Reliable Quantification of Ultratrace Selenium in Food, Beverages, and Water Samples by Cloud Point Extraction and Spectrometric Analysis
Abstract
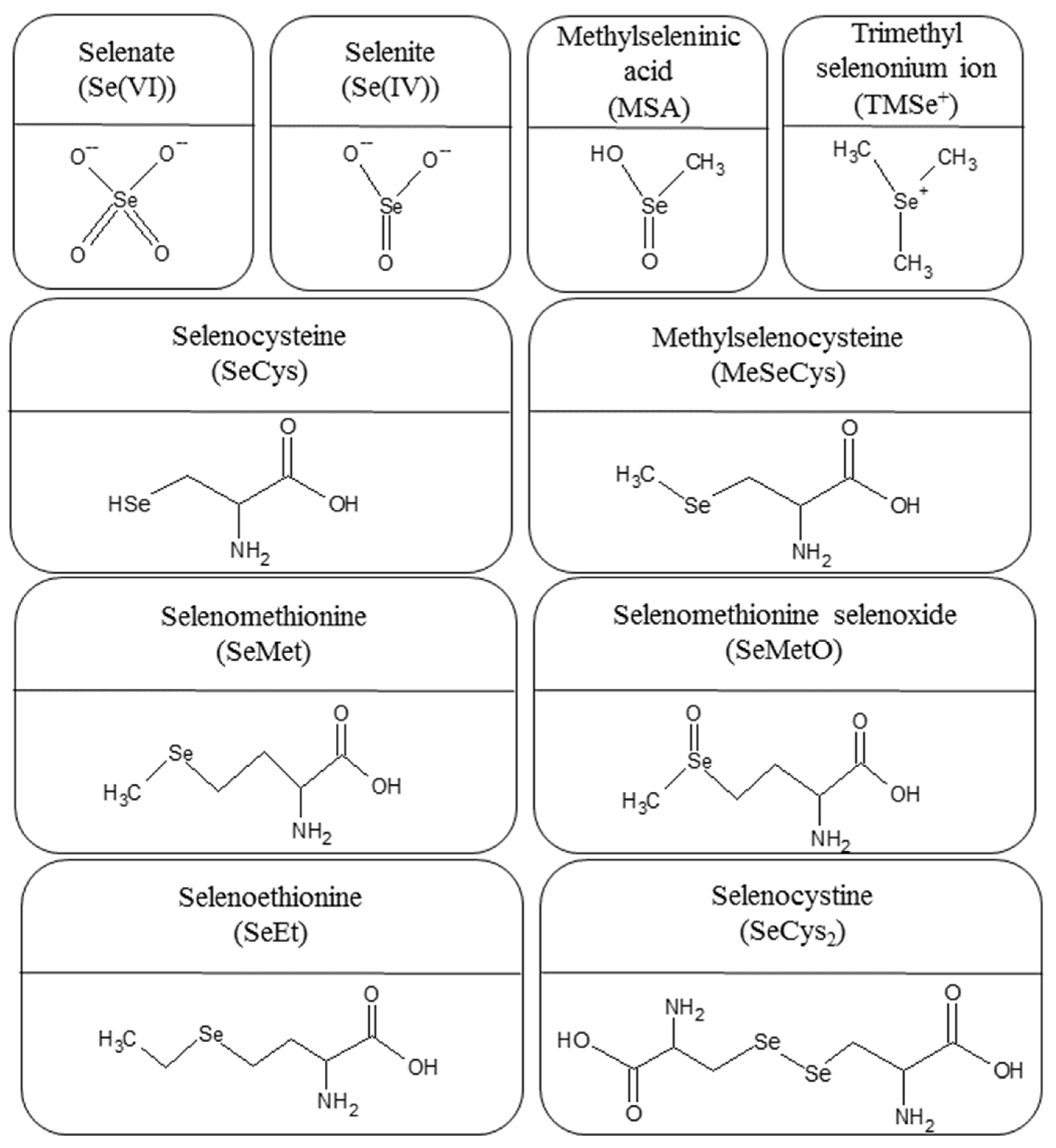
:1. Introduction
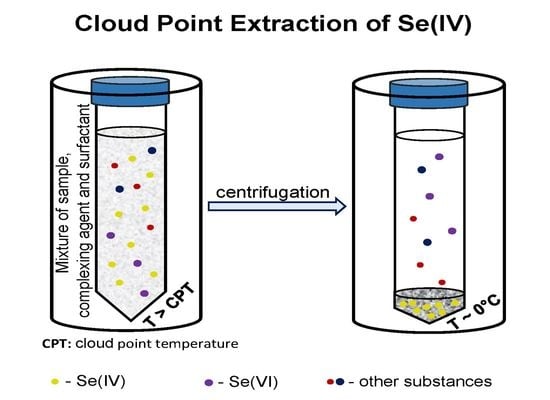
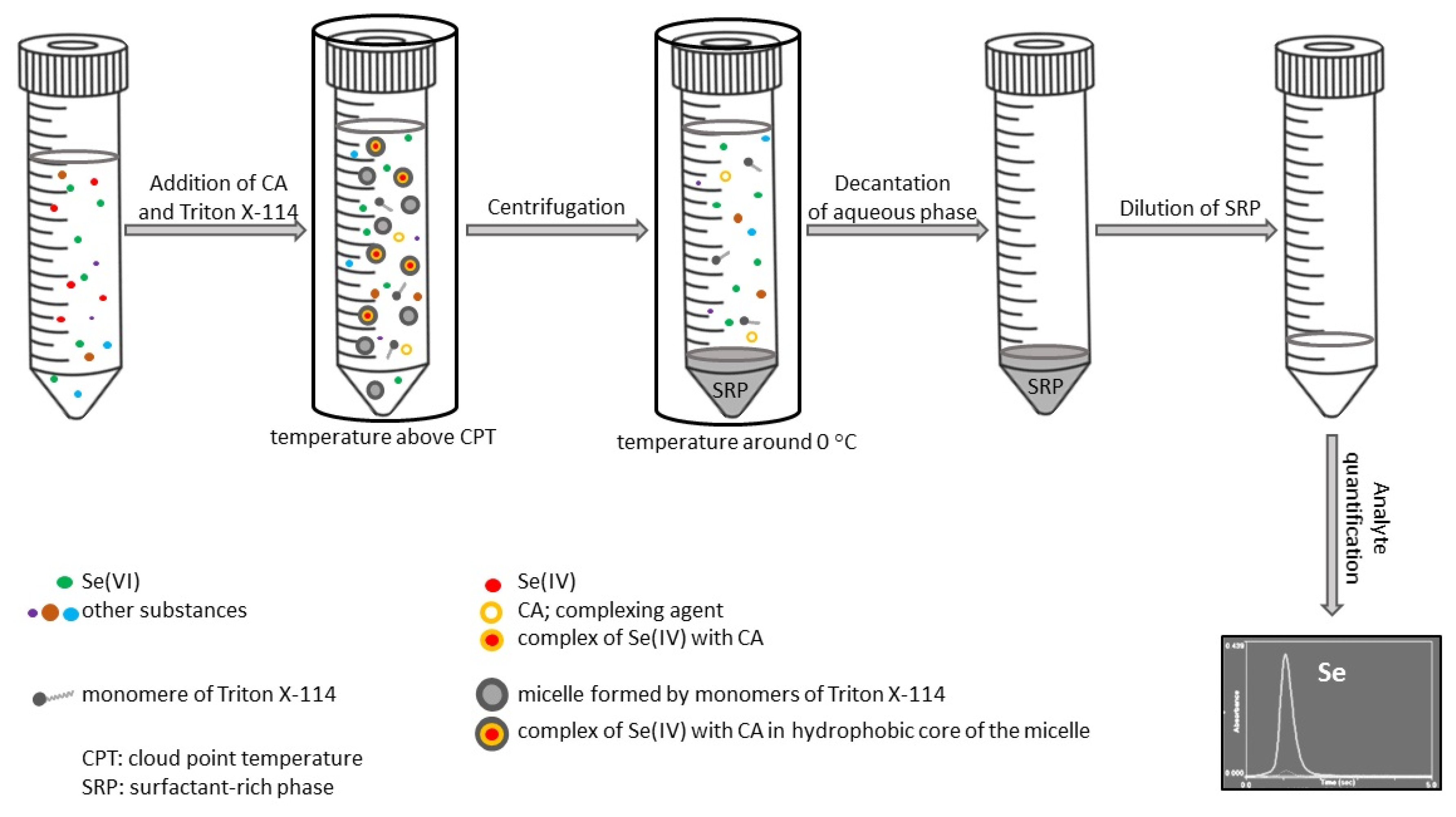
2. Cloud Point Extraction of Inorganic Ions
3. Different Approaches to Sample Preparation, Methodology, and Data Interpretation
| Sample | Extraction | Detection System | Extraction Agent | EF | LOD (µg/L) | RSD (%) | Reference |
|---|---|---|---|---|---|---|---|
| Water, beverage, food | UA-IL-DLLME | ETAAS | [C6mim][Tf2N] | 150 | 0.012 | 4.2 | [71] |
| Water, beverage, food | MEA-IL-DLLME | ETAAS | [C6mim][PF6] | na | 0.021 | 2.9 | [72] |
| Water, garlic extract | on-line IL-DLLME | ETAAS | CYPHOS® IL 101 | 20 | 0.015 | 5.1 | [73] |
| Water | DLLME | ETAAS | CCl4 | 70 | 0.050 | 4.5 | [74] |
| Water, food | SUPRAS-ME | ETAAS | 1-octanol | na | 0.10 | 4.3 | [75] |
| Food | VA-IL-LLME | ETAAS | C42H87O2P | 100 | 0.005 | 4.9 | [70] |
| Edible oil | IL-DLLME | ETAAS | [C12mim][Tf2N] | 140 | 0.03 * | 5.1 | [76] |
| Tea leaves, tea infusion | SFODME | ETV-ICP-MS | 1-undecanol | 500 | 0.00019 | 4.7 | [67] |
| Water | DLLME | ETV-ICP-MS | CHCl3 | 64.8 | 0.047 | 7.2 | [77] |
| Water | DLLME-SH-DES | UV-Vis | DES | 315 | 0.76 | 5.5 | [68] |
| Water, rice | IL-CI-AME | UV-Vis | [C4mim][PF6] | 25 | 1.5 | 1.2 | [69] |
| Water, beverage, food | VA-IL-DLLME | HG-AAS | [C8mim][Tf2N] | 120 | 0.0015 | 2.7 | [33] |
4. Summary
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rotruck, J.T.; Pope, A.L.; Ganther, H.E.; Swanson, A.B.; Hafeman, D.G.; Hoekstra, W.G. Selenium-biochemical role as a component of glutathione peroxidase. Science 1973, 179, 588–590. [Google Scholar] [CrossRef] [PubMed]
- Ali, W.; Benedetti, R.; Handzlik, J.; Zwergel, C.; Battistelli, C. The innovative potential of selenium-containing agents for fighting cancer and viral infections. Drug Discov. Today 2021, 26, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Matulová, M.; Bujdoš, M.; Miglierini, M.B.; Mitróova, Z.; Kubovčíková, M.; Urík, M. The effects of selenate on goethite synthesis and selenate sorption kinetics onto a goethite surface—A three-step process with an unexpected desorption phase. Chem. Geol. 2020, 556, 119852. [Google Scholar] [CrossRef]
- Farkas, B.; Vojtková, H.; Bujdoš, M.; Kolenčík, M.; Šebesta, M.; Matulová, M.; Duborská, E.; Danko, M.; Kim, H.; Kučová, K.; et al. Fungal mobilization of selenium in the presence of hausmannite and ferric oxyhydroxides. J. Fungi 2021, 7, 810. [Google Scholar] [CrossRef] [PubMed]
- Pyrzynska, K.; Sentkowska, A. Selenium in plant foods: Speciation analysis, bioavailability, and factors affecting composition. Crit. Rev. Food Sci. Nutr. 2021, 61, 1340–1352. [Google Scholar] [CrossRef] [PubMed]
- Dudová, J.; Bujdoš, M. Study of selenium sorption on iron oxide hydroxides. Chem. Listy 2015, 109, 770–774. [Google Scholar]
- WHO. Trace Elements in Human Nutrition and Health; WHO: Geneva, Switzerland, 1996. [Google Scholar]
- Kurt, C.; Demirbas, A.; Nawaz, M.A.; Chung, G.; Baloch, F.S.; Altunay, N. Determination of Se content of 78 sesame accessions with different geographical origin. J. Food Compos. Anal. 2020, 94, 103621. [Google Scholar] [CrossRef]
- Blazina, T.; Sun, Y.B.; Voegelin, A.; Lenz, M.; Berg, M.; Winkel, L.H.E. Terrestrial selenium distribution in China is potentially linked to monsoonal climate. Nat. Commun. 2014, 5, 5717. [Google Scholar] [CrossRef]
- Ferro, C.; Florindo, H.F.; Santos, H.A. Selenium nanoparticles for biomedical applications: From development and characterization to therapeutics. Adv. Healthc. Mater. 2021, 10, 2100598. [Google Scholar] [CrossRef]
- Jablonska, E.; Li, Q.; Reszka, E.; Wieczorek, E.; Tarhonska, K.; Wang, T. Therapeutic potential of selenium and selenium compounds in cervical cancer. Cancer Control 2021, 28, 1808. [Google Scholar] [CrossRef]
- Chen, N.; Yao, P.; Zhang, W.; Zhang, Y.T.; Xin, N.C.; Wei, H.D.; Zhang, T.H.; Zhao, C.H. Selenium nanoparticles: Enhanced nutrition and beyond. Crit. Rev. Food Sci. Nutr. 2022, 2022, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kieliszek, M.; Bano, I. Selenium as an important factor in various disease states—A review. Excli J. 2022, 21, 948–966. [Google Scholar] [CrossRef]
- Rataan, A.O.; Geary, S.M.; Zakharia, Y.; Rustum, Y.M.; Salem, A.K. Potential role of selenium in the treatment of cancer and viral infections. Int. J. Mol. Sci. 2022, 23, 2215. [Google Scholar] [CrossRef] [PubMed]
- Bañuelos, G.S.; Freeman, J.L.; Arroyo, I.S. Selenium content and speciation differences in selenium enriched soups made from selenium biofortified plants. J. Food Compos. Anal. 2022, 105, 104255. [Google Scholar] [CrossRef]
- Matulová, M.; Urík, M.; Bujdoš, M.; Duborská, E.; Cesnek, M.; Miglierini, M.B. Selenite sorption onto goethite: Isotherm and ion-competitive studies, and effect of pH on sorption kinetics. Chem. Pap. 2019, 73, 2975–2985. [Google Scholar] [CrossRef]
- Matulová, M.; Bujdoš, M.; Miglierini, M.B.; Cesnek, M.; Duborská, E.; Mosnáčková, K.; Vojtková, H.; Kmječ, T.; Dekan, J.; Matúš, P.; et al. The effect of high selenite and selenate concentrations on ferric oxyhydroxides transformation under alkaline conditions. Int. J. Mol. Sci. 2021, 22, 9955. [Google Scholar] [CrossRef] [PubMed]
- Haug, A.; Graham, R.D.; Christophersen, O.A.; Lyons, G.H. How to use the world’s scarce selenium resources efficiently to increase the selenium concentration in food. Microb. Ecol. Health Dis. 2007, 19, 209–228. [Google Scholar] [CrossRef]
- Tan, L.C.; Nancharaiah, Y.V.; van Hullebusch, E.D.; Lens, P.N.L. Selenium: Environmental significance, pollution, and biological treatment technologies. Biotechnol. Adv. 2016, 34, 886–907. [Google Scholar] [CrossRef]
- White, P.J. Selenium metabolism in plants. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2333–2342. [Google Scholar] [CrossRef]
- Ferreira, S.L.C.; Cerda, V.; Portugal, L.A.; Goncalves, L.B.; Neto, J.H.S.; Pereira, J.B.; Palacio, E. State of the art of the methods proposed for selenium speciation analysis by CVG-AFS. Trac-Trends Anal. Chem. 2022, 152, 116617. [Google Scholar] [CrossRef]
- Hagarová, I.; Žemberyová, M.; Bajčan, D. Sequential and single step extraction procedures used for fractionation of selenium in soil samples. Chem. Pap. 2005, 59, 93–98. [Google Scholar]
- Sobolev, O.I.; Gutyj, B.V.; Sobolieva, S.V.; Borshch, O.O.; Nedashkivsky, V.M.; Kachan, L.M.; Karkach, P.M.; Nedashkivska, N.V.; Poroshinska, O.A.; Stovbetska, L.S.; et al. Selenium in natural environment and food chains. A review. Ukr. J. Ecol. 2020, 10, 148–158. [Google Scholar] [CrossRef]
- Hagarová, I.; Nemček, L. Selenium in blood serum of healthy European population. Chem. Listy 2020, 114, 329–335. [Google Scholar]
- Žemberyová, M.; Hagarová, I. Determination of selenium in blood serum of children by electrothermal atomic absorption spectrometry. Chem. Listy 2005, 99, 34–39. [Google Scholar]
- Robberecht, H.; Van Cauwenbergh, R.; Hermans, N. Blood selenium levels and factors influencing concentration values. Trace Elem. Electrolytes 2012, 29, 172–188. [Google Scholar] [CrossRef]
- He, M.J.; Zhang, S.Q.; Mu, W.P.; Huang, Z.W. Selenium in infant formula milk. Asia Pac. J. Clin. Nutr. 2018, 27, 284–292. [Google Scholar] [CrossRef]
- Hogan, C.; Perkins, A.V. Selenoproteins in the human placenta: How essential is selenium to a healthy start to life? Nutrients 2022, 14, 628. [Google Scholar] [CrossRef]
- Fraczek, A.; Pasternak, K. Selenium in medicine and treatment. J. Elem. 2013, 18, 145–163. [Google Scholar] [CrossRef]
- Tobe, R.; Mihara, H. Delivery of selenium to selenophosphate synthetase for selenoprotein biosynthesis. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 2433–2440. [Google Scholar] [CrossRef]
- Schrauzer, G.N. Selenium and selenium-antagonistic elements in nutritional cancer prevention. Crit. Rev. Biotechnol. 2009, 29, 10–17. [Google Scholar] [CrossRef]
- Constantinescu-Aruxandei, D.; Frincu, R.M.; Capra, L.; Oancea, F. Selenium analysis and speciation in dietary supplements based on next-generation selenium ingredients. Nutrients 2018, 10, 1466. [Google Scholar] [CrossRef] [PubMed]
- Altunay, N.; Elik, A.; Katin, K. Optimization of vortex-assisted ionic liquid dispersive liquid-liquid microextraction by experimental design prior to hydride generation atomic absorption spectrometry for determination of selenium species in food, beverage and water samples. J. Food Compos. Anal. 2021, 99, 103871. [Google Scholar] [CrossRef]
- Beauchemin, D. Environmental analysis by inductively coupled plasma mass spectrometry. Mass Spectrom. Rev. 2010, 29, 560–592. [Google Scholar] [CrossRef] [PubMed]
- Balaram, V. Strategies to overcome interferences in elemental and isotopic geochemical analysis by quadrupole inductively coupled plasma mass spectrometry: A critical evaluation of the recent developments. Rapid Commun. Mass Spectrom. 2021, 35, 9065. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.; Karmakar, D.; Begum, S.N.; Ali, S.Y.; Patra, P.K. Recent trends in the analysis of trace elements in the field of environmental research: A review. Microchem. J. 2021, 165, 106086. [Google Scholar] [CrossRef]
- Wysocka, I. Determination of rare earth elements concentrations in natural waters—A review of ICP-MS measurement approaches. Talanta 2021, 221, 121636. [Google Scholar] [CrossRef]
- Li, D.Y.; Wang, X.N.; Huang, K.; Wang, Z.H. Multielemental determination of rare earth elements in seawater by inductively coupled plasma mass spectrometry (ICP-MS) after matrix separation and pre-concentration with crab shell particles. Front. Environ. Sci. 2021, 9, 781996. [Google Scholar] [CrossRef]
- Viana, J.L.M.; Menegario, A.A.; Fostier, A.H. Preparation of environmental samples for chemical speciation of metal/metalloids: A review of extraction techniques. Talanta 2021, 226, 122119. [Google Scholar] [CrossRef]
- Campillo, N.; Gavazov, K.; Viñas, P.; Hagarová, I.; Andruch, V. Liquid-phase microextraction: Update May 2016 to December 2018. Appl. Spectrosc. Rev. 2020, 55, 307–326. [Google Scholar] [CrossRef]
- Ibrahim, A.S.A.; Al-Farawati, R.; Hawas, U.; Shaban, Y. Recent microextraction techniques for determination and chemical speciation of selenium. Open Chem. 2017, 15, 103–122. [Google Scholar] [CrossRef]
- Nemček, L.; Hagarová, I. The recent strategies employed in chemical analysis of contaminated waters, sediments and soils as a part of the remediation process: Extraction. In Environmental Pollution and Remediation. Environmental and Microbial Biotechnology; Prasad, R., Ed.; Springer Nature: Singapore, 2021; pp. 131–173. [Google Scholar]
- Werner, J.; Grzeskowiak, T.; Zgola-Grzeskowiak, A.; Stanisz, E. Recent trends in microextraction techniques used in determination of arsenic species. Trac-Trends Anal. Chem. 2018, 105, 121–136. [Google Scholar] [CrossRef]
- Samaddar, P.; Sen, K. Cloud point extraction: A sustainable method of elemental preconcentration and speciation. J. Ind. Eng. Chem. 2014, 20, 1209–1219. [Google Scholar] [CrossRef]
- Hagarová, I.; Nemček, L. Recent advances in speciation analysis of trace antimony in environmental and biological samples based on cloud point extraction and spectrometric methods. In Sustainable Agriculture Reviews 52; Lichtfouse, E., Ed.; Springer Nature: Cham, Switzerland, 2021; pp. 49–77. [Google Scholar]
- Hagarová, I.; Urík, M. New approaches to the cloud point extraction: Utilizable for separation and preconcentration of trace metals. Curr. Anal. Chem. 2016, 12, 87–93. [Google Scholar] [CrossRef]
- Ghambarian, M.; Yamini, Y.; Saleh, A.; Shariati, S.; Yazdanfar, N. Taguchi OA(16) orthogonal array design for the optimization of cloud point extraction for selenium determination in environmental and biological samples by tungsten-modified tube electrothermal atomic absorption spectrometry. Talanta 2009, 78, 970–976. [Google Scholar] [CrossRef]
- Wang, M.; Zhong, Y.Z.; Qin, J.P.; Zhang, Z.H.; Li, S.; Yang, B.Y. Determination of total selenium in food samples by d-CPE and HG-AFS. Food Chem. 2017, 227, 329–334. [Google Scholar] [CrossRef]
- Sounderajan, S.; Kumar, G.K.; Udas, A.C. Cloud point extraction and electrothermal atomic absorption spectrometry of Se (IV)-3,3 ‘-diaminobenzidine for the estimation of trace amounts of Se (IV) and Se (VI) in environmental water samples and total selenium in animal blood and fish tissue samples. J. Hazard. Mater. 2010, 175, 666–672. [Google Scholar] [CrossRef]
- Chen, B.B.; Hu, B.; He, M. Cloud point extraction combined with electrothermal vaporization inductively coupled plasma mass spectrometry for the speciation of inorganic selenium in environmental water samples. Rapid Commun. Mass Spectrom. 2006, 20, 2894–2900. [Google Scholar] [CrossRef]
- Altunay, N.; Gürkan, R. Separation/preconcentration of ultra-trace levels of inorganic Sb and Se from different sample matrices by charge transfer sensitized ion-pairing using ultrasonic-assisted cloud point extraction prior to their speciation and determination by hydride generation AAS. Talanta 2016, 159, 344–355. [Google Scholar] [CrossRef]
- Wen, X.D.; Zhang, Y.Y.; Li, C.Y.; Fang, X.; Zhang, X.C. Comparison of rapidly synergistic cloud point extraction and ultrasound-assisted cloud point extraction for trace selenium coupled with spectrophotometric determination. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2014, 123, 200–205. [Google Scholar] [CrossRef]
- Depoi, F.D.; Pozebon, D. The use of cloud point extraction and hydride generation for improving the Sb and Se limits of detection in ICP OES. J. Braz. Chem. Soc. 2012, 23, 2211–2221. [Google Scholar] [CrossRef]
- Wen, S.P.; Zhu, X.S.; Wei, Y.X.; Wu, S.B. Cloud point extraction-inductively coupled plasma mass spectrometry for separation/analysis of aqueous-exchangeable and unaqueous-exchangeable selenium in tea samples. Food Anal. Methods 2013, 6, 506–511. [Google Scholar] [CrossRef]
- Ulusoy, H.I. Simple and useful method for determination of inorganic selenium species in real samples based on UV-VIS spectroscopy in a micellar medium. Anal. Methods 2015, 7, 953–960. [Google Scholar] [CrossRef]
- Li, Y.J.; Hu, B.; He, M.; Xiang, G.Q. Simultaneous speciation of inorganic selenium and antimony in water samples by electrothermal vaporization inductively coupled plasma mass spectrometry following selective cloud point extraction. Water Res. 2008, 42, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, G.J.; Wu, Q.H. Speciation of organic and inorganic selenium in selenium-enriched rice by graphite furnace atomic absorption spectrometry after cloud point extraction. Food Chem. 2013, 141, 66–71. [Google Scholar] [CrossRef]
- Altunay, N.; Gürkan, R. Determination of sub-ng g(-1) levels of total inorganic arsenic and selenium in foods by hydride-generation atomic absorption spectrometry after pre-concentration. Food Addit. Contam. Part A-Chem. Anal. Control. Expo. Risk Assess. 2017, 34, 390–403. [Google Scholar] [CrossRef]
- USEPA. National Primary Drinking Water Regulations, List of Contaminants and Their MCLs; EPA 816-F-09-0004; USEPA: Washington, DC, USA, 2009. [Google Scholar]
- USEPA. External Peer Review Draft-Aquatic Life Ambient Water Quality Criterion For Selenium-Freshwater 2014; EPA-820-F-14-005 2014; USEPA: Washington, DC, USA, 2014. [Google Scholar]
- Santos, S.; Ungureanu, G.; Boaventura, R.; Botelho, C. Selenium contaminated waters: An overview of analytical methods, treatment options and recent advances in sorption methods. Sci. Total Environ. 2015, 521, 246–260. [Google Scholar] [CrossRef]
- May, T.W.; Walther, M.J.; Petty, J.D.; Fairchild, J.F.; Lucero, J.; Delvaux, M.; Manring, J.; Armbruster, M.; Hartman, D. An evaluation of selenium concentrations in water, sediment, invertebrates, and fish from the Republican River Basin: 1997-1999. Environ. Monit. Assess. 2001, 72, 179–206. [Google Scholar] [CrossRef]
- Williams, P.N.; Lombi, E.; Sun, G.X.; Scheckel, K.; Zhu, Y.G.; Feng, X.B.; Zhu, J.M.; Carey, A.M.; Adomako, E.; Lawgali, Y.; et al. Selenium characterization in the global rice supply chain. Environ. Sci. Technol. 2009, 43, 6024–6030. [Google Scholar] [CrossRef]
- Dinh, Q.T.; Cui, Z.W.; Huang, J.; Tran, T.A.T.; Wang, D.; Yang, W.X.; Zhou, F.; Wang, M.K.; Yu, D.S.; Liang, D.L. Selenium distribution in the Chinese environment and its relationship with human health: A review. Environ. Int. 2018, 112, 294–309. [Google Scholar] [CrossRef]
- Tadayon, F.; Mehrandoost, S. Determination and speciation of Se(IV), Se(VI), and selenomethionine in Iranian walnut samples by spectrophotometry after cloud point extraction. J. Anal. Chem. 2015, 70, 1336–1341. [Google Scholar] [CrossRef]
- Güler, N.; Maden, M.; Bakirdere, S.; Ataman, O.Y.; Volkan, M. Speciation of selenium in vitamin tablets using spectrofluorometry following cloud point extraction. Food Chem. 2011, 129, 1793–1799. [Google Scholar] [CrossRef]
- Chen, S.Z.; Zhu, S.P.; Lu, D.B. Solidified floating organic drop microextraction for speciation of selenium and its distribution in selenium-rich tea leaves and tea infusion by electrothermal vapourisation inductively coupled plasma mass spectrometry. Food Chem. 2015, 169, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, B.; Feizbakhsh, A.; Konoz, E.; Faraji, H. Hydrophobic deep eutectic solvent based on centrifugation-free dispersive liquid-liquid microextraction for speciation of selenium in aqueous samples: One step closer to green analytical chemistry. Microchem. J. 2019, 148, 582–590. [Google Scholar] [CrossRef]
- Rahnama, R.; Abed, Z. Application of cold-induced aggregation microextraction as a fast, simple, and organic solvent-free method for the separation and preconcentration of Se(IV) in rice and various water samples. Environ. Monit. Assess. 2014, 186, 4209–4216. [Google Scholar] [CrossRef]
- Grijalba, A.C.; Martinis, E.M.; Wuilloud, R.G. Inorganic selenium speciation analysis in Allium and Brassica vegetables by ionic liquid assisted liquid-liquid microextraction with multivariate optimization. Food Chem. 2017, 219, 102–108. [Google Scholar] [CrossRef]
- Tuzen, M.; Pekiner, O.Z. Ultrasound-assisted ionic liquid dispersive liquid-liquid microextraction combined with graphite furnace atomic absorption spectrometric for selenium speciation in foods and beverages. Food Chem. 2015, 188, 619–624. [Google Scholar] [CrossRef]
- Wang, X.J.; Wu, L.; Cao, J.Q.; Hong, X.C.; Ye, R.; Chen, W.J.; Yuan, T. Magnetic effervescent tablet-assisted ionic liquid dispersive liquid-liquid microextraction of selenium for speciation in foods and beverages. Food Addit. Contam. Part A-Chem. Anal. Control. Expo. Risk Assess. 2016, 33, 1190–1199. [Google Scholar] [CrossRef]
- Martinis, E.M.; Escudero, L.B.; Berton, P.; Monasterio, R.P.; Filippini, M.F.; Wuilloud, R.G. Determination of inorganic selenium species in water and garlic samples with on-line ionic liquid dispersive microextraction and electrothermal atomic absorption spectrometry. Talanta 2011, 85, 2182–2188. [Google Scholar] [CrossRef]
- Bidari, A.; Jahromi, E.Z.; Assadi, Y.; Hosseini, M.R.M. Monitoring of selenium in water samples using dispersive liquid-liquid microextraction followed by iridium-modified tube graphite furnace atomic absorption spectrometry. Microchem. J. 2007, 87, 6–12. [Google Scholar] [CrossRef]
- Moradi, M.; Kashanaki, R.; Borhani, S.; Bigdeli, H.; Abbasi, N.; Kazemzadeh, A. Optimization of supramolecular solvent microextraction prior to graphite furnace atomic absorption spectrometry for total selenium determination in food and environmental samples. J. Mol. Liq. 2017, 232, 243–250. [Google Scholar] [CrossRef]
- López-García, I.; Vicente-Martínez, Y.; Hernández-Córdoba, M. Nonchromatographic speciation of selenium in edible oils using dispersive liquid-liquid microextraction and electrothermal atomic absorption spectrometry. J. Agric. Food Chem. 2013, 61, 9356–9361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duan, J.K.; He, M.; Chen, B.B.; Hu, B. Dispersive liquid liquid microextraction combined with electrothermal vaporization inductively coupled plasma mass spectrometry for the speciation of inorganic selenium in environmental water samples. Talanta 2013, 115, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Pyrzynska, K. Determination of selenium species in environmental samples. Microchim. Acta 2002, 140, 55–62. [Google Scholar] [CrossRef]
- Lima, L.W.; Stonehouse, G.C.; Walters, C.; El Mehdawi, A.F.; Fakra, S.C.; Pilon-Smits, E.A.H. Selenium accumulation, speciation and localization in Brazil nuts (Bertholletia excelsa H.B.K.). Plants 2019, 8, 289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Sample | Detection System | Complexing Reagent | Surfactant | Diluting Agent | EF | LOD (µg/L) | RSD (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Water, fish | ETAAS | DAB | TX-114 | HNO3 | 100 | 0.0025 | 3.6 | [49] |
| Water | ETAAS | OPD | TX-114 | Met-OH | 63.5 | 0.09 | 3.6 | [47] |
| Rice | ETAAS | Dithizone | TX-114 | Et-OH/HNO3 | 82 | 0.08 | 2.1 | [57] |
| Tea | ICP-MS | DDTC | TX-100 | HNO3 | 20 | 0.10 | 3.2 | [54] |
| Water | ETV-ICP-MS | DDTC | TX-114 | Et-OH | 50 | 0.05 | 3.5 | [56] |
| Water | ETV-ICP-MS | APDC | TX-114 | Met-OH | 39 | 0.008 | 3.9 | [50] |
| Walnut | UV-Vis | DAN | TX-114 | Met-OH | 20 | 9.00 | 1.6 | [65] |
| Water, hen egg | UV-Vis | DAHMP | TX-114/SDS | Et-OH | 50 | 6.06 | 2.8 | [55] |
| Water | UV-Vis | Dithizone | TX-114 (octanol) * | Met-OH | 124 | 0.20 | 4.3 | [52] |
| Water | UV-Vis | Dithizone | TX-114 | Met-OH | 103 | 0.30 | 3.2 | [52] |
| Water, nuts, white wine | HG-ICP-OES | DDTP | TX-114 | Met-OH/HCl | 8 | 0.10 | 4.4 | [53] |
| Various food | HG-AFS | APDC | TX-114 | HCl + H2O2 ** | 11.8 | 0.023 | 4.0 | [48] |
| Water, various food | HG-AAS | NRH+ | PONPE 7.5/CTAB | HCl | 155 | 0.00245 | 5.3 | [51] |
| Fish, various food | HG-AAS | Toluidine Red | Brij 35 | HNO3 | 140 | 0.0035 | 3.9 | [58] |
| Vitamin supplements | Fluorometry | DAN | TX-114 | H2O | 10 | 2.10 | 5.0 | [66] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hagarová, I.; Nemček, L. Reliable Quantification of Ultratrace Selenium in Food, Beverages, and Water Samples by Cloud Point Extraction and Spectrometric Analysis. Nutrients 2022, 14, 3530. https://doi.org/10.3390/nu14173530
Hagarová I, Nemček L. Reliable Quantification of Ultratrace Selenium in Food, Beverages, and Water Samples by Cloud Point Extraction and Spectrometric Analysis. Nutrients. 2022; 14(17):3530. https://doi.org/10.3390/nu14173530
Chicago/Turabian StyleHagarová, Ingrid, and Lucia Nemček. 2022. "Reliable Quantification of Ultratrace Selenium in Food, Beverages, and Water Samples by Cloud Point Extraction and Spectrometric Analysis" Nutrients 14, no. 17: 3530. https://doi.org/10.3390/nu14173530
APA StyleHagarová, I., & Nemček, L. (2022). Reliable Quantification of Ultratrace Selenium in Food, Beverages, and Water Samples by Cloud Point Extraction and Spectrometric Analysis. Nutrients, 14(17), 3530. https://doi.org/10.3390/nu14173530





.png)