0.93 | 0.08 | 1.42 | 0.04 * | 0.99 | 0.71 | 0.055 * | 0.51 |
| TG 16:0/18:2/22:6 | 920.7690 | 13.23 | C59H98O6 | 2.6 | 0.025 | 0.057 | 0.91 | 0.43 | 0.40 | 1.88 | 0.037 * | 1.59 | 0.48 | 0.013 | 1.08 |
| TG 16:1/18:1/18:1 | 874.7860 | 13.94 | C55H100O6 | 2.8 | 0.024 | 0.057 | 1.41 | 0.081 | 0.57 | 1.52 | 0.024 * | 1.12 | 0.93 | 0.57 | 0.49 |
| TG 16:1/18:1/18:2 | 872.7641 | 13.54 | C55H98O6 | 6.9 | 0.0098 | 0.044 | 1.33 | 0.035 | 0.38 | 1.53 | 0.020 * | 1.03 | 0.87 | 0.16 | 0.46 |
| TG 16:1/18:1/20:2 | 905.7567 | 13.94 | C57H102O6 | 7.0 | 0.014 | 0.047 | 1.35 | 0.030 | 0.46 | 1.42 | 0.021 * | 1.02 | 0.95 | 0.71 | 0.36 |
| TG 16:1/18:2/20:3 | 896.7702 | 13.56 | C57H98O6 | 15.7 | 0.014 | 0.047 | 1.16 | 0.22 | 0.28 | 1.60 | 0.019 * | 1.24 | 0.73 | 0.051 | 0.59 |
| TG 18:1/18:1/18:2 | 900.7971 | 13.56 | C57H102O6 | 13.8 | 0.011 | 0.044 | 1.09 | 0.41 | 0.19 | 1.61 | 0.013 * | 1.30 | 0.68 | 0.0082 | 0.60 |
| TG 18:1/18:2/22:0 | 958.8783 | 13.94 | C61H112O6 | 12.7 | 0.0064 | 0.035 | 1.32 | 0.013 | 0.44 | 1.38 | 0.019 * | 0.87 | 0.95 | 0.67 | 0.31 |
| TG 20:4/16:0/O-18:0 | 907.7515 | 12.44 | C57H104O5 | 4.6 | 0.0011 | 0.014 | 2.88 | 0.0017 | 1.45 | 2.50 | 0.014 * | 2.47 | 1.15 | 0.89 | 0.90 |
| TG 84:5 | 1367.23 | 14.69 | C87H164NO9 | 7.7 | 0.022 | 0.056 | 0.73 | 0.085 | 0.80 | 0.63 | 0.018 * | 1.23 | 1.15 | 0.46 | 0.31 |
| Glycerophospholipids | |||||||||||||||
| LPC 14:0/0:0 | 468.3082 | 1.68 | C22H46NO7P | 5.6 | 0.0046 | 0.028 | 1.07 | 0.43 | 0.13 | 0.76 | 0.014 * | 0.90 | 1.41 | 0.025 * | 0.53 |
| LPC 16:1/0:0 | 494.3239 | 1.84 | C24H48NO7P | 5.7 | 0.0025 | 0.019 | 1.12 | 0.27 | 0.23 | 0.68 | 0.013 * | 1.35 | 1.65 | 0.010 * | 0.81 |
| LPC 18:1/0:0 | 580.3615 | 2.70 | C26H52NO7P | 2.9 | 0.0050 | 0.051 | 1.11 | 0.11 | 0.37 | 0.86 | 0.031 | 0.70 | 1.29 | 0.011 * | 0.59 |
| LPC 20:3/0:0 | 546.3549 | 2.57 | C28H52NO7P | 6.2 | 2.2 × 10−4 | 0.0045 | 1.21 | 0.11 | 0.31 | 0.59 | 0.018 * | 1.65 | 2.04 | 0.0064 * | 1.07 |
| PC 16:0/16:1 | 732.5533 | 7.14 | C40H78NO8P | 1.6 | 0.010 | 0.044 | 1.06 | 0.67 | 0.22 | 0.73 | 0.025 * | 1.13 | 1.45 | 0.020 | 0.56 |
| PC 16:0/18:1 | 760.5852 | 8.86 | C42H82NO8P | 1.0 | 0.0069 | 0.035 | 1.09 | 0.68 | 0.21 | 0.80 | 0.020 * | 0.79 | 1.37 | 0.012 | 0.44 |
| PC 16:0/20:3 | 784.5846 | 8.55 | C44H82NO8P | 1.9 | 1.7 × 10−4 | 0.0045 | 1.15 | 0.26 | 0.29 | 0.44 | 0.013 * | 2.46 | 2.64 | 0.0064 * | 1.38 |
| PC 16:0/22:4 | 810.6010 | 8.28 | C46H84NO8P | 2.9 | 0.062 | 0.076 | 1.10 | 0.52 | 0.28 | 0.77 | 0.045 * | 0.77 | 1.43 | 0.078 | 0.46 |
| PC 16:0/22:4 | 810.6008 | 8.66 | C46H84NO8P | 2.8 | 0.0019 | 0.018 | 0.85 | 0.069 | 0.30 | 0.71 | 0.011 * | 1.02 | 1.20 | 0.039 | 0.25 |
| PC 18:0/20:2 | 814.6328 | 11.43 | C46H88NO8P | 3.9 | 0.013 | 0.047 | 0.89 | 0.22 | 0.25 | 0.77 | 0.018 * | 0.78 | 1.16 | 0.12 | 0.21 |
| PC 18:0/20:3 | 870.6224 | 11.29 | C46H86NO8P | 2.8 | 1.0 × 10−4 | 0.0016 | 1.03 | 0.92 | 0.20 | 0.42 | 0.0012 * | 1.73 | 2.44 | 0.0016 * | 1.19 |
| PC 18:0/20:4; 12OH | 826.5950 | 5.99 | C46H84NO9P | 12.6 | 0.13 | 0.14 | 1.03 | 0.91 | 1.51 | 1.46 | 0.042 * | 1.52 | 0.71 | 0.29 | 1.39 |
| PC 18:0/20:5 | 808.5845 | 8.06 | C46H82NO8P | 2.0 | 3.0 × 10−4 | 0.0051 | 0.81 | 0.13 | 0.33 | 0.54 | 0.012 * | 1.81 | 1.49 | 0.0083 * | 0.57 |
| PC 18:2/22:2 | 838.6314 | 11.06 | C48H88NO8P | 5.7 | 0.0013 | 0.015 | 0.74 | 0.040 * | 0.47 | 0.79 | 0.018 * | 0.74 | 0.95 | 0.45 | 0.12 |
| PC 22:2/16:1 | 812.6160 | 10.96 | C46H86NO8P | 4.2 | 1.5 × 10−4 | 0.0045 | 1.15 | 0.34 | 0.27 | 0.47 | 0.0033 * | 2.18 | 2.45 | 0.0064 * | 1.25 |
| PC 22:4/18:1 | 836.6157 | 10.25 | C48H86NO8P | 2.7 | 0.0024 | 0.019 | 0.82 | 0.092 | 0.30 | 0.59 | 0.0033 * | 1.61 | 1.39 | 0.028 * | 0.51 |
| PC O-18:1/0:0 | 508.3760 | 3.05 | C26H54NO6P | 5.0 | 0.030 | 0.059 | 1.45 | 0.030 | 0.60 | 0.99 | 0.61 | 0.43 | 1.47 | 0.026 | 0.63 |
| PC O-18:1/18:2 | 770.6050 | 9.31 | C44H84NO7P | 16.1 | 0.027 | 0.058 | 0.82 | 0.089 | 0.43 | 0.70 | 0.019 * | 0.96 | 1.18 | 0.71 | 0.19 |
| PS 20:0/22:6 | 864.5696 | 8.86 | C48H82NO10P | 18.7 | 0.016 | 0.049 | 1.09 | 0.34 | 0.16 | 0.83 | 0.048 * | 0.60 | 1.32 | 0.011 | 0.41 |
| PS 22:0/20:0 | 893.7000 | 13.55 | C48H94NO10P | 6.5 | 0.019 | 0.053 | 1.26 | 0.18 | 0.39 | 1.63 | 0.019 * | 1.30 | 0.77 | 0.15 | 0.57 |
| Sphingolipids | |||||||||||||||
| SM 16:0; O2/22:0 | 761.653 | 11.51 | C43H89N2O6P | 4.8 | 0.026 | 0.058 | 1.15 | 0.060 | 0.19 | 0.93 | 0.13 | 0.27 | 1.24 | 0.055 * | 0.30 |
| SM 18:0; O2/22:0 | 789.6857 | 11.94 | C45H93N2O6P | 6.2 | 0.0033 | 0.021 | 1.07 | 0.26 | 0.11 | 0.84 | 0.026 * | 0.52 | 1.27 | 0.028 * | 0.36 |
| SM 18:0; O2/26:0 | 845.7410 | 13.17 | C49H101N2O6P | 12.0 | 0.037 | 0.061 | 1.40 | 0.098 | 0.49 | 1.54 | 0.020 * | 1.29 | 0.90 | 0.31 | 0.47 |
| SM 18:1; O2/20:0 | 819.6590 | 11.64 | C43H89N2O6P | 5.3 | 0.018 | 0.11 | 1.11 | 0.21 | 0.37 | 0.85 | 0.016 | 0.69 | 1.31 | 0.020 * | 0.60 |
| SM 18:1; O2/21:0 | 811.6042 | 8.66 | C44H89N2O6P | 3.0 | 0.0026 | 0.019 | 0.87 | 0.11 | 0.27 | 0.73 | 0.011 * | 0.95 | 1.20 | 0.042 | 0.25 |
| SM 18:1; O2/22:0 | 787.6677 | 11.54 | C45H91N2O6P | 7.0 | 0.0070 | 0.035 | 1.36 | 0.018 | 0.43 | 0.92 | 0.29 | 0.31 | 1.47 | 0.046 * | 0.52 |
| SM 18:1; O2/24:0 | 815.7000 | 11.93 | C47H95N2O6P | 2.2 | 0.014 | 0.047 | 1.24 | 0.015 | 0.31 | 0.97 | 0.61 | 0.19 | 1.28 | 0.0070 | 0.35 |
| Sterol Lipids | |||||||||||||||
| 22:0-Glc-Cholesterol | 871.7420 | 13.25 | C55H98O7 | 2.4 | 0.030 | 0.059 | 1.10 | 0.67 | 0.21 | 1.45 | 0.020 * | 1.03 | 0.76 | 0.078 | 0.49 |
| 3-Deoxyvitamin D3 | 369.3510 | 12.52 | C27H44 | 4.9 | 0.047 | 0.067 | 1.16 | 0.49 | 1.58 | 1.88 | 0.030 * | 2.02 | 0.62 | 0.048 | 1.71 |
| CE 16:1 | 640.6020 | 14.57 | C43H74O2 | 3.6 | 0.020 | 0.054 | 0.93 | 0.68 | 0.42 | 0.61 | 0.0137 * | 1.76 | 1.53 | 0.079 | 0.60 |
| CE 18:3 | 664.6019 | 14.11 | C45H74O2 | 22.0 | 0.013 | 0.047 | 0.89 | 0.18 | 0.57 | 0.61 | 0.013 * | 1.61 | 1.46 | 0.40 | 0.53 |
| CE 20:3 | 713.5632 | 14.69 | C47H78O2 | 2.9 | 0.0066 | 0.035 | 0.89 | 0.19 | 0.24 | 0.77 | 0.013 * | 0.73 | 1.16 | 0.079 | 0.22 |
| Cholesteryl 11-hydroperoxy-eicosatetraenoate | 743.5375 | 12.41 | C47H76O4 | 3.2 | 1.0 × 10−4 | 0.0045 | 0.52 | 0.049 | 1.95 | 2.46 | 0.026 * | 2.72 | 0.21 | 0.0038 * | 1.97 |
| 1(3)-Glyceryl-6-keto-PGF1alpha/ 2-glyceryl-6-keto-PGF1α | 11-HEDE | 12-HETE | 12,20-DiHETE | 19R-hydroxy-PGE1 | 5S-HpEPE | 6-Hydroxypentadecanedioic acid | Methyl-FA 18:3;2OOH | |||||||||
| At birth | R | p | R | p | R | p | R | p | R | p | R | p | R | p | R | p |
| Weight | −0.82 | <0.0001 | 0.36 | 0.0279 | −0.56 | 0.0003 | −0.80 | <0.0001 | −0.91 | <0.0001 | −0.83 | <0.0001 | −0.82 | <0.0001 | −0.83 | <0.0001 |
| Length | −0.73 | <0.0001 | 0.40 | 0.0144 | −0.46 | 0.0042 | −0.70 | <0.0001 | −0.84 | <0.0001 | −0.74 | <0.0001 | −0.73 | <0.0001 | −0.74 | <0.0001 |
| BMI Z-score | −0.75 | <0.0001 | 0.39 | 0.0178 | −0.54 | 0.0007 | −0.72 | 0.0035 | −0.85 | <0.0001 | −0.75 | <0.0001 | −0.75 | <0.0001 | −0.77 | <0.0001 |
| HOMA-IR | 0.13 | 0.2034 | 0.49 | 0.0029 | −0.02 | 0.5755 | 0.08 | 0.6980 | 0.06 | 0.6797 | 0.09 | 0.5482 | 0.13 | 0.2323 | 0.11 | 0.4670 |
| At 4 months | ||||||||||||||||
| Weight | −0.59 | 0.0003 | 0.45 | 0.0087 | −0.23 | 0.2027 | −0.57 | 0.0006 | −0.70 | <0.0001 | −0.62 | 0.0001 | −0.61 | 0.0002 | −0.62 | 0.0001 |
| Length | −0.61 | 0.0002 | 0.28 | 0.1087 | −0.24 | 0.1761 | −0.56 | 0.0008 | −0.67 | <0.0001 | −0.63 | 0.0001 | −0.63 | <0.0001 | −0.60 | 0.0002 |
| BMI Z-score | −0.33 | 0.0642 | 0.57 | 0.0008 | −0.09 | 0.2725 | −0.35 | 0.0520 | −0.46 | 0.0088 | −0.37 | 0.0368 | −0.35 | 0.0543 | −0.39 | 0.0287 |
| HOMA-IR | 0.44 | 0.0033 | 0.13 | 0.3602 | 0.18 | 0.3165 | 0.37 | 0.0521 | 0.48 | 0.0026 | 0.42 | 0.0492 | 0.52 | <0.0001 | 0.46 | 0.0040 |
| At 12 months | ||||||||||||||||
| Weight | −0.38 | 0.0209 | 0.37 | 0.0251 | −0.26 | 0.1290 | −0.44 | 0.0070 | −0.46 | 0.0053 | −0.43 | 0.0086 | −0.34 | 0.0408 | −0.40 | 0.0171 |
| Length | −0.33 | 0.0464 | 0.17 | 0.3152 | −0.33 | 0.0520 | −0.40 | 0.0146 | −0.38 | 0.0212 | −0.35 | 0.0376 | −0.24 | 0.1559 | −0.37 | 0.0257 |
| BMI Z-score | −0.22 | 0.1995 | 0.43 | 0.0088 | −0.04 | 0.8369 | −0.24 | 0.1708 | −0.30 | 0.0817 | −0.28 | 0.1017 | −0.26 | 0.1275 | −0.22 | 0.2020 |
| HOMA-IR | 0.30 | 0.2893 | −0.10 | 0.3981 | 0.23 | 0.4132 | 0.26 | 0.3879 | 0.24 | 0.3960 | 0.32 | 0.1219 | 0.32 | 0.1039 | 0.28 | 0.1461 |
| Δ 0–4 months | ||||||||||||||||
| Weight | 0.05 | 0.7797 | 0.24 | 0.1723 | 0.28 | 0.1104 | 0.06 | 0.7188 | −0.00 | 0.9765 | 0.01 | 0.9170 | 0.01 | 0.9171 | 0.02 | 0.9133 |
| Length | 0.08 | 0.6620 | −0.15 | 0.3884 | 0.22 | 0.2218 | 0.11 | 0.5377 | 0.14 | 0.4316 | 0.06 | 0.7043 | 0.05 | 0.7556 | 0.10 | 0.5647 |
| BMI Z-score | 0.66 | <0.0001 | −0.12 | 0.5141 | 0.53 | 0.0018 | 0.63 | 0.0001 | 0.70 | <0.0001 | 0.65 | <0.0001 | 0.65 | <0.0001 | 0.64 | <0.0001 |
| HOMA-IR | 0.22 | 0.4699 | −0.24 | 0.4134 | 0.10 | 0.7117 | 0.18 | 0.5190 | 0.30 | 0.1867 | 0.22 | 0.3641 | 0.29 | 0.0911 | 0.25 | 0.2475 |
| Δ 0–12 months | ||||||||||||||||
| Weight | 0.11 | 0.5274 | 0.20 | 0.2305 | 0.07 | 0.6765 | 0.01 | 0.9130 | 0.08 | 0.6267 | 0.05 | 0.7468 | 0.16 | 0.3583 | 0.10 | 0.5524 |
| Length | 0.32 | 0.0553 | −0.18 | 0.2762 | 0.08 | 0.6426 | 0.23 | 0.1781 | 0.37 | 0.0245 | 0.32 | 0.0561 | 0.42 | 0.0108 | 0.30 | 0.0768 |
| BMI Z-score | 0.60 | 0.0002 | −0.12 | 0.6655 | 0.51 | 0.0020 | 0.56 | 0.0006 | 0.65 | <0.0001 | 0.56 | 0.0005 | 0.57 | 0.0004 | 0.62 | <0.0001 |
| HOMA-IR | 0.07 | 0.7758 | −0.20 | 0.0698 | 0.16 | 0.4879 | 0.06 | 0.8720 | 0.08 | 0.8618 | 0.11 | 0.2786 | 0.09 | 0.7417 | 0.07 | 0.7753 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Riano, C.; Santos, M.; Díaz, M.; García-Beltran, C.; Lerin, C.; Barbas, C.; Ibáñez, L.; Sánchez-Infantes, D. Birth Weight and Early Postnatal Outcomes: Association with the Cord Blood Lipidome. Nutrients 2022, 14, 3760. https://doi.org/10.3390/nu14183760
Gonzalez-Riano C, Santos M, Díaz M, García-Beltran C, Lerin C, Barbas C, Ibáñez L, Sánchez-Infantes D. Birth Weight and Early Postnatal Outcomes: Association with the Cord Blood Lipidome. Nutrients. 2022; 14(18):3760. https://doi.org/10.3390/nu14183760
Chicago/Turabian StyleGonzalez-Riano, Carolina, Marcelo Santos, Marta Díaz, Cristina García-Beltran, Carles Lerin, Coral Barbas, Lourdes Ibáñez, and David Sánchez-Infantes. 2022. "Birth Weight and Early Postnatal Outcomes: Association with the Cord Blood Lipidome" Nutrients 14, no. 18: 3760. https://doi.org/10.3390/nu14183760
APA StyleGonzalez-Riano, C., Santos, M., Díaz, M., García-Beltran, C., Lerin, C., Barbas, C., Ibáñez, L., & Sánchez-Infantes, D. (2022). Birth Weight and Early Postnatal Outcomes: Association with the Cord Blood Lipidome. Nutrients, 14(18), 3760. https://doi.org/10.3390/nu14183760





