Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Histology, Immunohistochemistry, and Immunofluorescence Analysis
2.3. Hematological Examination of Inflammatory Markers
2.4. RT–PCR
2.5. 16S rRNA Sequencing
2.6. Metabolic Profiling
2.7. Transcriptome Analysis
2.8. Statistical Analysis
3. Results
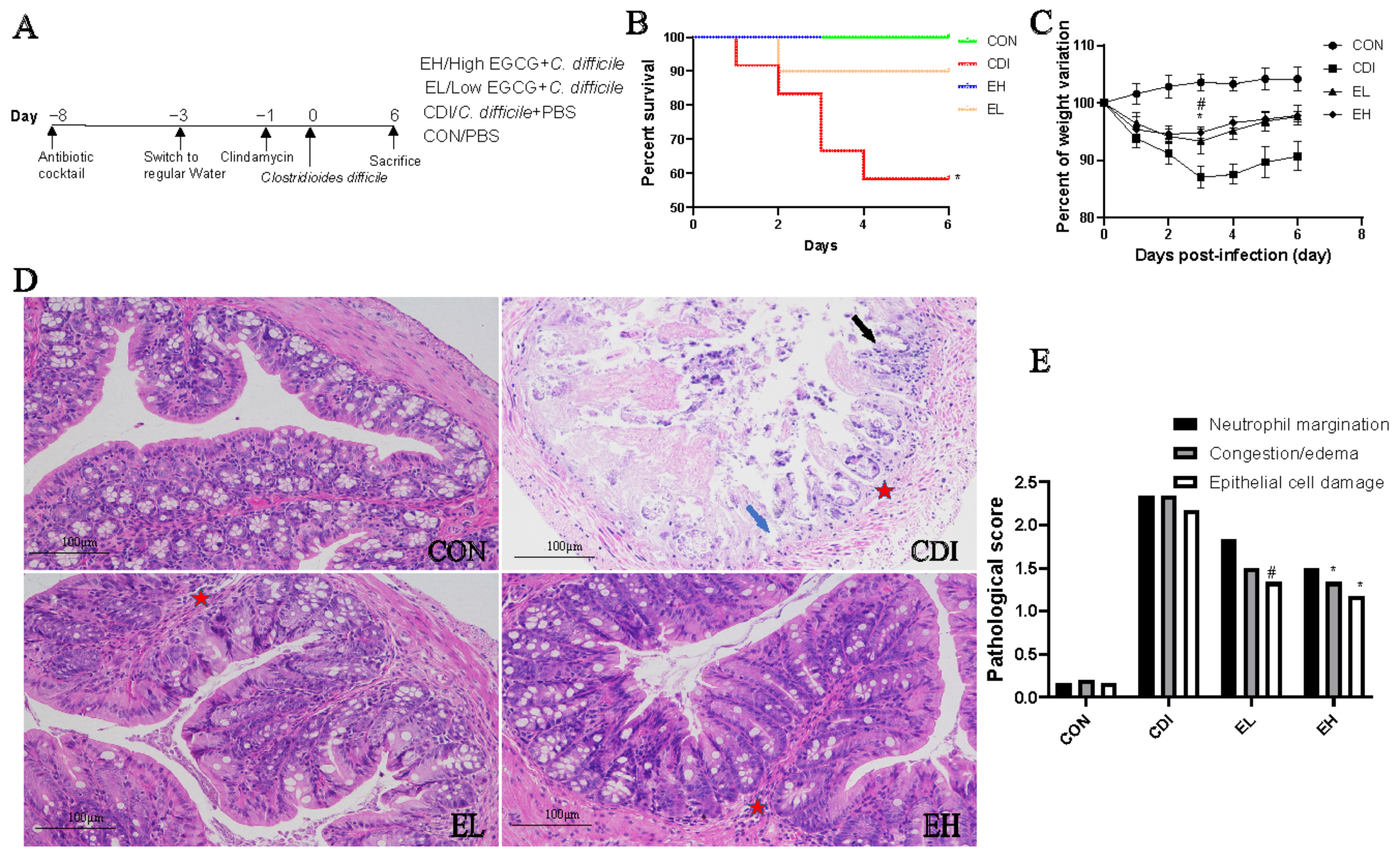
3.1. Oral EGCG Administration Reduces C. difficile-Induced Clinical Symptoms
3.2. Oral EGCG Inhibits C. difficile-Induced Intestinal Mucosal Disruption and Improves Barrier Function
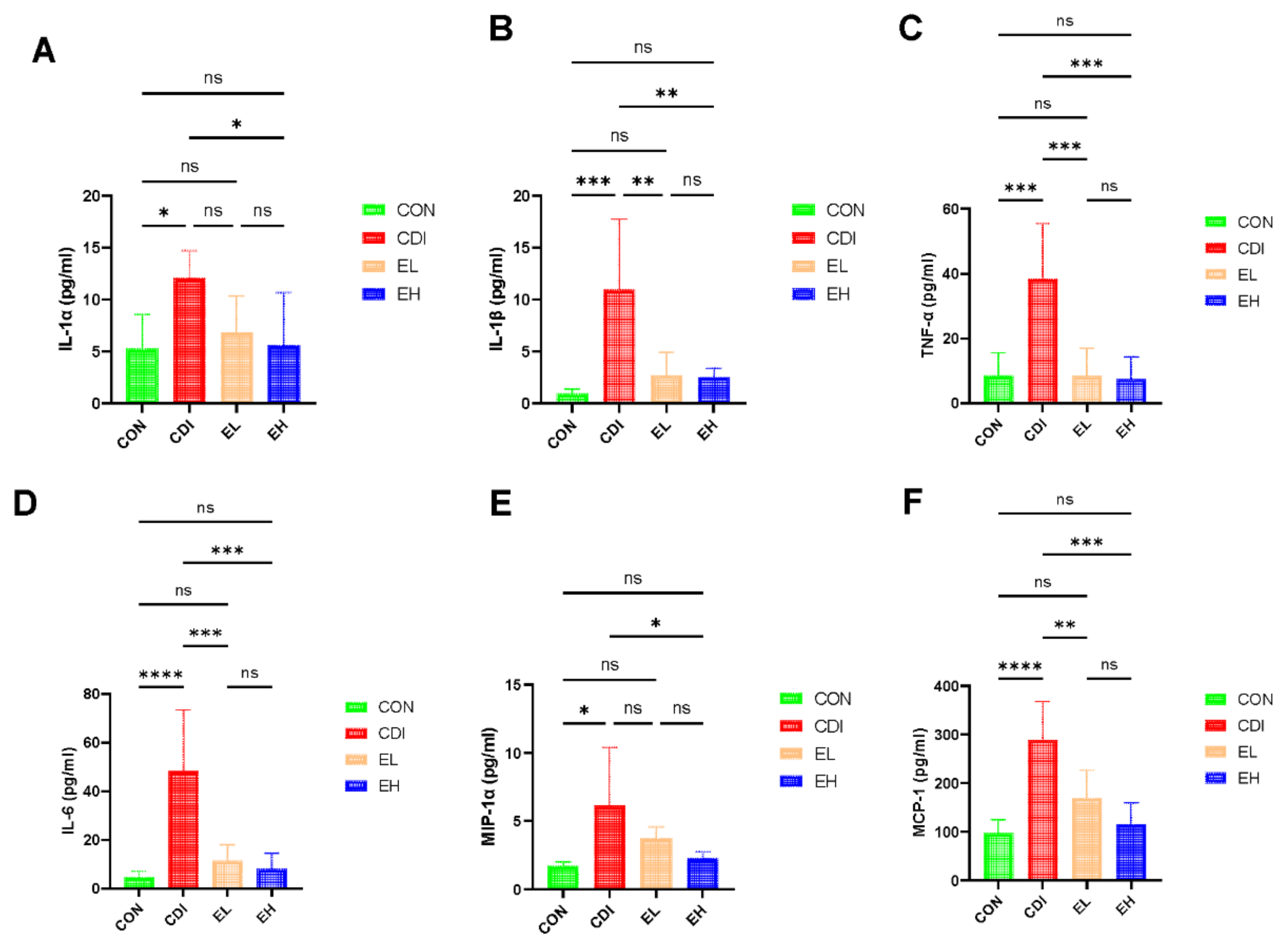
3.3. EGCG Reduces the Serum Inflammation in C. difficile Infection
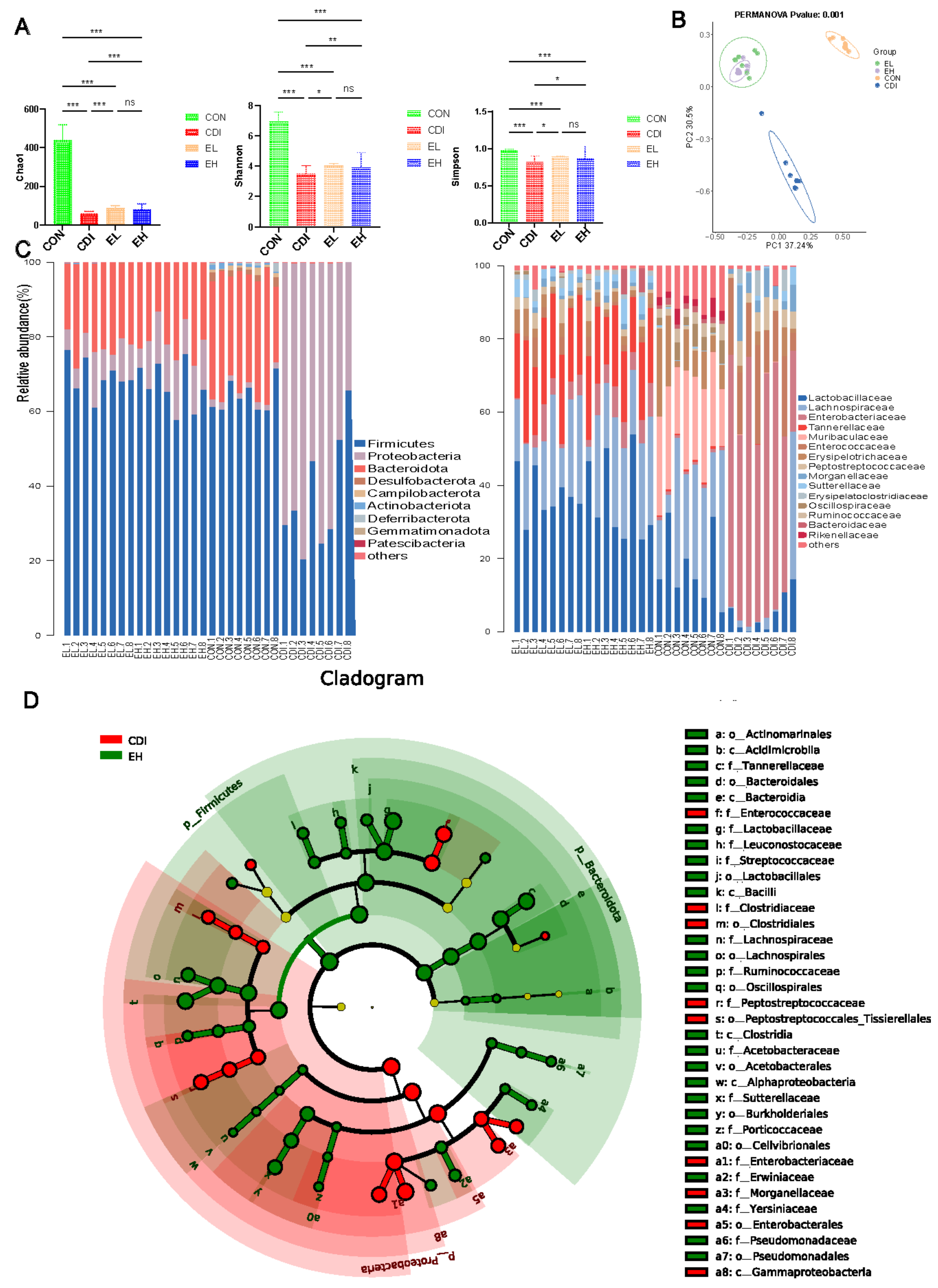
3.4. EGCG Regulates the Microbiota Composition in the Intestine
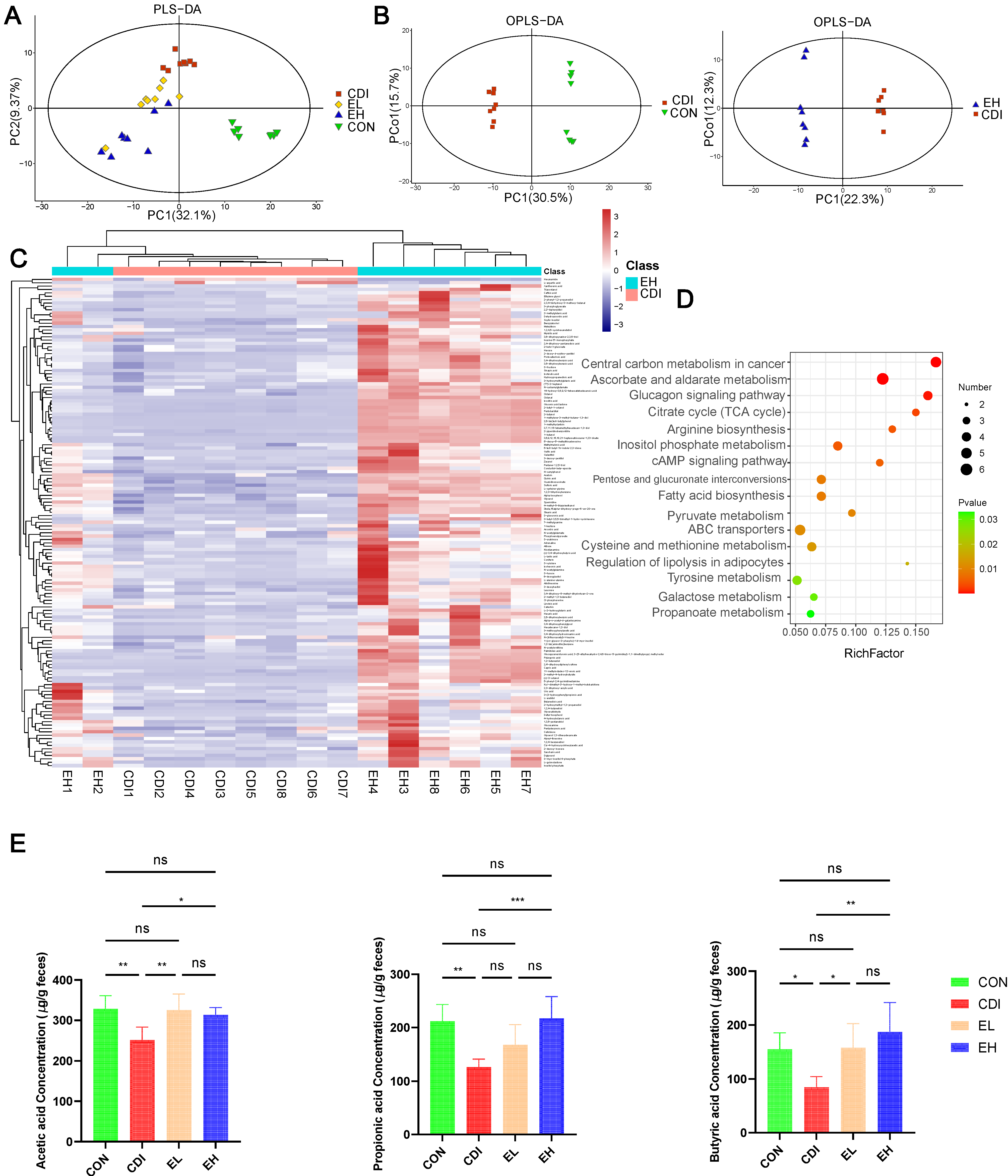
3.5. EGCG improves Metabolic Disorders and SCFAs Levels after C. difficile Infection
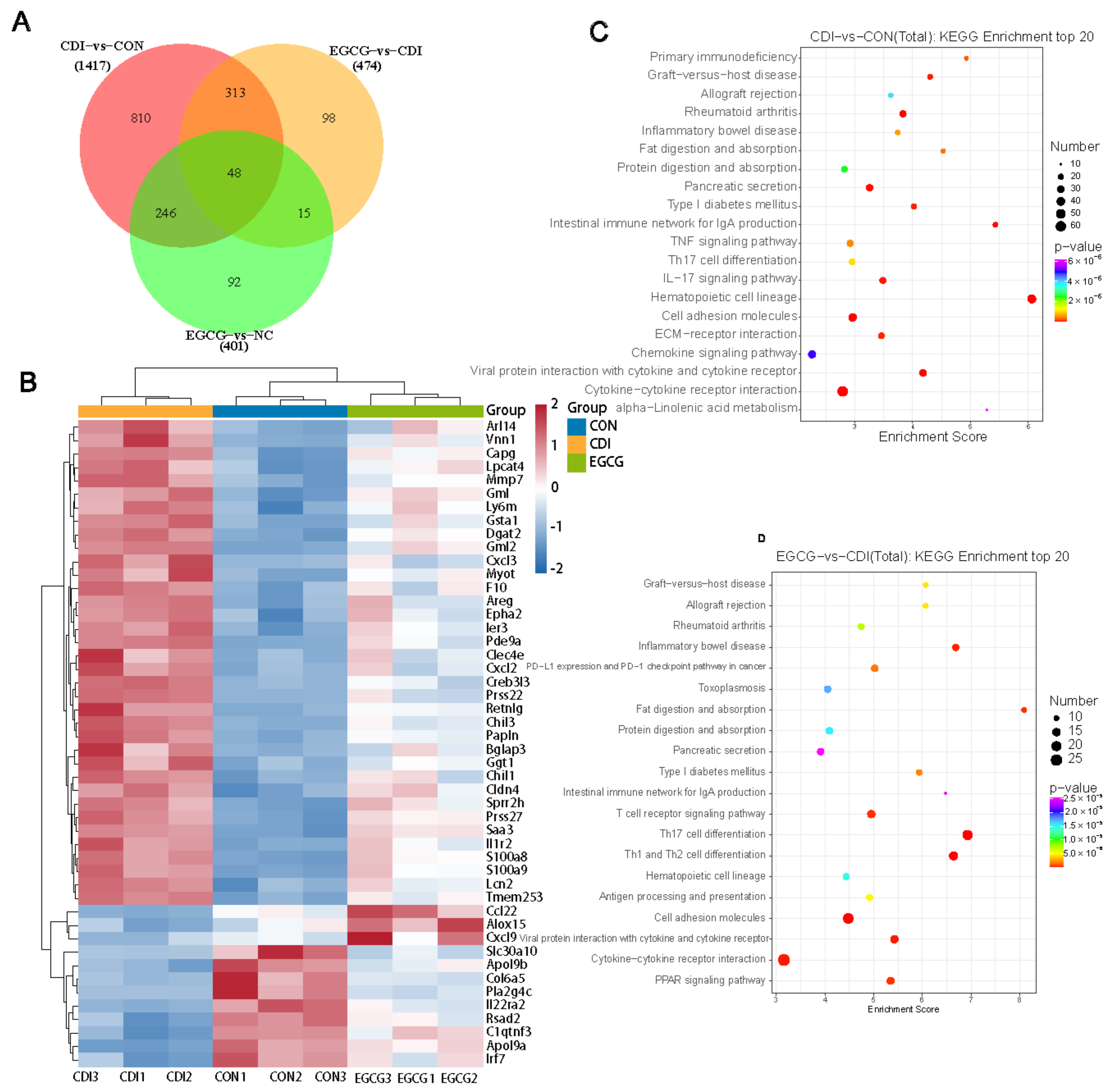
3.6. EGCG Reshapes the Transcriptome Profile of C. difficile-Infected Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, S.; Palazuelos-Munoz, S.; Balsells, E.M.; Nair, H.; Chit, A.; Kyaw, M.H. Cost of hospital management of Clostridium difficile infection in United States-a meta-analysis and modelling study. BMC Infect. Dis. 2016, 16, 447. [Google Scholar] [CrossRef] [PubMed]
- Ghantoji, S.S.; Sail, K.; Lairson, D.R.; DuPont, H.L.; Garey, K.W. Economic healthcare costs of Clostridium difficile infection: A systematic review. J. Hosp. Infect. 2010, 74, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Kriss, M.; Hazleton, K.Z.; Nusbacher, N.M.; Martin, C.G.; Lozupone, C.A. Low diversity gut microbiota dysbiosis: Drivers, functional implications and recovery. Curr. Opin. Microbiol. 2018, 44, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Huang, Y.D.; Chen, Y.B.; Lv, T.; Zhu, C.X.; Huang, J.R.; Li, L.J. Prediction of Clostridium difficile infection based on gut microbial traits in patients with Clostridium difficile colonization. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 298–300. [Google Scholar] [CrossRef]
- Weingarden, A.R.; Chen, C.; Zhang, N.; Graiziger, C.T.; Dosa, P.I.; Steer, C.J.; Shaughnessy, M.K.; Johnson, J.R.; Sadowsky, M.J.; Khoruts, A. Ursodeoxycholic Acid Inhibits Clostridium difficile Spore Germination and Vegetative Growth, and Prevents the Recurrence of Ileal Pouchitis Associated With the Infection. J. Clin. Gastroenterol. 2016, 50, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Leong, J.; Teo, J.Y.; Mitchell, J.W.; Gillette, M.U.; Han, B.; Lee, J.; Kong, H. Active Antioxidizing Particles for On-Demand Pressure-Driven Molecular Release. ACS Appl. Mater. Interfaces 2017, 9, 35642–35650. [Google Scholar] [CrossRef]
- Xing, L.; Zhang, H.; Qi, R.; Tsao, R.; Mine, Y. Recent Advances in the Understanding of the Health Benefits and Molecular Mechanisms Associated with Green Tea Polyphenols. J. Agric. Food Chem. 2019, 67, 1029–1043. [Google Scholar] [CrossRef]
- Zhong, Y.; Chiou, Y.S.; Pan, M.H.; Shahidi, F. Anti-inflammatory activity of lipophilic epigallocatechin gallate (EGCG) derivatives in LPS-stimulated murine macrophages. Food Chem. 2012, 134, 742–748. [Google Scholar] [CrossRef]
- Avadhani, K.S.; Manikkath, J.; Tiwari, M.; Chandrasekhar, M.; Godavarthi, A.; Vidya, S.M.; Hariharapura, R.C.; Kalthur, G.; Udupa, N.; Mutalik, S. Skin delivery of epigallocatechin-3-gallate (EGCG) and hyaluronic acid loaded nano-transfersomes for antioxidant and anti-aging effects in UV radiation induced skin damage. Drug Deliv. 2017, 24, 61–74. [Google Scholar] [CrossRef]
- Ahmad, N.; Mukhtar, H. Green tea polyphenols and cancer: Biologic mechanisms and practical implications. Nutr. Rev. 1999, 57, 78–83. [Google Scholar] [CrossRef]
- Renaud, J.; Nabavi, S.F.; Daglia, M.; Nabavi, S.M.; Martinoli, M.G. Epigallocatechin-3-Gallate, a Promising Molecule for Parkinson’s Disease? Rejuvenation Res. 2015, 18, 257–269. [Google Scholar] [CrossRef]
- Si, H.; Wang, X.; Zhang, L.; Parnell, L.D.; Admed, B.; LeRoith, T.; Ansah, T.A.; Zhang, L.; Li, J.; Ordovas, J.M.; et al. Dietary epicatechin improves survival and delays skeletal muscle degeneration in aged mice. FASEB J. 2019, 33, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; Qi, G.; Fan, R.; Qiao, Q.; Sun, Y.; Gao, Y.; Liu, X. EGCG ameliorates high-fat- and high-fructose-induced cognitive defects by regulating the IRS/AKT and ERK/CREB/BDNF signaling pathways in the CNS. FASEB J. 2017, 31, 4998–5011. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.; Oh, S.; Song, M.; Hong, Y.S.; Park, S.; Park, D.J.; Griffiths, M.W.; Oh, S. Inhibitory Effect of Epigallocatechin Gallate on the Virulence of Clostridium difficile PCR Ribotype 027. J. Food Sci. 2015, 80, M2925–M2931. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, S.; Li, T.; Li, N.; Han, D.; Zhang, B.; Xu, Z.Z.; Zhang, S.; Pang, J.; Wang, S.; et al. Gut microbiota from green tea polyphenol-dosed mice improves intestinal epithelial homeostasis and ameliorates experimental colitis. Microbiome 2021, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Remely, M.; Ferk, F.; Sterneder, S.; Setayesh, T.; Roth, S.; Kepcija, T.; Noorizadeh, R.; Rebhan, I.; Greunz, M.; Beckmann, J.; et al. EGCG Prevents High Fat Diet-Induced Changes in Gut Microbiota, Decreases of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Oxid. Med. Cell. Longev. 2017, 2017, 3079148. [Google Scholar] [CrossRef]
- Cai, S.; Xie, L.W.; Xu, J.Y.; Zhou, H.; Yang, C.; Tang, L.F.; Tian, Y.; Li, M. (-)-Epigallocatechin-3-Gallate (EGCG) Modulates the Composition of the Gut Microbiota to Protect Against Radiation-Induced Intestinal Injury in Mice. Front. Oncol. 2022, 12, 848107. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Chen, X.; Katchar, K.; Goldsmith, J.D.; Nanthakumar, N.; Cheknis, A.; Gerding, D.N.; Kelly, C.P. A mouse model of Clostridium difficile-associated disease. Gastroenterology 2008, 135, 1984–1992. [Google Scholar] [CrossRef]
- Yang, L.; Li, Y.; Wang, S.; Bian, X.; Jiang, X.; Wu, J.; Wang, K.; Wang, Q.; Xia, J.; Jiang, S.; et al. Western Diet Aggravated Carbon Tetrachloride-Induced Chronic Liver Injury by Disturbing Gut Microbiota and Bile Acid Metabolism. Mol. Nutr. Food Res. 2021, 65, e2000811. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.Z.; Li, Y.T.; Wu, W.R.; Shi, D.; Fang, D.Q.; Yang, L.Y.; Bian, X.Y.; Wu, J.J.; Wang, Q.; Jiang, X.W.; et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J. Gastroenterol. 2018, 24, 2468–2481. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Yang, L.; Wu, W.; Lv, L.; Jiang, X.; Wang, Q.; Wu, J.; Li, Y.; Ye, J.; Fang, D.; et al. Pediococcus pentosaceus LI05 alleviates DSS-induced colitis by modulating immunological profiles, the gut microbiota and short-chain fatty acid levels in a mouse model. Microb. Biotechnol. 2020, 13, 1228–1244. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Poxton, I.R.; McCoubrey, J.; Blair, G. The pathogenicity of Clostridium difficile. Clin. Microbiol. Infect. 2001, 7, 421–427. [Google Scholar] [CrossRef]
- Nakamoto, N.; Amiya, T.; Aoki, R.; Taniki, N.; Koda, Y.; Miyamoto, K.; Teratani, T.; Suzuki, T.; Chiba, S.; Chu, P.S.; et al. Commensal Lactobacillus Controls Immune Tolerance during Acute Liver Injury in Mice. Cell Rep. 2017, 21, 1215–1226. [Google Scholar] [CrossRef]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353.e1321. [Google Scholar] [CrossRef]
- Rathinam, V.A.K.; Zhao, Y.; Shao, F. Innate immunity to intracellular LPS. Nat. Immunol. 2019, 20, 527–533. [Google Scholar] [CrossRef]
- Kim, J.M.; Lee, J.Y.; Yoon, Y.M.; Oh, Y.K.; Youn, J.; Kim, Y.J. NF-kappa B activation pathway is essential for the chemokine expression in intestinal epithelial cells stimulated with Clostridium difficile toxin A. Scand. J. Immunol. 2006, 63, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Schnizlein, M.K.; Young, V.B. Capturing the environment of the Clostridioides difficile infection cycle. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Littman, D.R.; Pamer, E.G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 2011, 10, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Littmann, E.R.; Lee, J.J.; Denny, J.E.; Alam, Z.; Maslanka, J.R.; Zarin, I.; Matsuda, R.; Carter, R.A.; Susac, B.; Saffern, M.S.; et al. Host immunity modulates the efficacy of microbiota transplantation for treatment of Clostridioides difficile infection. Nat. Commun. 2021, 12, 755. [Google Scholar] [CrossRef] [PubMed]
- Petta, S.; Marchesini, G. Coffee and tea breaks for liver health. J. Hepatol. 2017, 67, 221–223. [Google Scholar] [CrossRef]
- Suzuki, T.; Hara, H. Role of flavonoids in intestinal tight junction regulation. J. Nutr. Biochem. 2011, 22, 401–408. [Google Scholar] [CrossRef]
- Hooper, L.V.; Wong, M.H.; Thelin, A.; Hansson, L.; Falk, P.G.; Gordon, J.I. Molecular analysis of commensal host-microbial relationships in the intestine. Science 2001, 291, 881–884. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Zhang, X.; Lu, H.; Lv, T.; Shen, P.; Lv, L.; Zheng, B.; Jiang, X.; Li, L. Identification of key taxa that favor intestinal colonization of Clostridium difficile in an adult Chinese population. Microbes Infect. 2016, 18, 30–38. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Li, D.; Ho, C.T.; Li, J.; Wan, X. The absorption, distribution, metabolism and excretion of procyanidins. Food Funct. 2016, 7, 1273–1281. [Google Scholar] [CrossRef]
- Li, Y.; Gao, X.; Lou, Y. Interactions of tea polyphenols with intestinal microbiota and their implication for cellular signal conditioning mechanism. J. Food Biochem. 2019, 43, e12953. [Google Scholar] [CrossRef]
- Wheeldon, L.J.; Worthington, T.; Lambert, P.A. Histidine acts as a co-germinant with glycine and taurocholate for Clostridium difficile spores. J. Appl. Microbiol. 2011, 110, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Rau, M.; Rehman, A.; Dittrich, M.; Groen, A.K.; Hermanns, H.M.; Seyfried, F.; Beyersdorf, N.; Dandekar, T.; Rosenstiel, P.; Geier, A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 2018, 6, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Ruotolo, R.; Minato, I.; La Vitola, P.; Artioli, L.; Curti, C.; Franceschi, V.; Brindani, N.; Amidani, D.; Colombo, L.; Salmona, M.; et al. Flavonoid-Derived Human Phenyl-gamma-Valerolactone Metabolites Selectively Detoxify Amyloid-beta Oligomers and Prevent Memory Impairment in a Mouse Model of Alzheimer’s Disease. Mol. Nutr. Food Res. 2020, 64, e1900890. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N.; Liddle, C.; Atkins, A.R.; Downes, M.; Evans, R.M. PPARgamma signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef]
- Dong, C. TH17 cells in development: An updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008, 8, 337–348. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.H.G. IL-17 and IL-17-producing cells in protection versus pathology. Nat. Rev. Immunol. 2022. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Shen, J.; Xu, Q.; Xiang, Q.; Chen, Y.; Lv, L.; Zheng, B.; Wang, Q.; Wang, S.; Li, L. Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection. Nutrients 2022, 14, 3756. https://doi.org/10.3390/nu14183756
Wu Z, Shen J, Xu Q, Xiang Q, Chen Y, Lv L, Zheng B, Wang Q, Wang S, Li L. Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection. Nutrients. 2022; 14(18):3756. https://doi.org/10.3390/nu14183756
Chicago/Turabian StyleWu, Zhengjie, Jian Shen, Qiaomai Xu, Qiangqiang Xiang, Yunbo Chen, Longxian Lv, Beiwen Zheng, Qiangqiang Wang, Shuting Wang, and Lanjuan Li. 2022. "Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection" Nutrients 14, no. 18: 3756. https://doi.org/10.3390/nu14183756
APA StyleWu, Z., Shen, J., Xu, Q., Xiang, Q., Chen, Y., Lv, L., Zheng, B., Wang, Q., Wang, S., & Li, L. (2022). Epigallocatechin-3-Gallate Improves Intestinal Gut Microbiota Homeostasis and Ameliorates Clostridioides difficile Infection. Nutrients, 14(18), 3756. https://doi.org/10.3390/nu14183756



