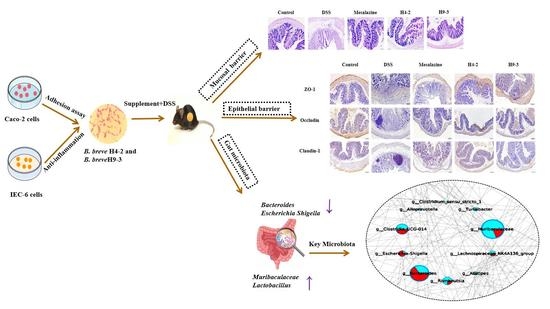
Bifidobacterium breve Alleviates DSS-Induced Colitis in Mice by Maintaining the Mucosal and Epithelial Barriers and Modulating Gut Microbes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Bacterial Strains and Cells
2.2. Analysis of Probiotic Characteristics of Strains
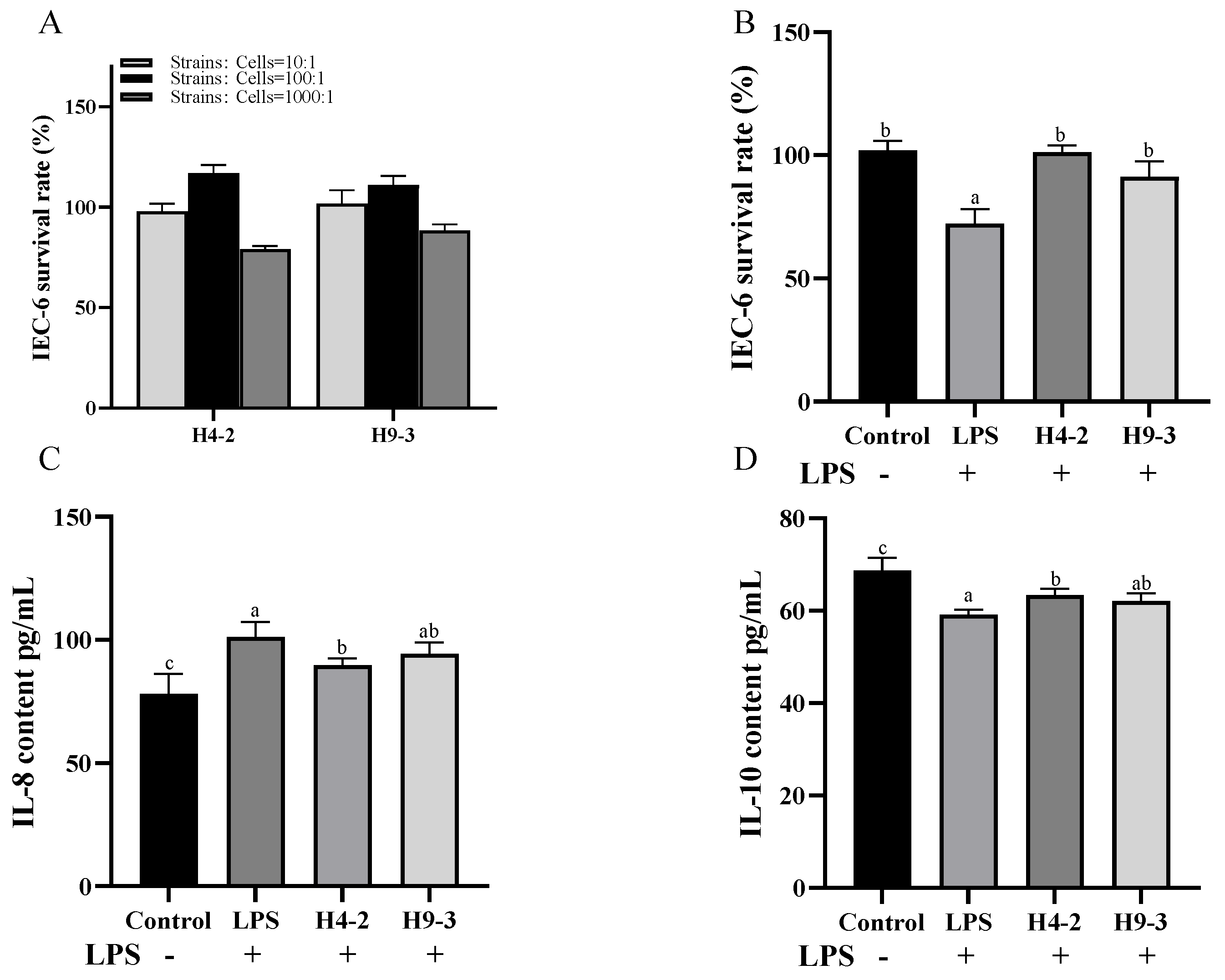
2.3. Assays of the Cell Proliferation and Cytokine Level Test
2.4. Animals and Treatments
2.5. Inflammatory Markers and Histopathology
2.6. Determination of D-Lactic Acid in Serum
2.7. Assessment of Myeloperoxidase (MPO) Activity
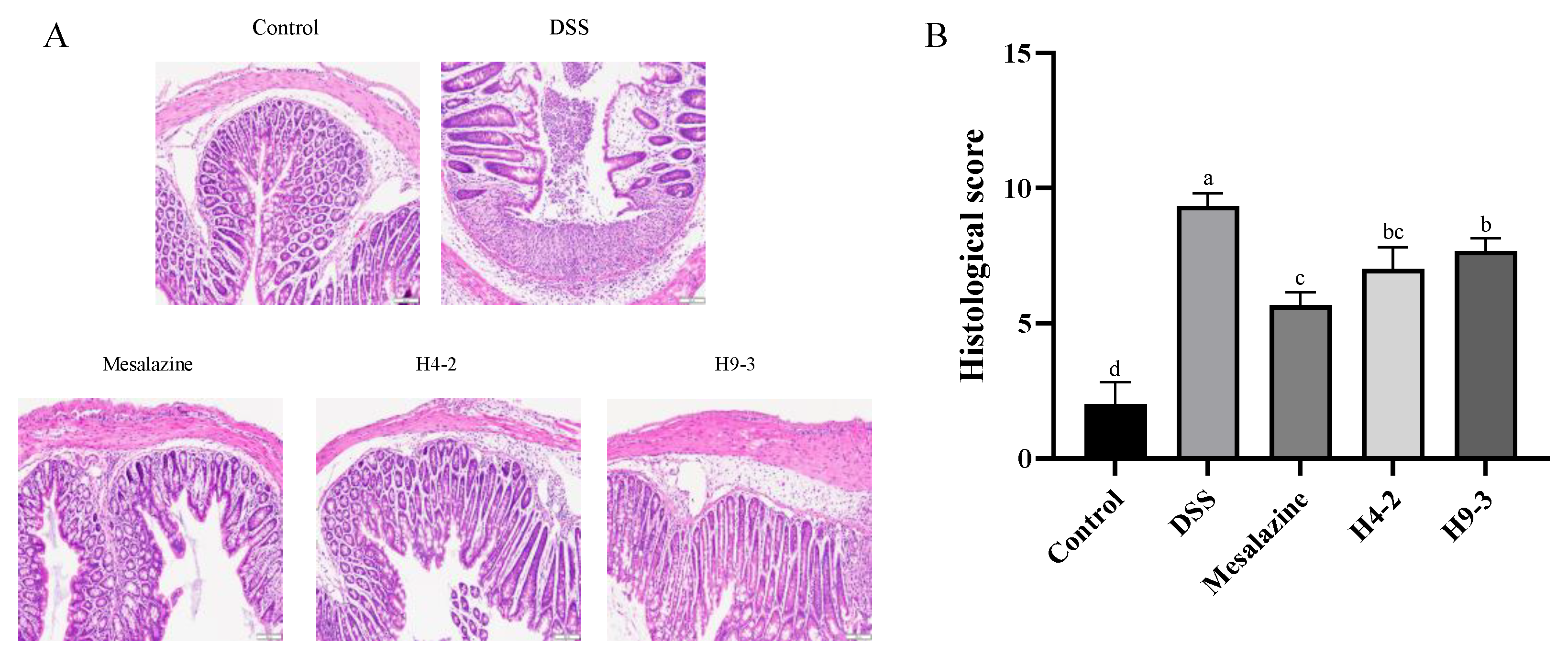
2.8. Evaluation of the Histological Score
2.9. Determination of Biochemical Indices in Serum and Colon
2.10. Immunohistochemical Staining and Scoring
2.11. Quantitative Real-Time PCR
2.12. SCFAs Quantification
2.13. Gut Microbiota Analysis
2.14. Statistical Analysis
3. Results
3.1. Gastrointestinal Tolerance and Adhesion Characteristics of Strains
3.2. Intervention with B. breve Reduced Inflammation in LPS Induced IEC-6 Cells
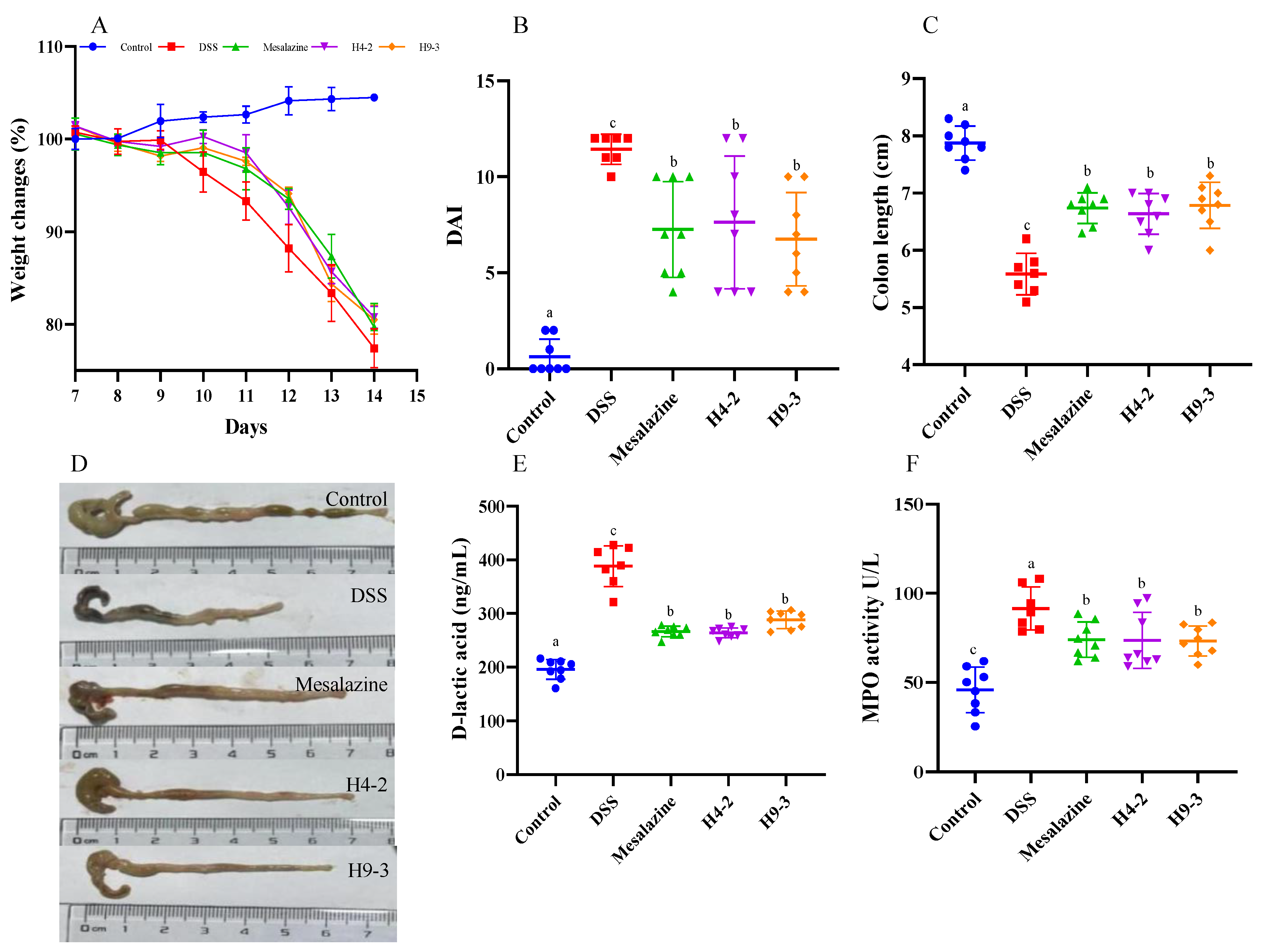
3.3. B. breve Improved Colitis Symptoms
3.4. B. breve Recovered the Damage in Colonic Tissue
3.5. B. breve Recovered Mucus Disruption and Goblet Cells Exhaustion
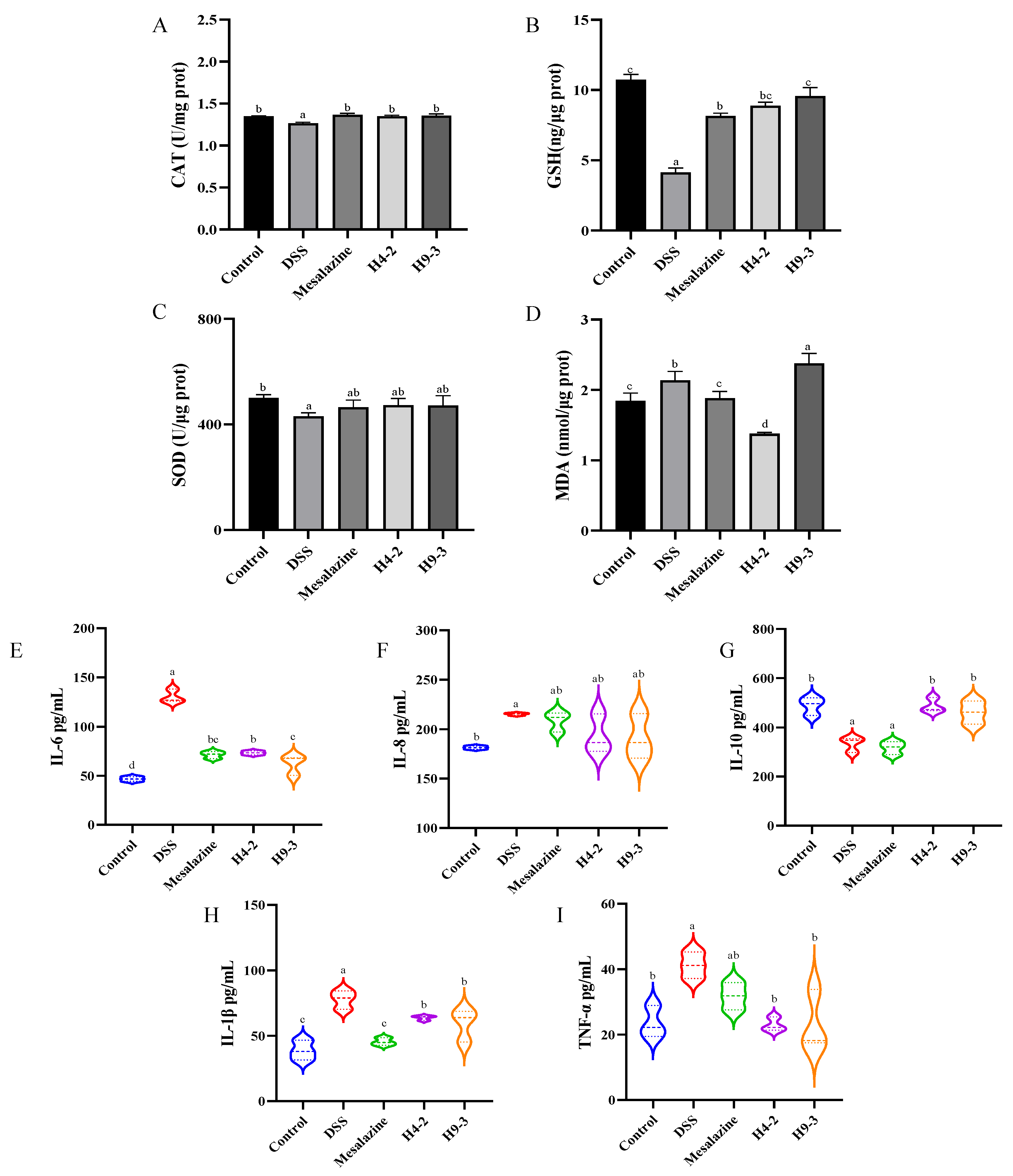
3.6. B. breve Improved Serum and Colon Biochemical Functions
3.7. B. breve Protected the Epithelial Barrier
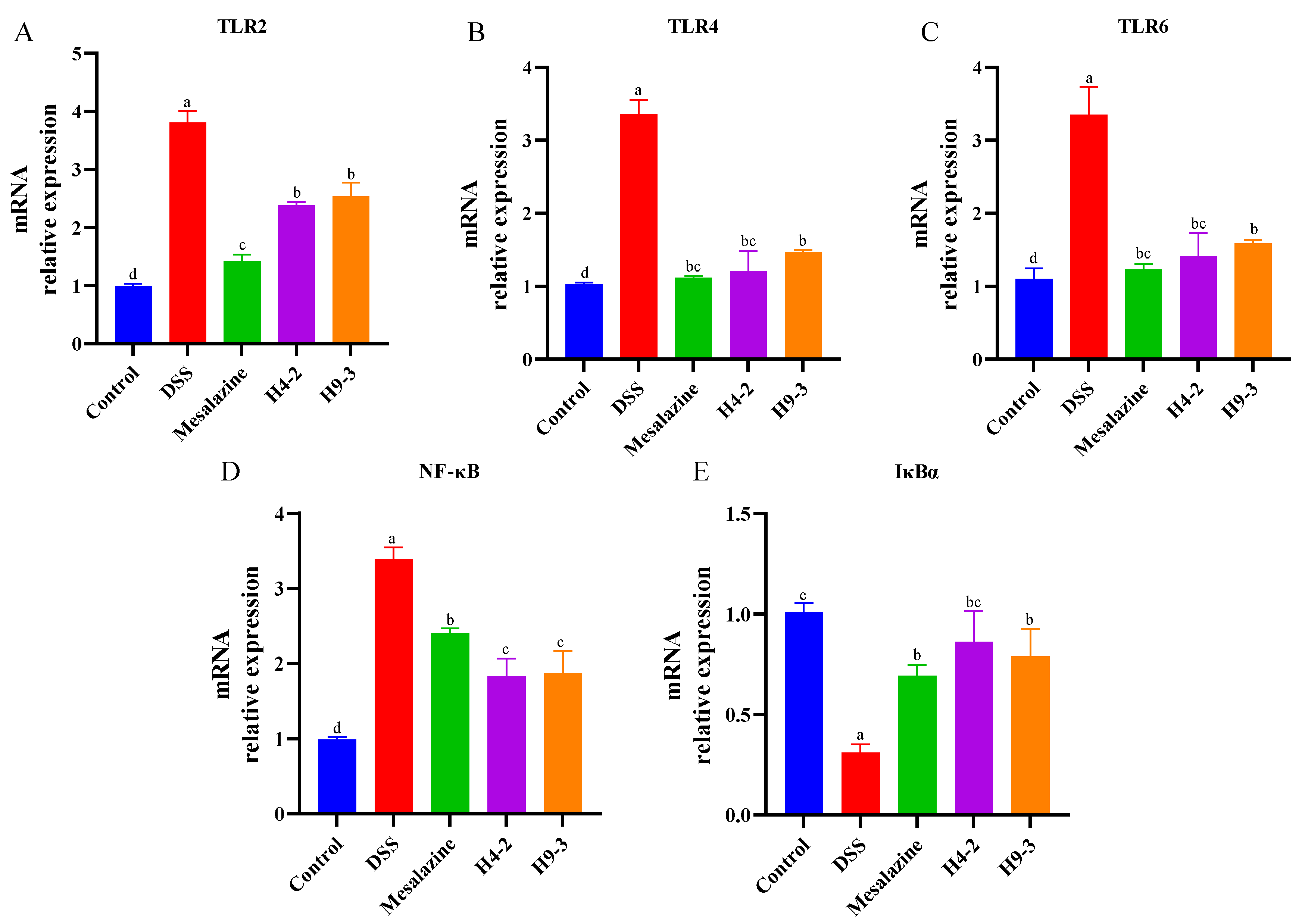
3.8. B. breve Regulated Toll-like Receptors (TLRs) and NF-κB Signaling Pathways
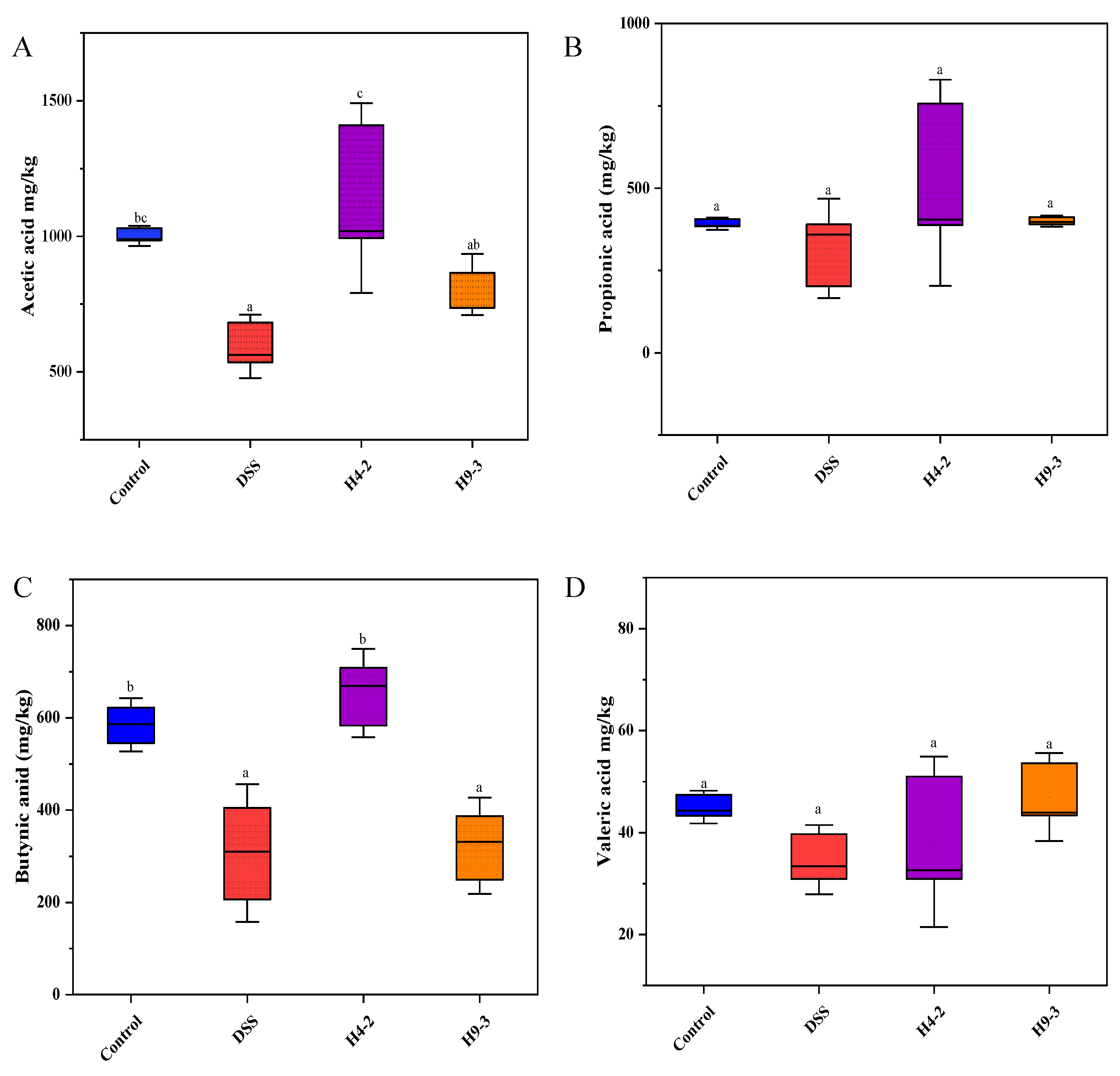
3.9. B. breve Improved SCFAs Level
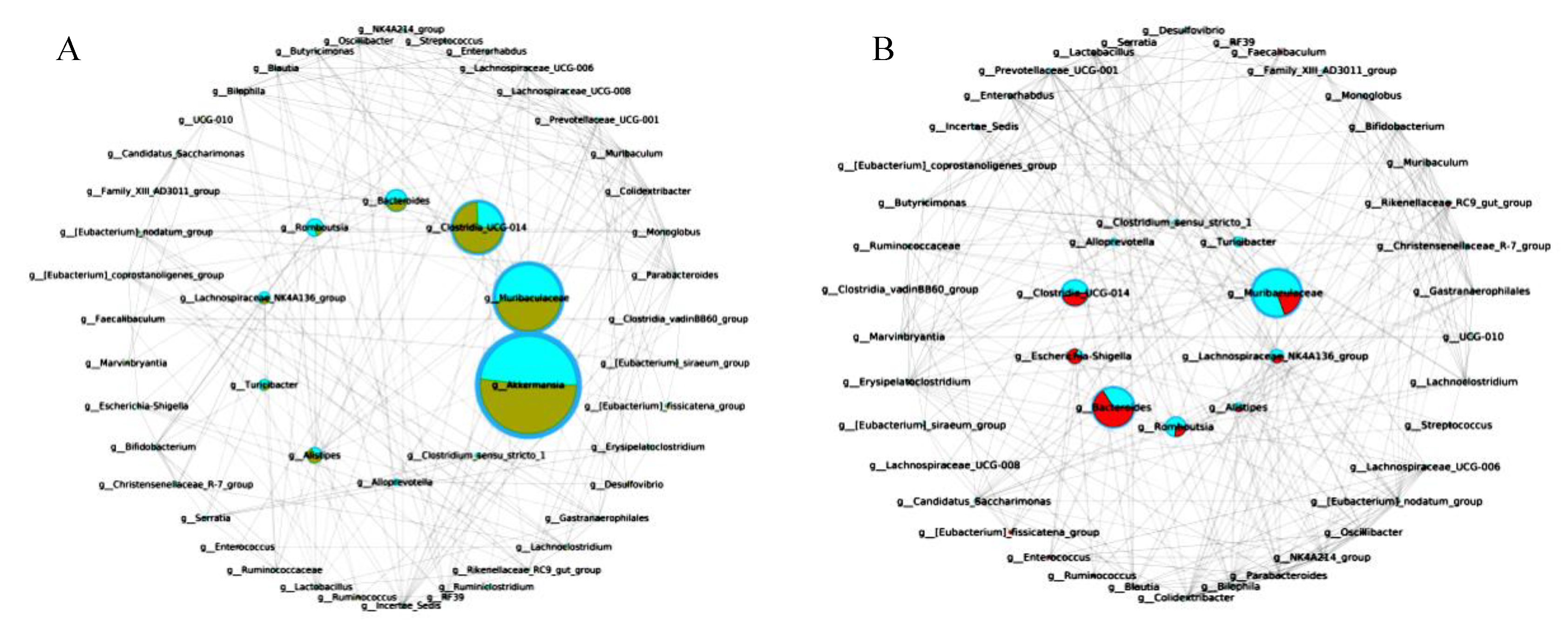
3.10. B. breve Regulated Gut Microbiota
3.11. The Key Microbiota Associated with Colitis
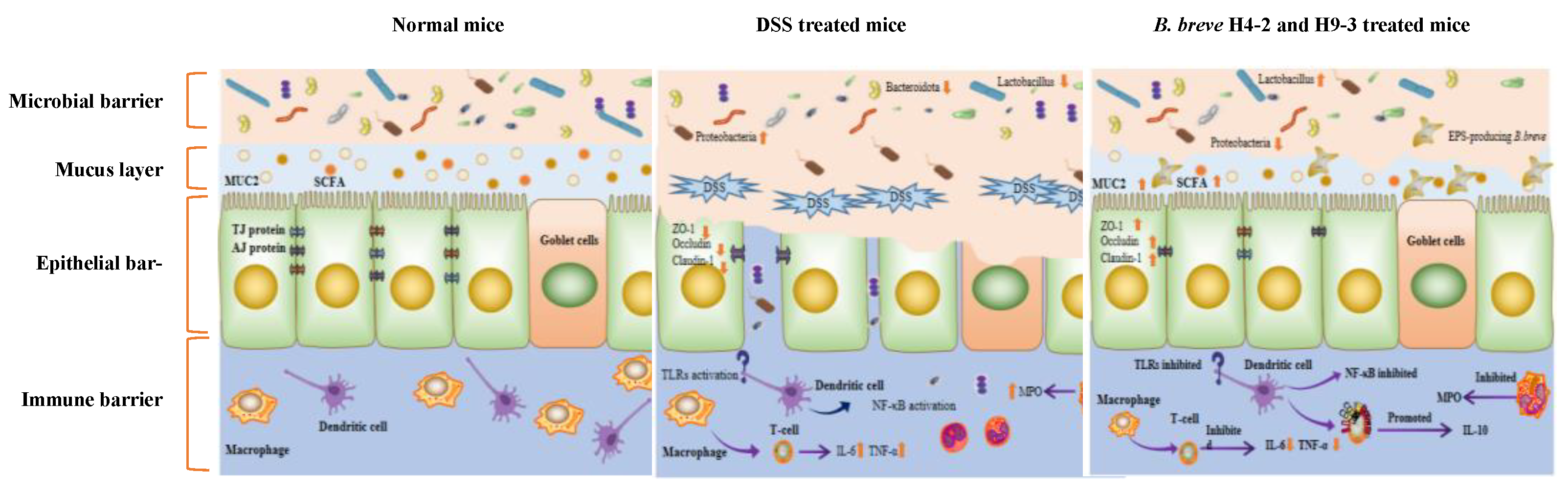
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akutko, K.; Stawarski, A. Probiotics, Prebiotics and Synbiotics in Inflammatory Bowel Diseases. J. Clin. Med. 2021, 10, 2466. [Google Scholar] [CrossRef]
- Alatab, S.; Sepanlou, S.; Ikuta, K.S.; Vahedi, H.; Naghavi, M. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef]
- Dahlhamer, J.M.; Zammitti, E.P.; Ward, B.W.; Wheaton, A.G.; Croft, J.B. Prevalence of Inflammatory Bowel Disease among Adults Aged ≥ 18 Years—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1166–1169. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hu, B.; Cheng, Y.; Guo, Y.; Yao, W.; Qian, H. In-depth analysis of the mechanisms of aloe polysaccharides on mitigating subacute colitis in mice via microbiota informatics. Carbohydr. Polym. 2021, 265, 118041. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sun, K.; Wu, Y.; Yang, Y.; Patrick, T.; Wu, Z. Interactions between Intestinal Microbiota and Host Immune Response in Inflammatory Bowel Disease. Front. Immunol. 2017, 8, 942. [Google Scholar] [CrossRef]
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; Mciver, L.J. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305. [Google Scholar] [CrossRef]
- Clarke, K.; Chintanaboina, J. Allergic and Immunologic Perspectives of Inflammatory Bowel Disease. Clin. Rev. Allergy Immunol. 2019, 57, 179–193. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; Leleiko, N.; Snapper, S.B. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Franzosa, E.A.; Lloyd-Price, J.; Mciver, L.J.; Huttenhower, C. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat. Microbiol. 2018, 3, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Kudelka, M.R.; Stowell, S.R.; Cummings, R.D.; Neish, A.S. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 597–617. [Google Scholar] [CrossRef] [PubMed]
- Theodoratou, E.; Campbell, H.; Ventham, N.T.; Kolarich, D.; Pučić-Baković, M.; Zoldoš, V.; Fernandes, D.; Pemberton, I.K.; Rudan, I.; Kennedy, N.A. The role of glycosylation in IBD. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 588–600. [Google Scholar] [CrossRef]
- Caballero-Franco, C.; Keller, K.; De Simone, C.; Chadee, K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G315–G322. [Google Scholar]
- Ivanov, A.I.; Naydenov, N.G. Dynamics and regulation of epithelial adherens junctions: Recent discoveries and controversies. Int. Rev. Cell Mol. Biol. 2013, 303, 27–99. [Google Scholar]
- Shen, L.; Weber, C.R.; Raleigh, D.R.; Yu, D.; Turner, J.R. Tight junction pore and leak pathways: A dynamic duo. Annu. Rev. Physiol. 2011, 73, 283–309. [Google Scholar] [CrossRef]
- Zhao, Z.; Chen, L.; Zhao, Y.; Wang, C.; Duan, C.; Yang, G.; Niu, C.; Li, S. Lactobacillus plantarum NA136 ameliorates nonalcoholic fatty liver disease by modulating gut microbiota, improving intestinal barrier integrity, and attenuating inflammation. Appl. Microbiol. Biotechnol. 2020, 104, 5273–5282. [Google Scholar] [CrossRef]
- Suzuki, T.; Yoshida, S.; Hara, H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br. J. Nutr. 2008, 100, 297–305. [Google Scholar] [CrossRef]
- Forbes, A.; Escher, J.; Hebuterne, X.; Klek, S.; Krznaric, Z.; Schneider, S.; Shamir, R.; Stardelova, K.; Wierdsma, N.; Wiskin, A.E.; et al. ESPEN guideline: Clinical nutrition in inflammatory bowel disease. Clin. Nutr. 2017, 36, 321–347. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Odamaki, T.; Mitsuyama, E.; Sugahara, H.; Xiao, J.; Osawa, R. Age-Related Changes in the Composition of Gut Bifidobacterium Species. Curr. Microbiol. 2017, 74, 987–995. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jin, Y.; Stanton, C.; Ross, R.; Wang, Z.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Dose-response efficacy and mechanisms of orally administered CLA-producing Bifidobacterium breve CCFM683 on DSS-induced colitis in mice. J. Funct. Foods 2020, 75, 104245. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, Y.; Stanton, C.; Ross, R.; Zhao, J.; Zhang, H.; Yang, B.; Chen, W. Alleviation effects of Bifidobacterium breve on DSS-induced colitis depends on intestinal tract barrier maintenance and gut microbiota modulation. Eur. J. Nutr. 2021, 60, 369–387. [Google Scholar] [CrossRef]
- Du, M.; Xie, X.; Yang, S.; Li, Y.; Jiang, T.; Yang, J.; Li, L.; Huang, Y.; Wu, Q.; Chen, W.; et al. Lysozyme-like Protein Produced by Bifidobacterium longum Regulates Human Gut Microbiota Using In Vitro Models. Molecules 2021, 26, 6480. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Niu, M.; Song, D.; Song, X.; Zhao, J.; Wu, Y.; Lu, B.; Niu, G. Preparation, partial characterization and biological activity of exopolysaccharidesproduced from Lactobacillus fermentum S1. J. Biosci. Bineng. 2020, 129, 206–214. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.F.; Li, Q.; Tian, K.; Xu, L.; Liu, G.; Guo, C. Exopolysaccharide, Isolated from a Novel Strain Bifidobacterium breve lw01 Possess an Anticancer Effect on Head and Neck Cancer—Genetic and Biochemical Evidences. Front. Microbiol. 2019, 10, 1044. [Google Scholar] [CrossRef]
- Yuan, L.; Chu, B.; Chen, S.; Li, Y.; Liu, N.; Zhu, Y.; Zhou, D. Exopolysaccharides from Bifidobacterium animalis Ameliorate Escherichia coli-Induced IPEC-J2 Cell Damage via Inhibiting Apoptosis and Restoring Autophagy. Microorganisms 2021, 9, 2363. [Google Scholar] [CrossRef]
- Yan, S.; Zhao, G.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Production of exopolysaccharide by Bifidobacterium longum isolated from elderly and infant feces and analysis of priming glycosyltransferase genes. RSC Adv. 2017, 7, 31736–31744. [Google Scholar] [CrossRef]
- Saadat, Y.; Khosroushahi, A.; Gargari, B. A comprehensive review of anticancer, immunomodulatory and health beneficial effects of the lactic acid bacteria exopolysaccharides. Carbohydr. Polym. 2019, 217, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Rook, G.; Bäckhed, F.; Levin, B.; McFall-Ngai, M.; McLean, A. Evolution, human-microbe interactions, and life history plasticity. Lancet 2017, 390, 521–530. [Google Scholar] [CrossRef]
- Hughes, K.R.; Harnisch, L.C.; Alcon-Giner, C.; Mitra, S.; Wright, C.J.; Ketskemety, J.; Van Sinderen, D.; Watson, A.J.M.; Hall, L.J. Bifidobacterium breve reduces apoptotic epithelial cell shedding in an exopolysaccharide and MyD88-dependent manner. Open Biol. 2017, 7, 160155. [Google Scholar] [CrossRef] [Green Version]
- Püngel, D.; Treveil, A.; Dalby, M.J.; Caim, S.; Hall, L.J. Bifidobacterium breve UCC2003 Exopolysaccharide Modulates the Early Life Microbiota by Acting as a Potential Dietary Substrate. Nutrients 2020, 12, 948. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Yang, B.; Zhao, J.; Zhao, J.; Stanton, C.; Ross, R.P.; Zhang, H.; Chen, W. A ropy exopolysaccharide producing strain Bifidobacterium longum subsp. longum YS108R alleviates DSS-induced colitis by maintenance of the mucosal barrier and gut microbiota modulation. Food Funct. 2019, 10, 1595–1608. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, F.; Zhao, L.; Meng, X.; Li, B. Potential Immunomodulatory Activity of a Selected Strain Bifidobacterium bifidum H3-R2 as Evidenced In Vitro and in Immunosuppressed Mice. Front. Microbiol. 2020, 11, 2089. [Google Scholar] [CrossRef]
- Philippe, D.; Heupel, E.; Blum-Sperisen, S.; Riedel, C.U. Treatment with Bifidobacterium bifidum 17 partially protects mice from Th1-driven inflammation in a chemically induced model of colitis. Int. J. Food Microbiol. 2011, 149, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tao, H.; Huang, H.; Xiao, Y.; Wu, X.; Li, M.; Shen, J.; Xiao, Z.; Zhao, Y.; Du, F. The dietary supplement Rhodiola crenulata extract alleviates dextran sulfate sodium-induced colitis in mice through anti-inflammation, mediating gut barrier integrity and reshaping the gut microbiome. Food Funct. 2021, 12, 3142–3158. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014, 9, e96801. [Google Scholar] [CrossRef]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.; Fierer, N.; Peña, A.; Goodrich, J.; Huttley, G.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Dharmaprakash, V.; Nighot, P.; Guo, S.; Nighot, M.; Do, T.; Ma, T.Y. Bifidobacterium bifidum Enhances the Intestinal Epithelial Tight Junction Barrier and Protects against Intestinal Inflammation by Targeting the Toll-like Receptor-2 Pathway in an NF-κB-Independent Manner. Int. J. Mol. Sci. 2021, 22, 8070. [Google Scholar] [CrossRef]
- Lai, J.L.; Liu, Y.H.; Liu, C.; Qi, M.P.; Liu, R.N.; Zhu, X.F.; Zhou, Q.G.; Chen, Y.Y.; Guo, A.Z.; Hu, C.M. Indirubin Inhibits LPS-Induced Inflammation via TLR4 Abrogation Mediated by the NF-kB and MAPK Signaling Pathways. Inflammation 2017, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Brenner, M.; Yang, W.L.; Wang, P. Recombinant human MFG-E8 ameliorates colon damage in DSS- and TNBS-induced colitis in mice. Lab. Investig. J. Technol. Methods Pathol. 2015, 95, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Xie, Q.; Yue, Y.; Chen, Q.; Zhao, L.; Evivie, S.E.; Li, B.; Huo, G. Gut microbiota modulation and anti-inflammatory properties of mixed lactobacilli in dextran sodium sulfate-induced colitis in mice. Food Funct. 2021, 12, 5130–5143. [Google Scholar] [CrossRef]
- Chao, L.; Zheng, P.; Xia, L.; Yong, Y.; Zhao, Z. Calycosin attenuates dextran sulfate sodium (DSS)-induced experimental colitis. Iran. J. Basic Med. Sci. 2017, 20, 1056–1062. [Google Scholar] [PubMed]
- Hartley, M.G.; Hudson, M.J.; Swarbrick, E.T.; Hill, M.J.; Grace, R.H. The rectal mucosa-associated microflora in patients with ulcerative colitis. J. Med. Microbiol. 1992, 36, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Hansson, G.C. Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 2012, 15, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.M.I. Lactic acid bacteria: Microbiological and functional aspects. Rev. Bras. Ciênc. Farm. 2006, 42, 473. [Google Scholar] [CrossRef]
- Lee, S.H. Intestinal Permeability Regulation by Tight Junction: Implication on Inflammatory Bowel Diseases. Intest. Res. 2015, 13, 11–18. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, B.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacterium pseudocatenulatum Ameliorates DSS-Induced Colitis by Maintaining Intestinal Mechanical Barrier, Blocking Proinflammatory Cytokines, Inhibiting TLR4/NF-κB Signaling, and Altering Gut Microbiota. J. Agric. Food Chem. 2021, 69, 1496–1512. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, S.; Cui, J.; Guo, T.; Zhang, J.; Li, B. Alleviative Effects of Exopolysaccharide Produced by Lactobacillus helveticus KLDS1.8701 on Dextran Sulfate Sodium-Induced Colitis in Mice. Microorganisms 2021, 9, 2086. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Ye, L.; Niu, Z.; Fang, W. Anti-inflammatory effects of Vicenin-2 on dextran sulfate sodium-induced colitis in mice. Drug Dev. Res. 2019, 80, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, E.C.; Murphy, C.; O’Neill, L.; Creagh, E.M. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 2010, 3, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Bocker, U.; Yezerskyy, O.; Feick, P.; Manigold, T.; Panja, A.; Kalina, U.; Herweck, F.; Rossol, S.; Singer, M.V. Responsiveness of intestinal epithelial cell lines to lipopolysaccharide is correlated with Toll-like receptor 4 but not Toll-like receptor 2 or CD14 expression. Int. J. Colorectal Dis. 2003, 18, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Rozková, D.; Novotná, L.; Pytlík, R.; Hochová, I.; Spísek, R. Toll-like receptors on B-CLL cells: Expression and functional consequences of their stimulation. Int. J. Cancer 2010, 126, 1132–1143. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Nighot, P.; Nighot, M.; Haque, M.; Rawat, M.; Ma, T.Y. Lactobacillus acidophilus Induces a Strain-specific and Toll-Like Receptor 2-Dependent Enhancement of Intestinal Epithelial Tight Junction Barrier and Protection against Intestinal Inflammation. Am. J. Pathol. 2021, 191, 872–884. [Google Scholar] [CrossRef]
- Gu, M.J.; Song, S.K.; Lee, I.K.; Ko, S.; Han, S.E.; Bae, S.; Ji, S.Y.; Park, B.C.; Song, K.D.; Lee, H.K.; et al. Barrier protection via Toll-like receptor 2 signaling in porcine intestinal epithelial cells damaged by deoxynivalnol. Vet. Res. 2016, 47, 25. [Google Scholar] [CrossRef]
- Rocha-Ramírez, L.M.; Pérez-Solano, R.A.; Castaón-Alonso, S.L.; Guerrero, S.S.M.; Eslava, C. Probiotic Lactobacillus Strains Stimulate the Inflammatory Response and Activate Human Macrophages. J. Immunol. Res. 2017, 2017, 4607491. [Google Scholar] [CrossRef]
- Llewellyn, A.; Foey, A. Probiotic Modulation of Innate Cell Pathogen Sensing and Signaling Events. Nutrients 2017, 9, 1156. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.; Wang, Z.; Zhao, Y.; Peng, X. gga-miR-146c Activates TLR6/MyD88/NF-κB Pathway through Targeting MMP16 to Prevent Mycoplasma Gallisepticum (HS Strain) Infection in Chickens. Cells 2019, 8, 501. [Google Scholar] [CrossRef]
- Tayyeb, J.Z.; Popeijus, H.E.; Mensink, R.P.; Konings, M.C.; Mokhtar, F.B.; Plat, J. Short-Chain Fatty Acids (Except Hexanoic Acid) Lower NF-kB Transactivation, Which Rescues Inflammation-Induced Decreased Apolipoprotein A-I Transcription in HepG2 Cells. Int. J. Mol. Sci. 2020, 21, 5088. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Lührs, H.; Gerke, T.; Müller, J.G.; Melcher, R.; Schauber, J.; Boxberger, F.; Scheppach, W.; Menzel, T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 2002, 37, 458–466. [Google Scholar] [CrossRef] [PubMed]
- He, X.X.; Li, Y.H.; Yan, P.G.; Meng, X.C.; Chen, C.Y.; Li, K.M.; Li, J.N. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative colitis. World J. Gastroenterol. 2021, 27, 4722–4737. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, M.J.; Wang, S.; Ross, R.P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Lactobacillus ruminis alleviates DSS-induced colitis by inflammatory cytokines and gut microbiota modulation. Foods 2021, 10, 1349. [Google Scholar] [CrossRef] [PubMed]
- Undi, R.B.; Sarvothaman, S.; Narasaiah, K.; Gutti, U.; Gutti, R.K. Toll-like receptor 2 signalling: Significance in megakaryocyte development through wnt signalling cross-talk and cytokine induction. Cytokine 2016, 83, 245–249. [Google Scholar] [CrossRef]
- Jin, S.; Zhao, D.; Cai, C.; Song, D.; Shen, J.; Xu, A.; Qiao, Y.; Ran, Z.; Zheng, Q. Low-dose penicillin exposure in early life decreases Th17 and the susceptibility to DSS colitis in mice through gut microbiota modification. Sci. Rep. 2017, 7, 43662. [Google Scholar] [CrossRef]
- Yeom, Y.; Kim, B.S.; Kim, S.J.; Kim, Y. Sasa quelpaertensis leaf extract regulates microbial dysbiosis by modulating the composition and diversity of the microbiota in dextran sulfate sodium-induced colitis mice. BMC Complementary Altern. Med. 2016, 16, 481. [Google Scholar] [CrossRef]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Du, P.; Xie, Q.; Wang, N.; Li, H.; Smith, E.E.; Li, C.; Liu, F.; Huo, G.; Li, B. Protective effects of tryptophan-catabolizing Lactobacillus plantarum KLDS 1.0386 against dextran sodium sulfate-induced colitis in mice. Food Funct. 2020, 11, 10736–10747. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, G.; Sun, C.; Xiong, K.; Yao, T.; Su, Y.; Fang, H. TAK-242 ameliorates DSS-induced colitis by regulating the gut microbiota and the JAK2/STAT3 signaling pathway. Microb. Cell Factories 2020, 19, 158. [Google Scholar] [CrossRef] [PubMed]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Li, Y.; Yu, R.Q.; Zhou, S.L.; Wang, X.G.; Le, G.W.; Jin, Q.; Xiao, H.; Sun, J. Composition and immuno-stimulatory properties of extracellular DNA from mouse gut flora. World J. Gastroenterol. 2017, 23, 7830–7839. [Google Scholar] [CrossRef]
- Wang, Z.; Elekwachi, C.; Jiao, J.; Wang, M.; Tang, S.; Zhou, C.; Tan, Z.; Forster, R.J. Changes in Metabolically Active Bacterial Community during Rumen Development, and Their Alteration by Rhubarb Root Powder Revealed by 16S rRNA Amplicon Sequencing. Front. Microbiol. 2017, 8, 159. [Google Scholar] [CrossRef]
- Ye, J.; Zhao, Y.; Chen, X.; Zhou, H.; Yang, Y.; Zhang, X.; Huang, Y.; Zhang, N.; Lui, E.M.K.; Xiao, M. Pu-erh tea ameliorates obesity and modulates gut microbiota in high fat diet fed mice. Food Res. Int. 2021, 144, 110360. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, K.; Qi, W.; Zhou, Y.; Hong, T.; Xiong, T.; Xie, M.; Nie, S. Exopolysaccharides from Lactobacillus plantarum NCU116 Enhances Colonic Mucosal Homeostasis by Controlling Epithelial Cell Differentiation and c-Jun/Muc2 Signaling. J. Agric. Food Chem. 2019, 67, 9831–9839. [Google Scholar] [CrossRef]
- Zeng, W.; He, D.; Xing, Y.; Liu, J.; Su, N.; Zhang, C.; Wang, Y.; Xing, X. Internal connections between dietary intake and gut microbiota homeostasis in disease progression of ulcerative colitis: A review. Food Sci. Hum. Wellness 2021, 10, 119–130. [Google Scholar] [CrossRef]



| Group | Daily Gavage Treatment (0.2 mL) | 1–7 Days | 8–14 Days |
|---|---|---|---|
| Control | Saline (0.9%) | Free drinking sterilized water | Free drinking sterilized water |
| DSS | Saline (0.9%) | Free drinking sterilized water | Free drinking 2.5% DSS water |
| Mesalazine | 75 mg/kg/day mesalazine | Free drinking sterilized water | Free drinking 2.5% DSS water |
| H4-2 | 5 × 109 cfu/mL H4-2 | Free drinking sterilized water | Free drinking 2.5% DSS water |
| H9-3 | 5 × 109 cfu/mL H9-3 | Free drinking sterilized water | Free drinking 2.5% DSS water |
| Score | Weight Loss (%) | Stool Consistency | Blood in Feces |
|---|---|---|---|
| 0 | none | normal | negative (no bleeding) |
| 1 | 1.0–5.0 | loose stools | negative |
| 2 | 5.0–10.0 | loose stools | hemoccult positive (slight) |
| 3 | 10.0–15.0 | diarrhea (slight) | hemoccult positive |
| 4 | 15.0–20.0 | diarrhea (watery diarrhea) | gross bleeding |
| Genes | Primer Sequences (5′→3′) |
|---|---|
| GAPDH | Forward:5′-TCAAGAAGGTGGTGAAGCAG-3′ Reverse: 5′-AAGGTGGAAGAGTGGGAGTTG-3′ |
| MUC1 | Forward:5′-CATTCCAGACCACAATGGCTCCTC-3′ Reverse: 5′-ATTGACTTGGCACTGAAGGCTGAG-3′ |
| MUC2 | Forward:5′-TGGTCCAGGGTTTCTTACTCC-3′ Reverse: 5′-TGATGAGGTGGCAGACAGGAGAC-3′ |
| ZO-1 | Forward: 5′-CTTCTCTTGCTGGCCCTAAAC-3′ Reverse: 5′-TGGCTTCACTTGAGGTTTCTG-3′ |
| Claudin-1 | Forward: 5′- TCTACGAGGGACTGTGGATG-3′ Reverse: 5′-TCAGATTCAGCTAGGAGTCG-3′ |
| Occludin | Forward: 5′- CACACTTGCTTGGGACAGAG-3′ Reverse: 5′-TAGCCATAGCCTCCATAGCC-3′ |
| TLR-2 | Forward: 5′-GACTCTTCACTTAAGCGAGTCT-3′ Reverse: 5′-AACCTGGCCAAGTTAGTATCTC-3′ |
| TLR-4 | Forward: 5′-GCCATCATTATGAGTGCCAATT-3′ Reverse: 5′-AGGGATAAGAACGCTGAGAATT-3′ |
| TLR6 | Forward: 5′-CAACTTAACGATAACTGAGAG-3′ Reverse: 5′-CCAGAGAGGACATATTCTTAG-3′ |
| NF-κB | Forward: 5′-GCAAGCGTATCCCAAGAAGAGGTG-3′ Reverse: 5′-GCGATGCCTGCTACCACTCATTAC-3′ |
| IκBα | Forward: 5′-TGGTGTGACTGTGGATCTCTGGAG-3′ Reverse: 5′-GGCTGGCTTCTCTGTGGTGATTC-3′ |
| Strains | Survival Rate in Simulated Gastrointestinal Tract (%) | Adhesion Rate (%) |
|---|---|---|
| H4-2 | 71.05 ± 1.04 b | 48.66 ± 0.72 b |
| H9-3 | 64.30 ± 0.96 a | 45.33 ± 0.17 a |
| BB12 | 64.55 ± 0.50 a | 44.77± 1.56 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, M.-M.; Guo, H.-X.; Cai, J.-W.; Meng, X.-C. Bifidobacterium breve Alleviates DSS-Induced Colitis in Mice by Maintaining the Mucosal and Epithelial Barriers and Modulating Gut Microbes. Nutrients 2022, 14, 3671. https://doi.org/10.3390/nu14183671
Niu M-M, Guo H-X, Cai J-W, Meng X-C. Bifidobacterium breve Alleviates DSS-Induced Colitis in Mice by Maintaining the Mucosal and Epithelial Barriers and Modulating Gut Microbes. Nutrients. 2022; 14(18):3671. https://doi.org/10.3390/nu14183671
Chicago/Turabian StyleNiu, Meng-Meng, Huan-Xin Guo, Jun-Wu Cai, and Xiang-Chen Meng. 2022. "Bifidobacterium breve Alleviates DSS-Induced Colitis in Mice by Maintaining the Mucosal and Epithelial Barriers and Modulating Gut Microbes" Nutrients 14, no. 18: 3671. https://doi.org/10.3390/nu14183671
APA StyleNiu, M.-M., Guo, H.-X., Cai, J.-W., & Meng, X.-C. (2022). Bifidobacterium breve Alleviates DSS-Induced Colitis in Mice by Maintaining the Mucosal and Epithelial Barriers and Modulating Gut Microbes. Nutrients, 14(18), 3671. https://doi.org/10.3390/nu14183671