Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. Habitual Dietary Intake Assessment
2.3. Biochemical Determinations
2.4. Stool Sampling
2.5. 16S rRNA Sequencing
2.6. Sequence Processing
2.7. Bioinformatic Analysis
2.8. Statistical Analysis
3. Results
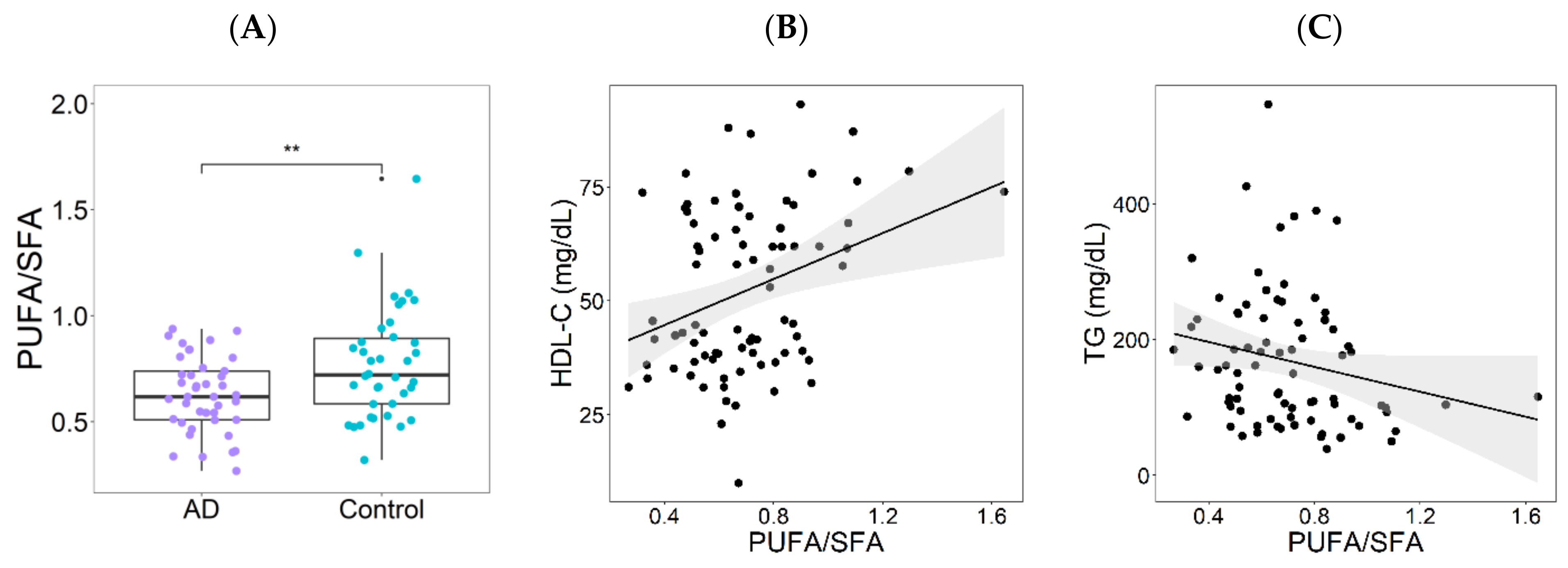
3.1. Dietary Patterns in the Atherogenic Dyslipidemia and Control Groups
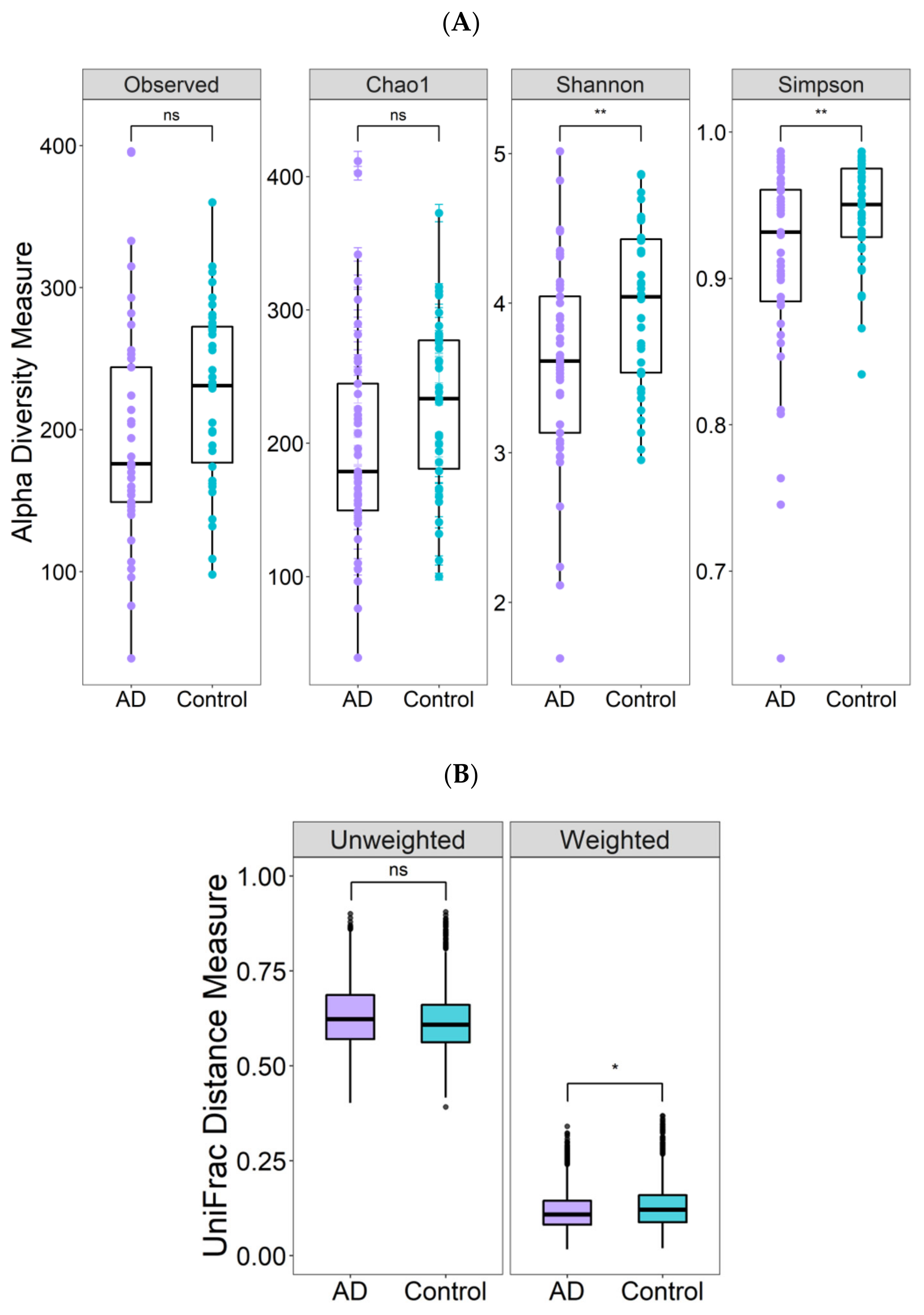
3.2. Differences in Gut Microbiota Diversity
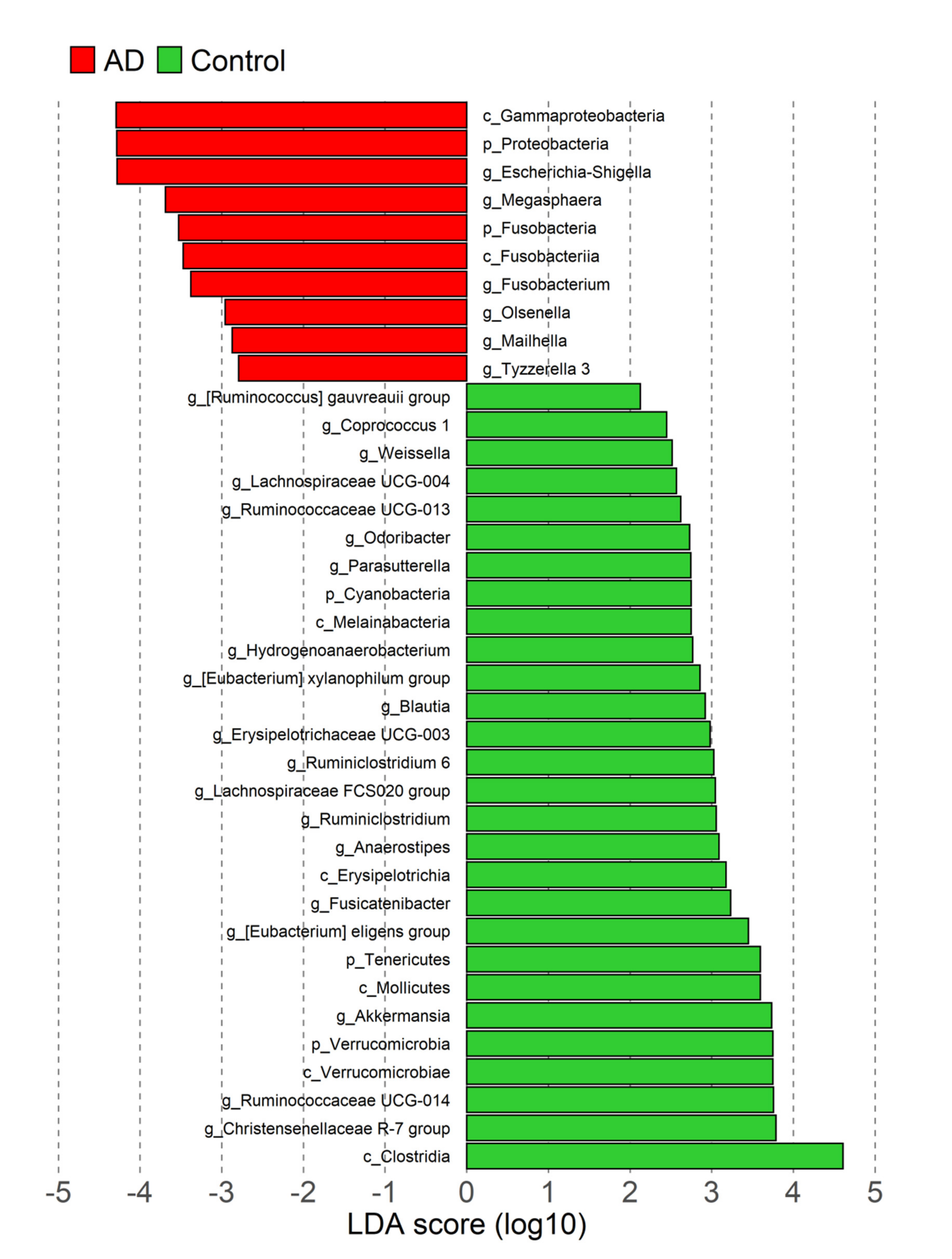
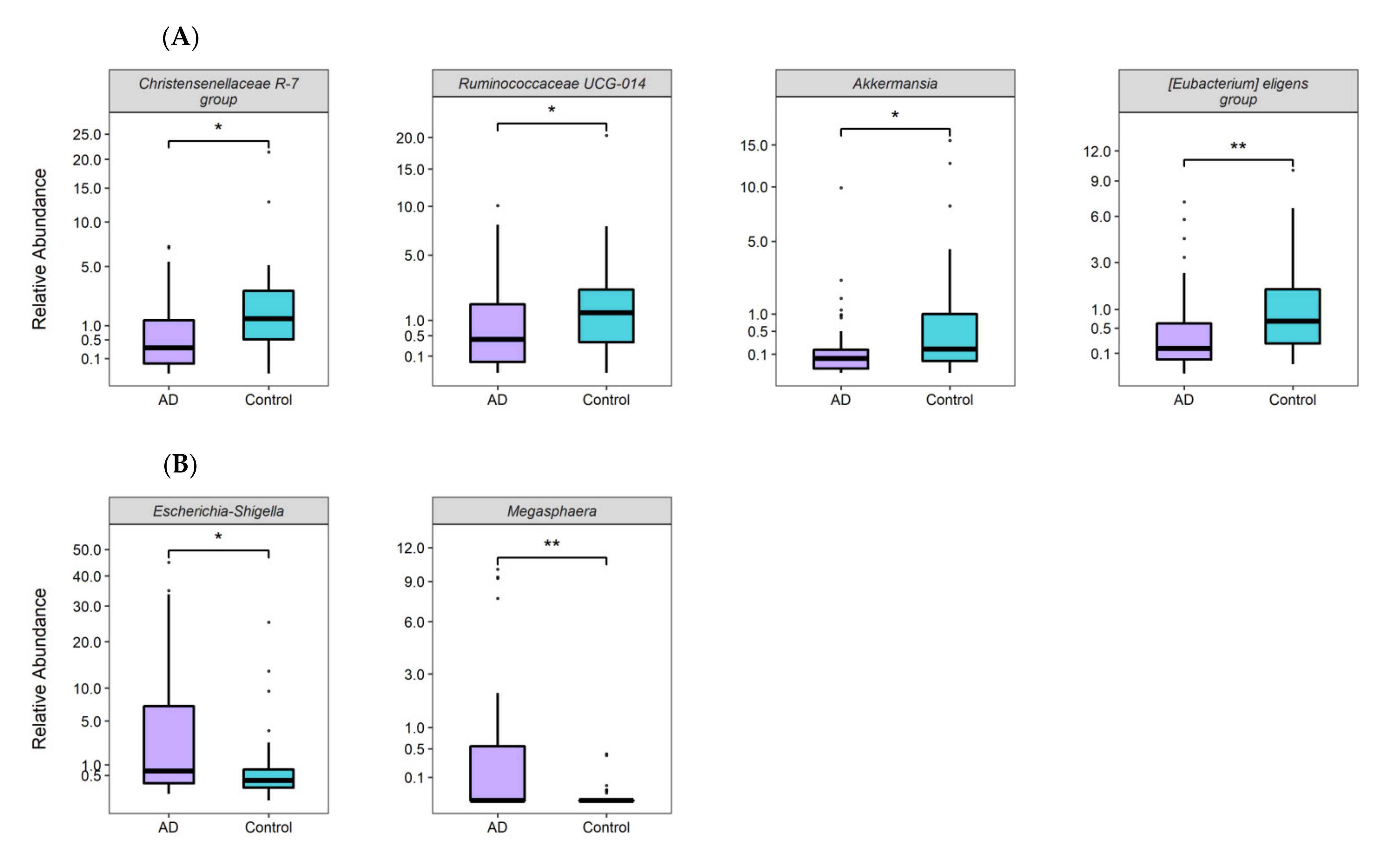
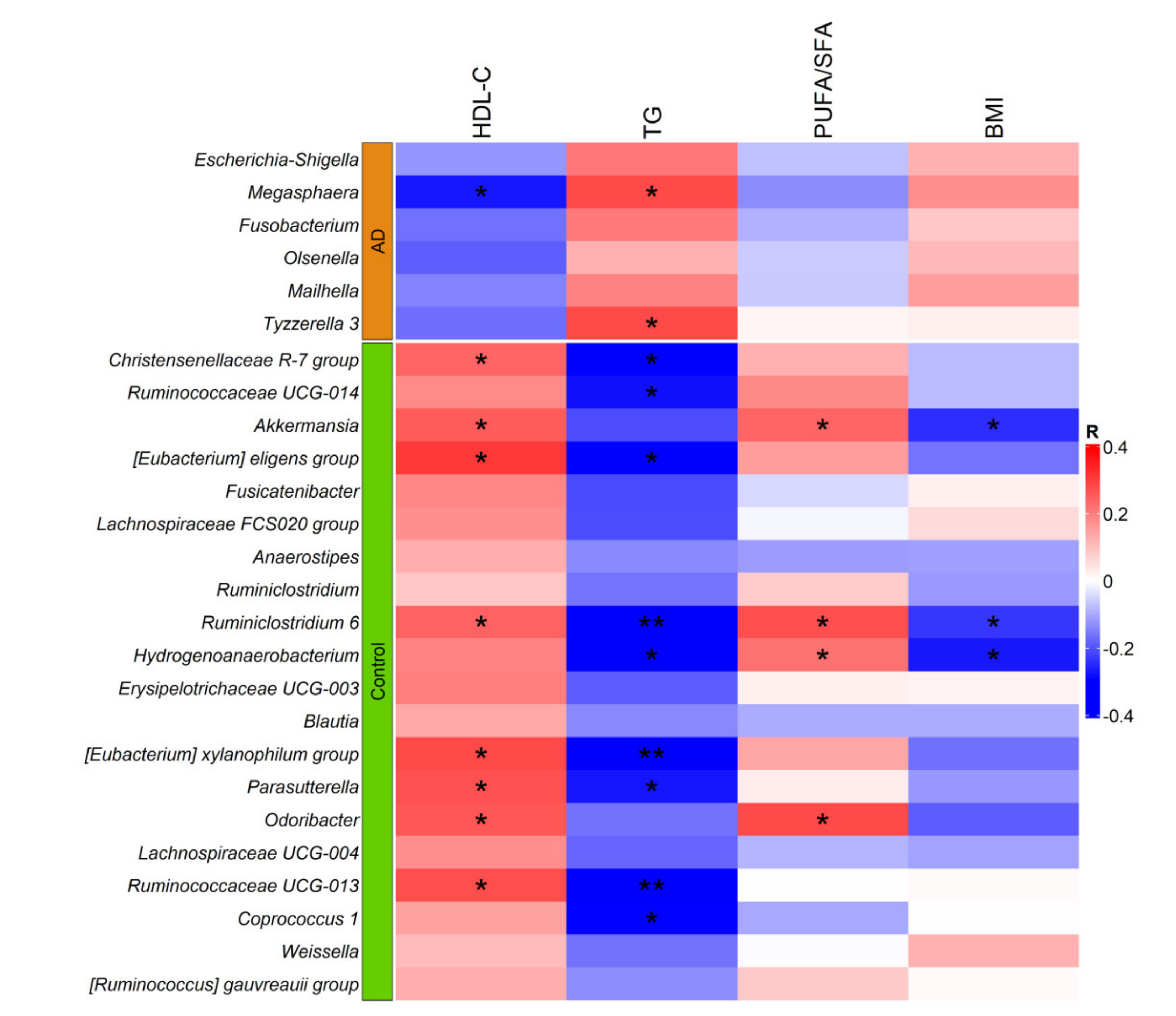
3.3. Taxonomic Gut Microbiota Differences
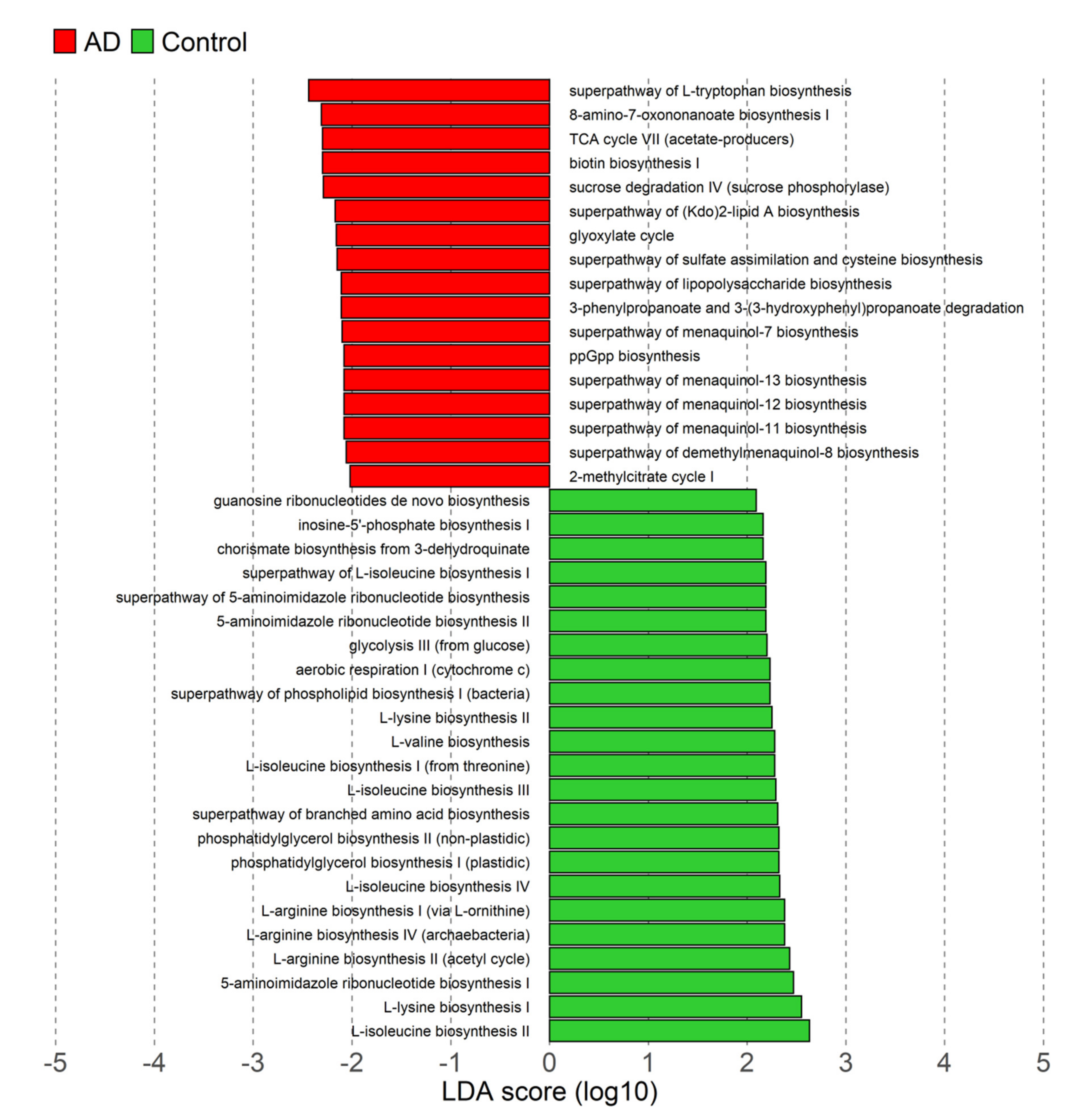
3.4. Differences in Microbiota Functional Profiles
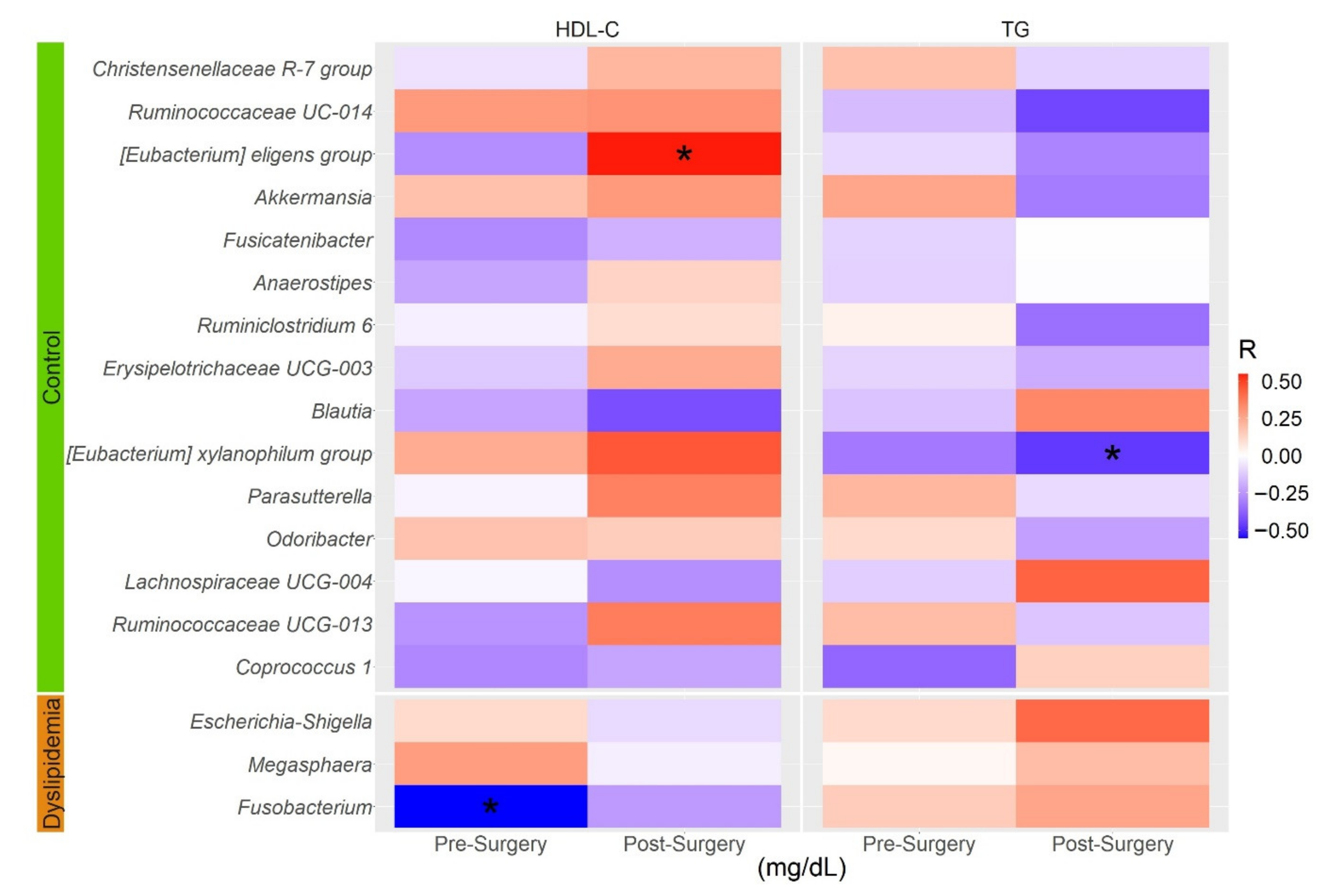
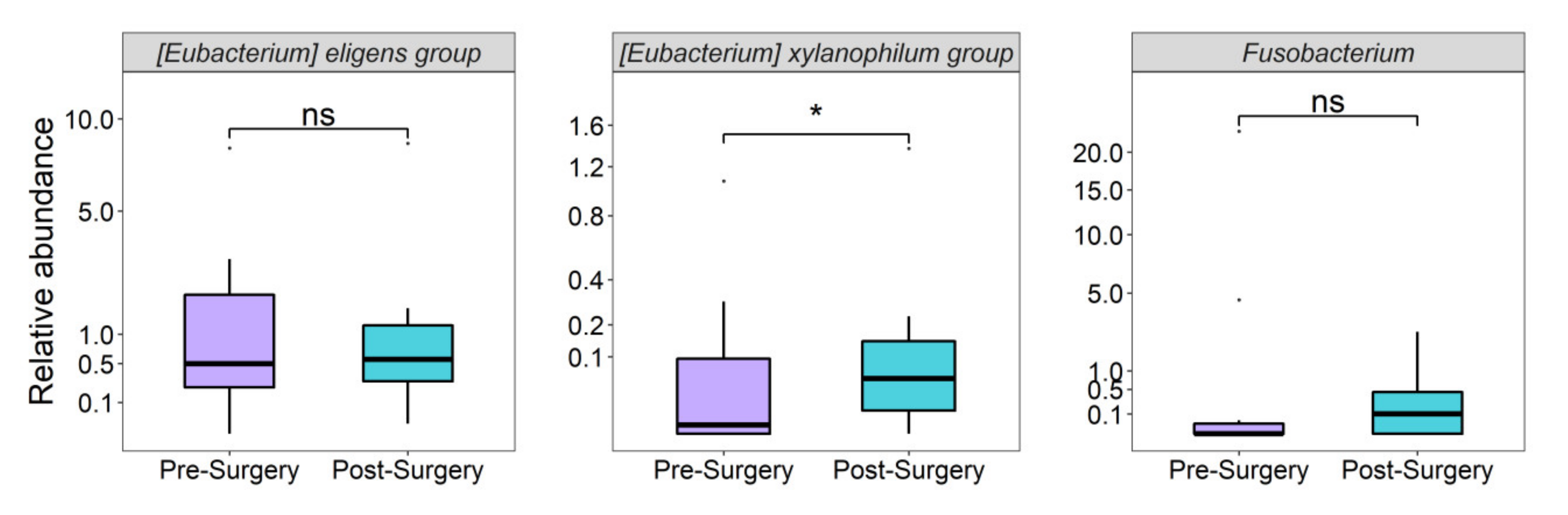
3.5. Gut Microbiota Associated with TG and HDL-C Levels before and after RYGB Surgery
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Brown, T.M.; Bittner, V. Biomarkers of atherosclerosis: Clinical applications. Curr. Cardiol. Rep. 2008, 10, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Aday, A.W.; Everett, B.M. Dyslipidemia Profiles in Patients with Peripheral Artery Disease. Curr. Cardiol. Rep. 2019, 21, 42. [Google Scholar] [CrossRef] [PubMed]
- Carroll, M.; Kit, B.; Lacher, D. Trends in elevated triglyceride in adults: United States, 2001–2012. NCHS Data Brief. 2015, 198, 198. [Google Scholar]
- Mahdy Ali, K.; Wonnerth, A.; Huber, K.; Wojta, J. Cardiovascular disease risk reduction by raising HDL cholesterol--current therapies and future opportunities. Br. J. Pharmacol. 2012, 167, 1177–1194. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Salinas, C.A.; Olaiz, G.; Valles, V.; Torres, J.M.; Gómez Pérez, F.J.; Rull, J.A.; Rojas, R.; Franco, A.; Sepulveda, J. High prevalence of low HDL cholesterol concentrations and mixed hyperlipidemia in a Mexican nationwide survey. J. Lipid Res. 2001, 42, 1298–1307. [Google Scholar] [CrossRef]
- Hernández-Alcaraz, C.; Aguilar-Salinas, C.A.; Mendoza-Herrera, K.; Pedroza-Tobías, A.; Villalpando, S.; Shamah-Levy, T.; Rivera-Dommarco, J.; Hernández-Ávila, M.; Barquera, S. Dyslipidemia prevalence, awareness, treatment and control in Mexico: Results of the Ensanut 2012. Salud. Publica Mex. 2020, 62, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, C.M.; Olsson, A.G.; Cook, T.J.; Mercuri, M.F.; Pedersen, T.R.; Kjekshus, J. Influence of low high-density lipoprotein cholesterol and elevated triglyceride on coronary heart disease events and response to simvastatin therapy in 4S. Circulation 2001, 104, 3046–3051. [Google Scholar] [CrossRef]
- Caselli, C.; De Caterina, R.; Smit, J.M.; Campolo, J.; El Mahdiui, M.; Ragusa, R.; Clemente, A.; Sampietro, T.; Clerico, A.; Liga, R.; et al. Triglycerides and low HDL cholesterol predict coronary heart disease risk in patients with stable angina. Sci. Rep. 2021, 11, 20714. [Google Scholar] [CrossRef] [PubMed]
- Suburu, J.; Gu, Z.; Chen, H.; Chen, W.; Zhang, H.; Chen, Y.Q. Fatty acid metabolism: Implications for diet, genetic variation, and disease. Food Biosci. 2013, 4, 1–12. [Google Scholar] [CrossRef]
- Wu, J.H.Y.; Micha, R.; Mozaffarian, D. Dietary fats and cardiometabolic disease: Mechanisms and effects on risk factors and outcomes. Nat. Rev. Cardiol 2019, 16, 581–601. [Google Scholar] [CrossRef] [PubMed]
- Pigsborg, K.; Gürdeniz, G.; Rangel-Huerta, O.D.; Holven, K.B.; Dragsted, L.O.; Ulven, S.M. Effects of changing from a diet with saturated fat to a diet with n-6 polyunsaturated fat on the serum metabolome in relation to cardiovascular disease risk factors. Eur. J. Nutr. 2022, 61, 2079–2089. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bonder, M.J.; Cenit, M.C.; Tigchelaar, E.F.; Maatman, A.; Dekens, J.A.; Brandsma, E.; Marczynska, J.; Imhann, F.; Weersma, R.K.; et al. The Gut Microbiome Contributes to a Substantial Proportion of the Variation in Blood Lipids. Circ. Res. 2015, 117, 817–824. [Google Scholar] [CrossRef] [PubMed]
- Rebolledo, C.; Cuevas, A.; Zambrano, T.; Acuña, J.J.; Jorquera, M.A.; Saavedra, K.; Martínez, C.; Lanas, F.; Serón, P.; Salazar, L.A.; et al. Bacterial Community Profile of the Gut Microbiota Differs between Hypercholesterolemic Subjects and Controls. Biomed. Res. Int. 2017, 2017, 8127814. [Google Scholar] [CrossRef] [PubMed]
- Vojinovic, D.; Radjabzadeh, D.; Kurilshikov, A.; Amin, N.; Wijmenga, C.; Franke, L.; Ikram, M.A.; Uitterlinden, A.G.; Zhernakova, A.; Fu, J.; et al. Relationship between gut microbiota and circulating metabolites in population-based cohorts. Nat. Commun. 2019, 10, 5813. [Google Scholar] [CrossRef]
- Le Roy, T.; Lécuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barceló, J.; Huby, T.; Guerin, M.; et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef] [PubMed]
- Yun, K.E.; Kim, J.; Kim, M.H.; Park, E.; Kim, H.L.; Chang, Y.; Ryu, S.; Kim, H.N. Major Lipids, Apolipoproteins, and Alterations of Gut Microbiota. J. Clin. Med. 2020, 9, 1589. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64. [Google Scholar] [CrossRef]
- Granado-Serrano, A.B.; Martín-Garí, M.; Sánchez, V.; Riart Solans, M.; Berdún, R.; Ludwig, I.A.; Rubió, L.; Vilaprinyó, E.; Portero-Otín, M.; Serrano, J.C.E. Faecal bacterial and short-chain fatty acids signature in hypercholesterolemia. Sci. Rep. 2019, 9, 1772. [Google Scholar] [CrossRef] [PubMed]
- Bamba, V.; Rader, D.J. Obesity and atherogenic dyslipidemia. Gastroenterology 2007, 132, 2181–2190. [Google Scholar] [CrossRef] [PubMed]
- Vekic, J.; Zeljkovic, A.; Stefanovic, A.; Jelic-Ivanovic, Z.; Spasojevic-Kalimanovska, V. Obesity and dyslipidemia. Metabolism 2019, 92, 71–81. [Google Scholar] [CrossRef]
- Chawla, S.; Tessarolo Silva, F.; Amaral Medeiros, S.; Mekary, R.A.; Radenkovic, D. The Effect of Low-Fat and Low-Carbohydrate Diets on Weight Loss and Lipid Levels: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 3774. [Google Scholar] [CrossRef] [PubMed]
- Vest, A.R.; Heneghan, H.M.; Agarwal, S.; Schauer, P.R.; Young, J.B. Bariatric surgery and cardiovascular outcomes: A systematic review. Heart 2012, 98, 1763–1777. [Google Scholar] [CrossRef] [PubMed]
- Steinert, R.E.; Rehman, A.; Souto Lima, E.J.; Agamennone, V.; Schuren, F.H.J.; Gero, D.; Schreiner, P.; Vonlanthen, R.; Ismaeil, A.; Tzafos, S.; et al. Roux-en-Y gastric bypass surgery changes fungal and bacterial microbiota in morbidly obese patients-A pilot study. PLoS ONE 2020, 15, e0236936. [Google Scholar] [CrossRef] [PubMed]
- Denova-Gutiérrez, E.; Flores, Y.N.; Gallegos-Carrillo, K.; Ramírez-Palacios, P.; Rivera-Paredez, B.; Muñoz-Aguirre, P.; Velázquez-Cruz, R.; Torres-Ibarra, L.; Meneses-León, J.; Méndez-Hernández, P.; et al. Health workers cohort study: Methods and study design. Salud. Publica Mex. 2016, 58, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, J.N.; Flores-Lopez, R.; López-Contreras, B.E.; Villamil-Ramirez, H.; Guzman-Muñoz, D.; Macias-Kauffer, L.R.; León-Mimila, P.; Granados-Portillo, O.; Del-Rio-Navarro, B.E.; Gómez-Perez, F.J.; et al. Effect of Gut Microbial Enterotypes on the Association between Habitual Dietary Fiber Intake and Insulin Resistance Markers in Mexican Children and Adults. Nutrients 2021, 13, 3892. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Avila, M.; Romieu, I.; Parra, S.; Hernández-Avila, J.; Madrigal, H.; Willett, W. Validity and reproducibility of a food frequency questionnaire to assess dietary intake of women living in Mexico City. Salud. Publica Mex. 1998, 40, 133–140. [Google Scholar] [CrossRef]
- Hernández-Ávila, J.E.; González-Avilés, L.; Rosales-Mendoza, E. Manual de usuario. In SNUT Sistema de Evaluación de Hábitos Nutricionales y Consumo de Nutrimentos; Instituto Nacional de Salud Pública: Cuernavaca, México, 2003. [Google Scholar]
- Capuano, E.; Oliviero, T.; Fogliano, V.; Pellegrini, N. Role of the food matrix and digestion on calculation of the actual energy content of food. Nutr. Rev. 2018, 76, 274–289. [Google Scholar] [CrossRef]
- National Cholesterol Education Program (NCEP). Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation 2002, 106, 3143–3421. [Google Scholar]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2010, 33 (Suppl. S1), S62–S69. [Google Scholar] [CrossRef]
- Moran-Ramos, S.; Lopez-Contreras, B.E.; Villarruel-Vazquez, R.; Ocampo-Medina, E.; Macias-Kauffer, L.; Martinez-Medina, J.N.; Villamil-Ramirez, H.; León-Mimila, P.; Del Rio-Navarro, B.E.; Ibarra-Gonzalez, I.; et al. Environmental and intrinsic factors shaping gut microbiota composition and diversity and its relation to metabolic health in children and early adolescents: A population-based study. Gut. Microbes 2020, 11, 900–917. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. VVegan: Community Ecology Package. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 25 July 2022).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome. Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J. MaAsLin 2: Multivariable Association in Population-Scale Meta-Omics Studies. R/Bioconductor Package. 2020. Available online: http://huttenhower.sph.harvard.edu/maaslin2 (accessed on 25 July 2022).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Midford, P.E.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Trumbo, P.; Schlicker, S.; Yates, A.A.; Poos, M.; Food and Nutrition Board of the Institute of Medicine, T.e.N.A. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein and amino acids. J. Am. Diet Assoc. 2002, 102, 1621–1630. [Google Scholar] [CrossRef]
- Kim, M.H.; Yun, K.E.; Kim, J.; Park, E.; Chang, Y.; Ryu, S.; Kim, H.L.; Kim, H.N. Gut microbiota and metabolic health among overweight and obese individuals. Sci. Rep. 2020, 10, 19417. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Astbury, S.; Le Roy, C.; Spector, T.D.; Valdes, A.M. The prebiotic effects of omega-3 fatty acid supplementation: A six-week randomised intervent.tion trial. Gut Microbes 2021, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [PubMed]
- López-Contreras, B.E.; Morán-Ramos, S.; Villarruel-Vázquez, R.; Macías-Kauffer, L.; Villamil-Ramírez, H.; León-Mimila, P.; Vega-Badillo, J.; Sánchez-Muñoz, F.; Llanos-Moreno, L.E.; Canizalez-Román, A.; et al. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr. Obes. 2018, 13, 381–388. [Google Scholar] [CrossRef]
- Waters, J.L.; Ley, R.E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 2019, 17, 83. [Google Scholar] [CrossRef]
- Brahe, L.K.; Le Chatelier, E.; Prifti, E.; Pons, N.; Kennedy, S.; Hansen, T.; Pedersen, O.; Astrup, A.; Ehrlich, S.D.; Larsen, L.H. Specific gut microbiota features and metabolic markers in postmenopausal women with obesity. Nutr. Diabetes 2015, 5, e159. [Google Scholar] [CrossRef]
- Martin-Gallausiaux, C.; Marinelli, L.; Blottière, H.M.; Larraufie, P.; Lapaque, N. SCFA: Mechanisms and functional importance in the gut. Proc. Nutr. Soc. 2021, 80, 37–49. [Google Scholar] [CrossRef]
- Moreno-Indias, I.; Tinahones, F.J. Impaired adipose tissue expandability and lipogenic capacities as ones of the main causes of metabolic disorders. J. Diabetes Res. 2015, 2015, 970375. [Google Scholar] [CrossRef]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.; Ijaz, U.Z.; Gallagher, E.; Horton, F.; Ellis, R.J.; Jaiyeola, E.; Duparc, T.; Russell-Jones, D.; Hinton, P.; Cani, P.D.; et al. Fecal Enterobacteriales enrichment is associated with increased in vivo intestinal permeability in humans. Physiol. Rep. 2018, 6, e13649. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Feingold, K.R.; Hardardottir, I.; Memon, R.; Krul, E.J.; Moser, A.H.; Taylor, J.M.; Grunfeld, C. Effect of endotoxin on cholesterol biosynthesis and distribution in serum lipoproteins in Syrian hamsters. J. Lipid Res. 1993, 34, 2147–2158. [Google Scholar] [CrossRef]
- Graessler, J.; Qin, Y.; Zhong, H.; Zhang, J.; Licinio, J.; Wong, M.L.; Xu, A.; Chavakis, T.; Bornstein, A.B.; Ehrhart-Bornstein, M.; et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: Correlation with inflammatory and metabolic parameters. Pharm. J. 2013, 13, 514–522. [Google Scholar] [CrossRef]
- Sedighi, M.; Razavi, S.; Navab-Moghadam, F.; Khamseh, M.E.; Alaei-Shahmiri, F.; Mehrtash, A.; Amirmozafari, N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017, 111, 362–369. [Google Scholar] [CrossRef]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef]
- Buckley, J.D.; Howe, P.R. Long-chain omega-3 polyunsaturated fatty acids may be beneficial for reducing obesity-a review. Nutrients 2010, 2, 1212–1230. [Google Scholar] [CrossRef]
- Lim, M.Y.; You, H.J.; Yoon, H.S.; Kwon, B.; Lee, J.Y.; Lee, S.; Song, Y.M.; Lee, K.; Sung, J.; Ko, G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 2017, 66, 1031–1038. [Google Scholar] [CrossRef]
- Furet, J.P.; Kong, L.C.; Tap, J.; Poitou, C.; Basdevant, A.; Bouillot, J.L.; Mariat, D.; Corthier, G.; Doré, J.; Henegar, C.; et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: Links with metabolic and low-grade inflammation markers. Diabetes 2010, 59, 3049–3057. [Google Scholar] [CrossRef]
- Nie, X.; Chen, J.; Ma, X.; Ni, Y.; Shen, Y.; Yu, H.; Panagiotou, G.; Bao, Y. A metagenome-wide association study of gut microbiome and visceral fat accumulation. Comput. Struct. Biotechnol. J. 2020, 18, 2596–2609. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhao, Y.; Zhou, C.; Zhao, Q.; Zhong, H.; Zhu, X.; Fu, T.; Pan, L.; Shang, Q.; Yu, G. Dietary Polysaccharide from Enteromorpha clathrata Attenuates Obesity and Increases the Intestinal Abundance of Butyrate-Producing Bacterium, Eubacterium xylanophilum, in Mice Fed a High-Fat Diet. Polymers 2021, 13, 3286. [Google Scholar] [CrossRef] [PubMed]
- Liou, A.P.; Paziuk, M.; Luevano, J.M.; Machineni, S.; Turnbaugh, P.J.; Kaplan, L.M. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 2013, 5, 178ra141. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef]
- Amiri, P.; Hosseini, S.A.; Ghaffari, S.; Tutunchi, H.; Mosharkesh, E.; Asghari, S.; Roshanravan, N. Role of Butyrate, a Gut Microbiota Derived Metabolite, in Cardiovascular Diseases: A comprehensive narrative review. Front. Pharmacol. 2021, 12, 837509. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Belda, E.; Nielsen, T.; Aron-Wisnewsky, J.; Chakaroun, R.; Forslund, S.K.; Assmann, K.; Valles-Colomer, M.; Nguyen, T.T.D.; et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 2020, 581, 310–315. [Google Scholar] [CrossRef]
| Trait | Atherogenic Dyslipidemia | Control | p |
|---|---|---|---|
| (n = 41) | (n = 38) | ||
| Female, n (%) | 31 (75.6) | 32 (84.2) | 0.342 |
| Age, years | 59.0 (48.0–69.5) | 55.0 (35.0–64.0) | 0.160 |
| BMI, kg/m2 | 27.9 (26.3–30.7) | 24.1 (21.9–26.9) | 1.0 × 10−5 |
| HDL-C, mg/dL | 38.0 (33.0–41.8) | 67.9 (61.9–73.9) | 2.1 × 10−14 |
| Triglycerides, mg/dL | 229.0 (183.5–267.5) | 90.0 (71.3–108.0) | 2.1 × 10−14 |
| Non HDL-C, mg/dL | 139.0 (122.6–163.5) | 137.7 (115.9–160.9) | 0.312 |
| Total cholesterol, mg/dL | 179.0 (161.0–197.5) | 208.0 (180.0–229.3) | 0.004 |
| Fasting glucose, mg/dL | 99.0 (92.5–109.5) | 92.5 (85.8–97.3) | 0.002 |
| Diabetes, n (%) | 6 (14.6) | 3 (7.9) | 0.207 |
| Hypolipidemic treatment, n (%) | 9 (22.0) | 0 (0) | 0.002 |
| Trait | Pre-Surgery | Post-Surgery | p |
|---|---|---|---|
| (n = 20) | (n = 20) | ||
| Female, n (%) | 13 (65.0) | - | - |
| Age, years | 40.0 (31.3–44.8) | - | - |
| BMI, kg/m2 | 45.7 (42.3–51.9) | 32.9 (28.7–36.2) | 5.0 × 10−6 |
| HDL-C, mg/dL | 35.0 (31.3–41.8) | 45.0 (37.0–49.0) | 0.001 |
| Triglycerides, mg/dL | 159.5 (104.3–180.8) | 100.0 (67.0–144.0) | 0.011 |
| Total cholesterol, mg/dL | 154.5 (136.3–186.5) | 86.0 (78.0–96.0) | 0.097 |
| Hypolipidemic treatment, n (%) | 4 (20.0) | 1 (5.0) | 0.151 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Montoya, P.; Cerqueda-García, D.; Rodríguez-Flores, M.; López-Contreras, B.; Villamil-Ramírez, H.; Morán-Ramos, S.; Molina-Cruz, S.; Rivera-Paredez, B.; Antuna-Puente, B.; Velázquez-Cruz, R.; et al. Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery. Nutrients 2022, 14, 3545. https://doi.org/10.3390/nu14173545
López-Montoya P, Cerqueda-García D, Rodríguez-Flores M, López-Contreras B, Villamil-Ramírez H, Morán-Ramos S, Molina-Cruz S, Rivera-Paredez B, Antuna-Puente B, Velázquez-Cruz R, et al. Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery. Nutrients. 2022; 14(17):3545. https://doi.org/10.3390/nu14173545
Chicago/Turabian StyleLópez-Montoya, Priscilla, Daniel Cerqueda-García, Marcela Rodríguez-Flores, Blanca López-Contreras, Hugo Villamil-Ramírez, Sofía Morán-Ramos, Selene Molina-Cruz, Berenice Rivera-Paredez, Bárbara Antuna-Puente, Rafael Velázquez-Cruz, and et al. 2022. "Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery" Nutrients 14, no. 17: 3545. https://doi.org/10.3390/nu14173545
APA StyleLópez-Montoya, P., Cerqueda-García, D., Rodríguez-Flores, M., López-Contreras, B., Villamil-Ramírez, H., Morán-Ramos, S., Molina-Cruz, S., Rivera-Paredez, B., Antuna-Puente, B., Velázquez-Cruz, R., Villarreal-Molina, T., & Canizales-Quinteros, S. (2022). Association of Gut Microbiota with Atherogenic Dyslipidemia, and Its Impact on Serum Lipid Levels after Bariatric Surgery. Nutrients, 14(17), 3545. https://doi.org/10.3390/nu14173545





