A Comparison of the Impact of Restrictive Diets on the Gastrointestinal Tract of Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. qRT-PCR
2.3. Western Blot
2.4. Protein Array
2.5. Quantification of NFκB
2.6. Histology
2.6.1. Hematoxylin and Eosin Staining
2.6.2. Goblet Cells Staining
2.7. Imaging
2.8. Metabolomics
2.8.1. SCFA and MCFA Detection
2.8.2. Bile Acids Quantification
2.8.3. Untargeted Metabolomics
2.9. Statistics
3. Results
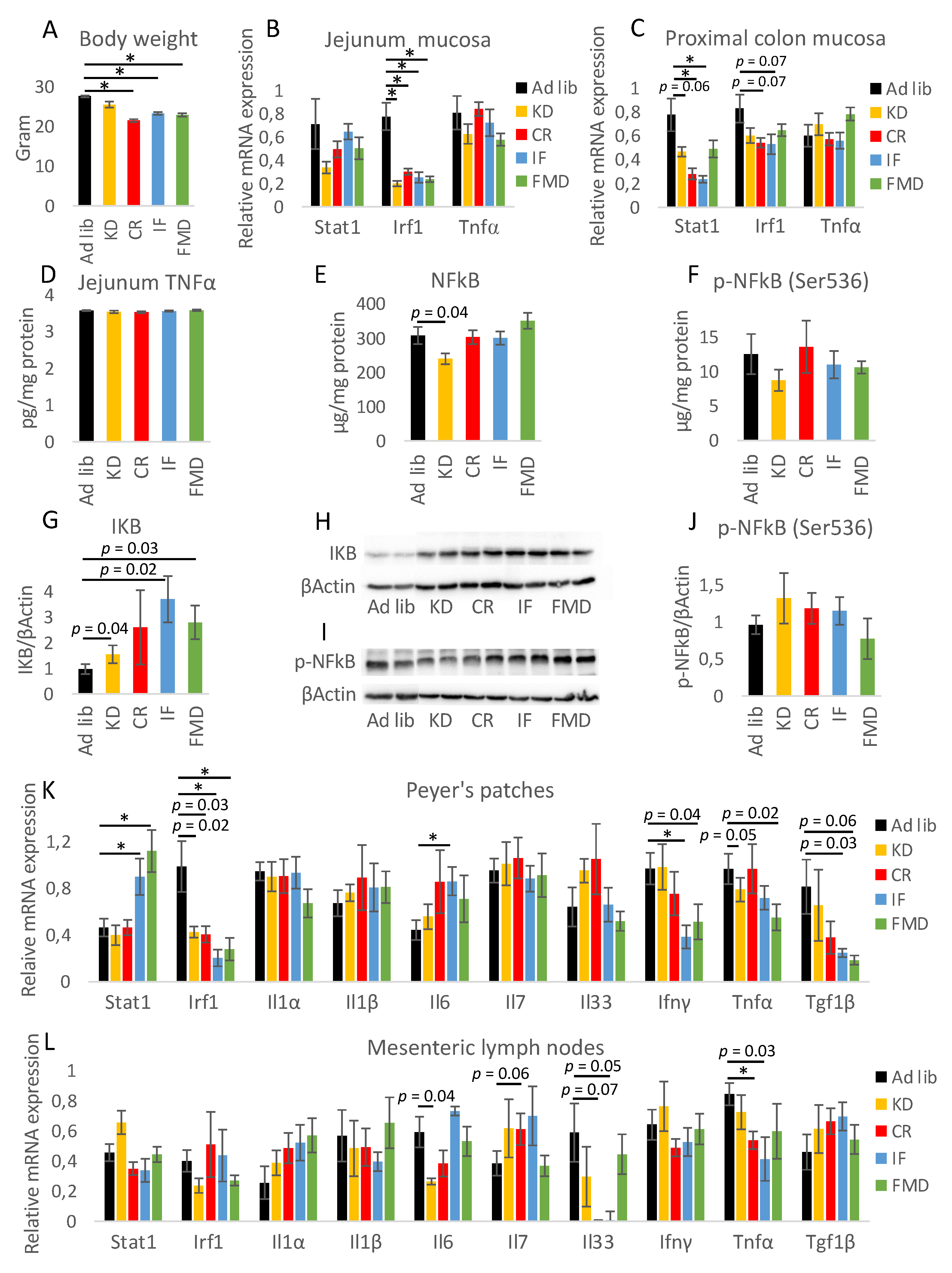
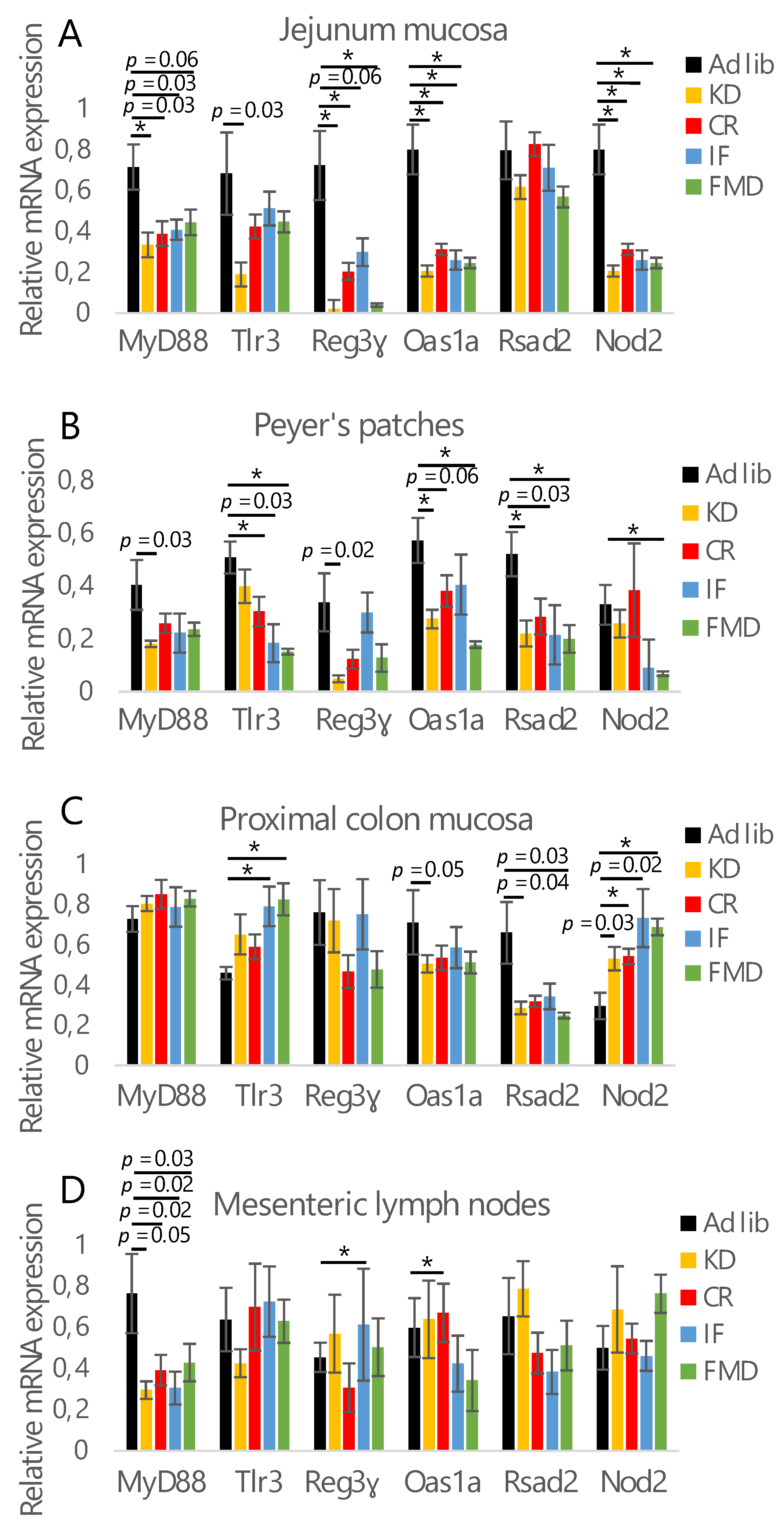
3.1. All Restrictive Diets Modulate the Expression of Inflammatory and Antibacterial Factors
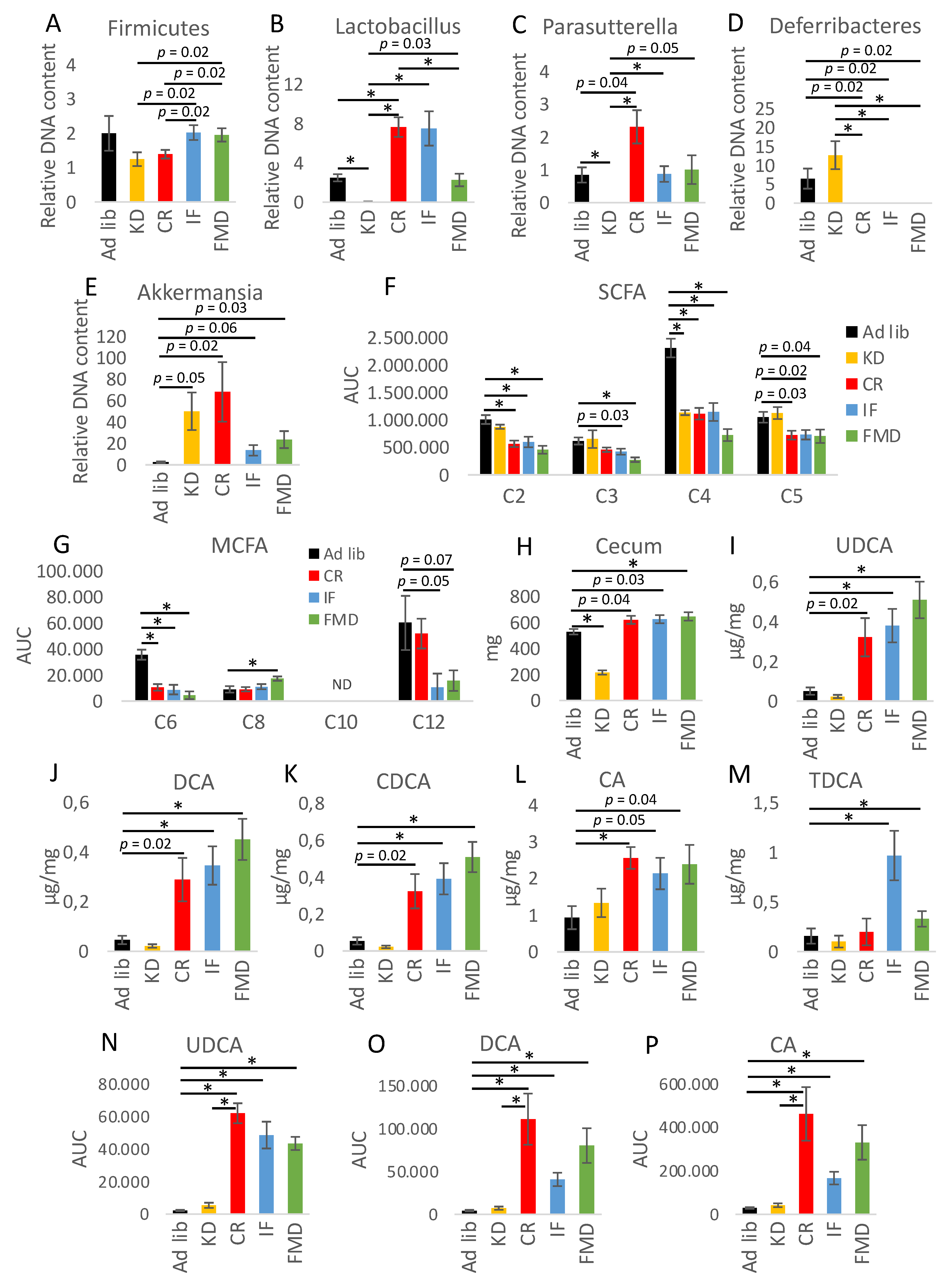
3.2. Microbiota Composition Changes Depend on Energy and Macronutrients Restriction
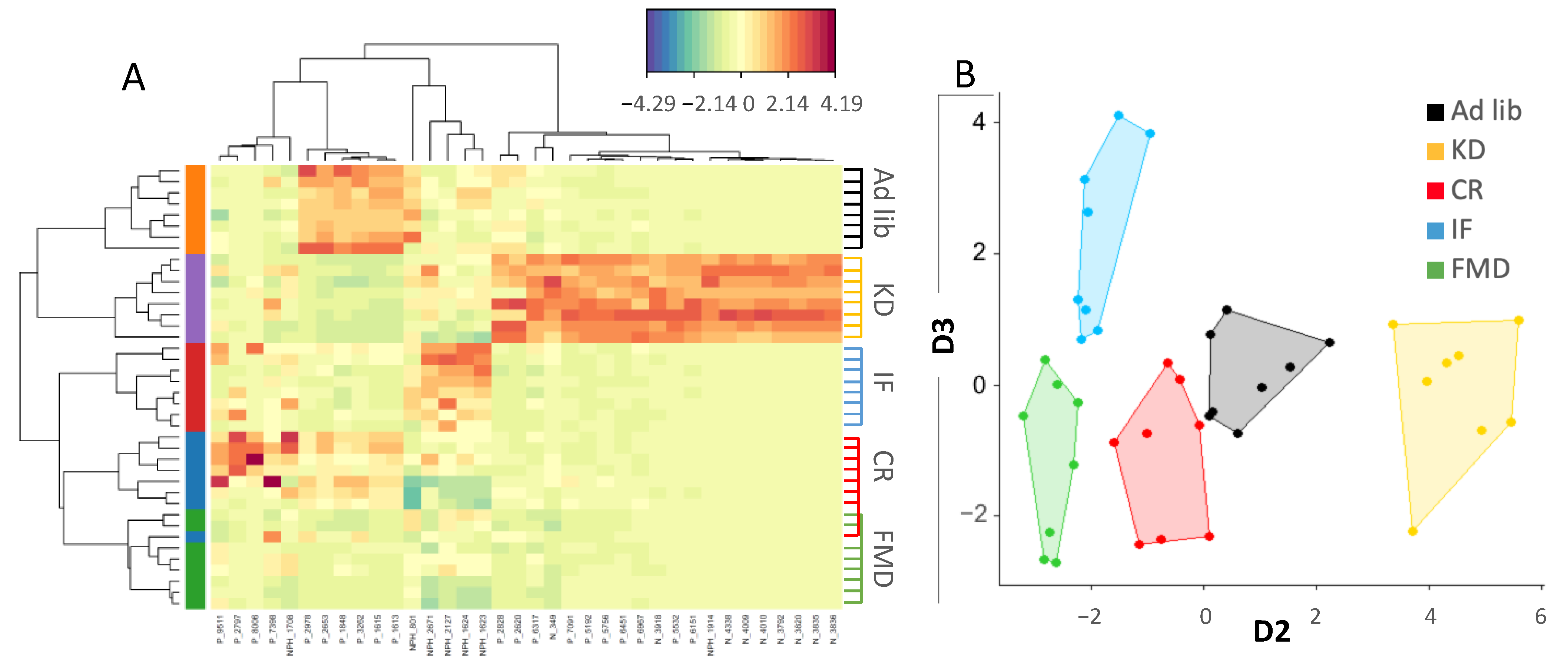
3.3. Metabolites Composition Changes Depend on Energy and Macronutrients Restriction
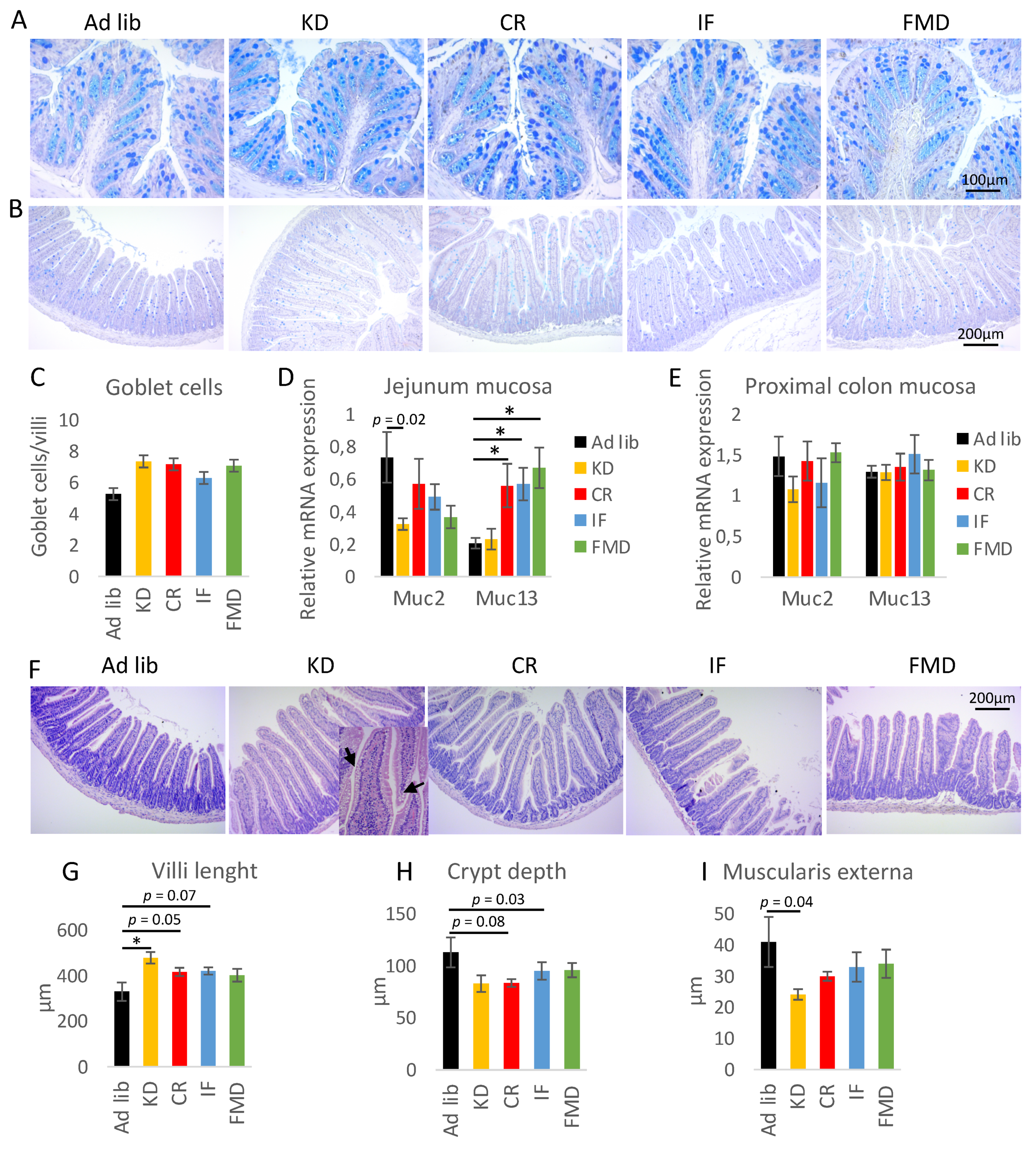
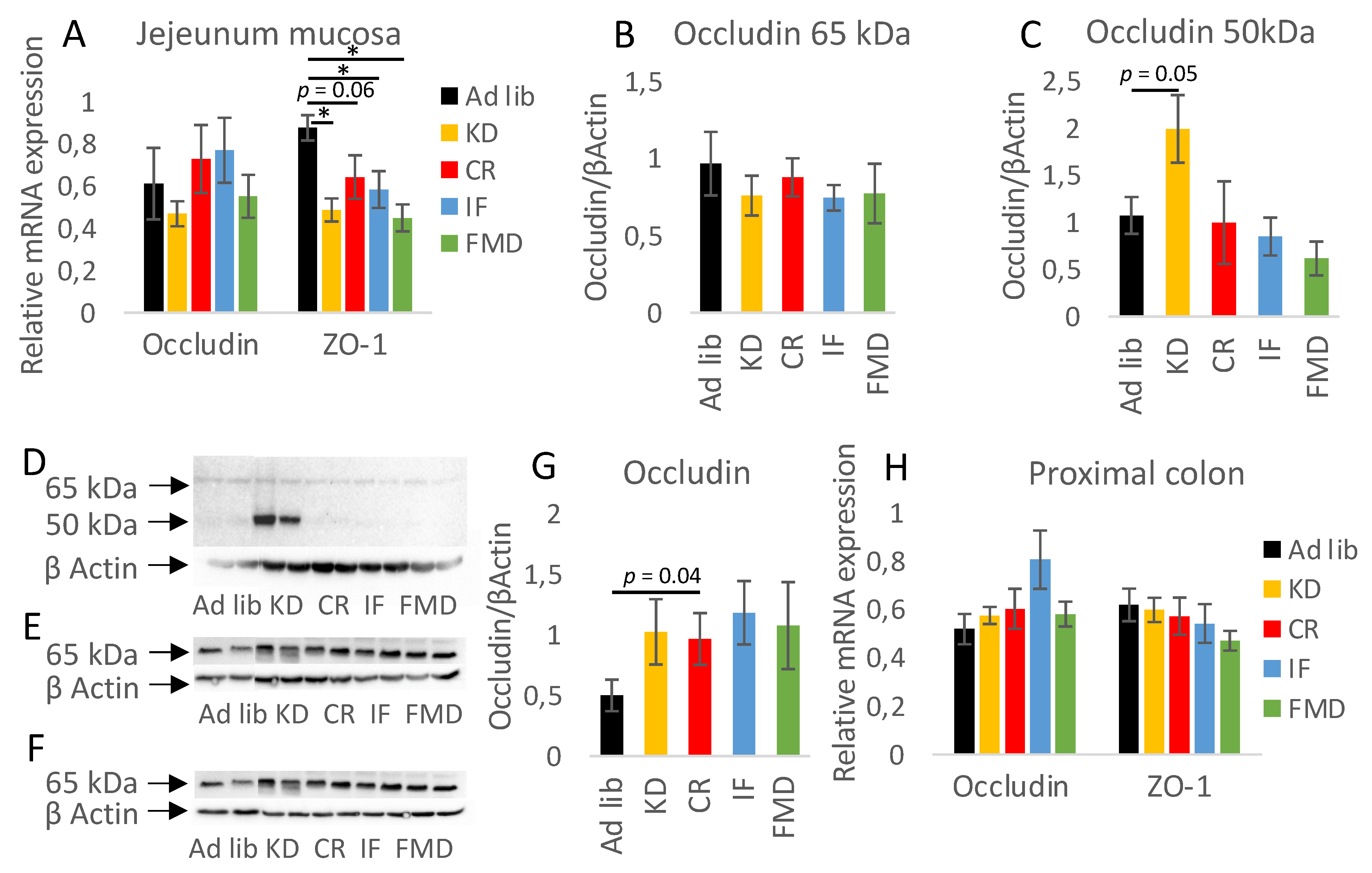
3.4. Restrictive Diets Affect Mucin Production and Intestine Morphology
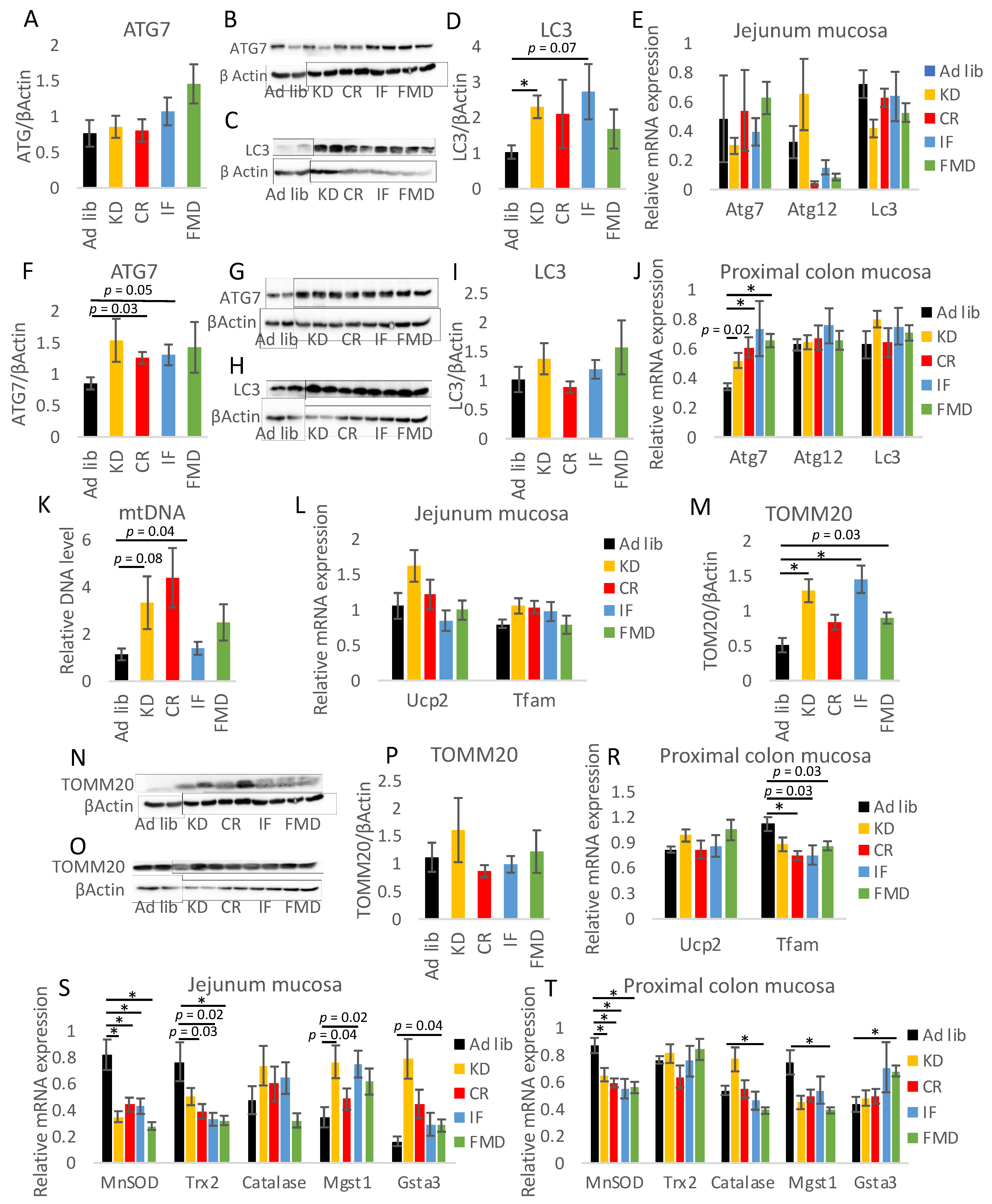
3.5. Modulation of Autophagy, Mitochondria, and Oxidative Stress by Restrictive Diets
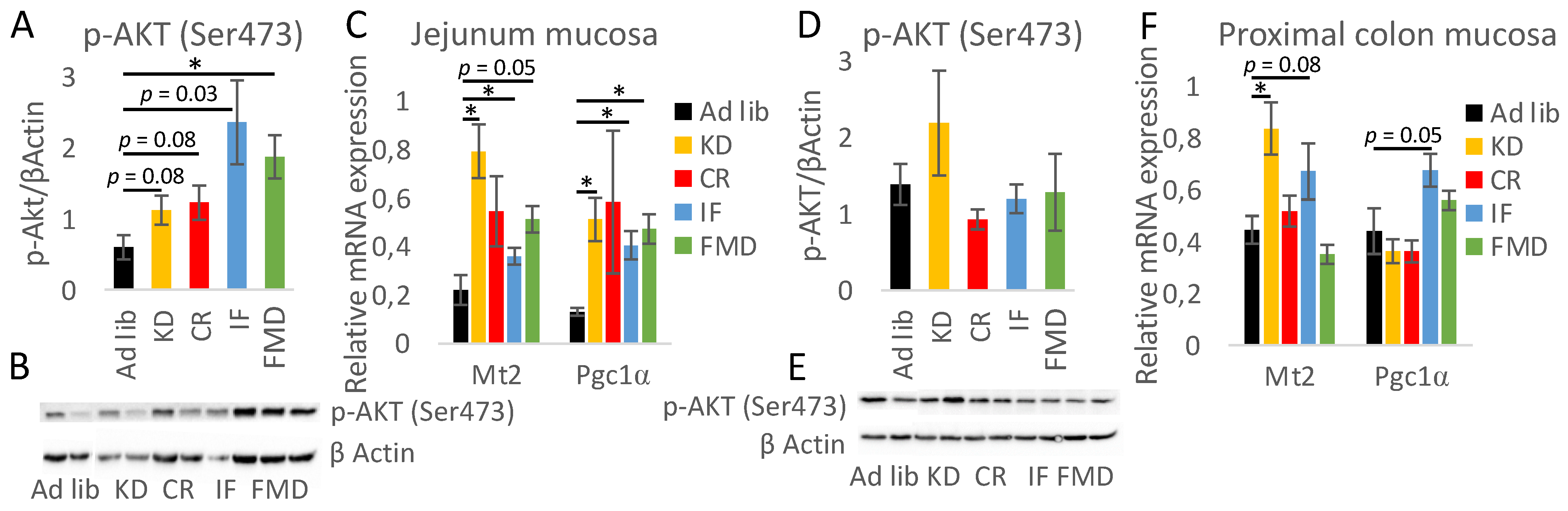
3.6. The Impact of Restrictive Diets on the Markers of Energy Limitation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molodecky, N.A.; Soon, I.S.; Rabi, D.M.; Ghali, W.A.; Ferris, M.; Chernoff, G.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Barkema, H.W.; et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012, 142, 46–54.e2. [Google Scholar] [CrossRef] [Green Version]
- Skypala, I.; Vlieg-Boerstra, B. Food intolerance and allergy: Increased incidence or contemporary inadequate diets? Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Chiba, M.; Nakane, K.; Komatsu, M. Westernized Diet is the Most Ubiquitous Environmental Factor in Inflammatory Bowel Disease. Perm. J. 2019, 23, 18–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Brandhorst, S.; Choi, I.Y.; Wei, M.; Cheng, C.W.; Sedrakyan, S.; Navarrete, G.; Dubeau, L.; Yap, L.P.; Park, R.; Vinciguerra, M.; et al. A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab. 2015, 22, 86–99. [Google Scholar] [CrossRef] [Green Version]
- Lara-Padilla, E.; Godínez-Victoria, M.; Drago-Serrano, M.E.; Reyna-Garfias, H.; Arciniega-Martínez, I.M.; Abarca-Rojano, E.; Cruz-Hernández, T.R.; Campos-Rodríguez, R. Intermittent fasting modulates IgA levels in the small intestine under intense stress: A mouse model. J. Neuroimmunol. 2015, 285, 22–30. [Google Scholar] [CrossRef]
- Masoro, E.J. Overview of caloric restriction and ageing. Mech. Ageing Dev. 2005, 126, 913–922. [Google Scholar] [CrossRef]
- Koga, A.; Kimura, S. Influence of restricted diet on the cell cycle in the crypt of mouse small intestine. J. Nutr. Sci. Vitaminol. 1980, 26, 33–38. [Google Scholar] [CrossRef]
- Suarez-Souto, M.A.; Lara-Padilla, E.; Reyna-Garfias, H.; Viloria, M.; López-Sánchez, P.; Rivera-Aguilar, V.; Miliar-García, Á.; Kormanovski, A.; Domínguez-López, M.L.; Campos-Rodríguez, R. Caloric restriction modifies both innate and adaptive immunity in the mouse small intestine. J. Physiol. Biochem. 2012, 68, 163–173. [Google Scholar] [CrossRef]
- Casirola, D.M.; Rifkin, B.; Tsai, W.; Ferraris, R.P. Adaptations of intestinal nutrient transport to chronic caloric restriction in mice. Am. J. Physiol. 1996, 271, G192–G200. [Google Scholar] [CrossRef]
- Duszka, K.; Ellero-Simatos, S.; Ow, G.S.; Defernez, M.; Paramalingam, E.; Tett, A.; Ying, S.; König, J.; Narbad, A.; Kuznetsov, V.A.; et al. Complementary intestinal mucosa and microbiota responses to caloric restriction. Sci. Rep. 2018, 8, 11338. [Google Scholar] [CrossRef] [PubMed]
- Goodrick, C.L.; Ingram, D.K.; Reynolds, M.A.; Freeman, J.R.; Cider, N. Effects of intermittent feeding upon body weight and lifespan in inbred mice: Interaction of genotype and age. Mech. Ageing Dev. 1990, 55, 69–87. [Google Scholar] [CrossRef]
- Okada, T.; Otsubo, T.; Hagiwara, T.; Inazuka, F.; Kobayashi, E.; Fukuda, S.; Inoue, T.; Higuchi, K.; Kawamura, Y.I.; Dohi, T. Intermittent fasting prompted recovery from dextran sulfate sodium-induced colitis in mice. J. Clin. Biochem. Nutr. 2017, 61, 100–107. [Google Scholar] [CrossRef]
- Rangan, P.; Choi, I.; Wei, M.; Navarrete, G.; Guen, E.; Brandhorst, S.; Enyati, N.; Pasia, G.; Maesincee, D.; Ocon, V.; et al. Fasting-Mimicking Diet Modulates Microbiota and Promotes Intestinal Regeneration to Reduce Inflammatory Bowel Disease Pathology. Cell Rep. 2019, 26, 2704–2719e.6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Bai, M.; Ling, Z.; Lin, Y.; Wang, S.; Chen, Y. Intermittent administration of a fasting-mimicking diet reduces intestinal inflammation and promotes repair to ameliorate inflammatory bowel disease in mice. J. Nutr. Biochem. 2021, 96, 108785. [Google Scholar] [CrossRef]
- Cheng, C.W.; Villani, V.; Buono, R.; Wei, M.; Kumar, S.; Yilmaz, O.H.; Cohen, P.; Sneddon, J.B.; Perin, L.; Longo, V.D.; et al. Fasting-Mimicking Diet Promotes Ngn3-Driven beta-Cell Regeneration to Reverse Diabetes. Cell 2017, 168, 775–788.e12. [Google Scholar] [CrossRef] [Green Version]
- Caffa, I.; Spagnolo, V.; Vernieri, C.; Valdemarin, F.; Becherini, P.; Wei, M.; Brandhorst, S.; Zucal, C.; Driehuis, E.; Ferrando, L.; et al. Fasting-mimicking diet and hormone therapy induce breast cancer regression. Nature 2020, 583, 620–624. [Google Scholar] [CrossRef]
- Zhou, Z.L.; Jia, X.-B.; Sun, M.-F.; Zhu, Y.-L.; Qiao, C.-M.; Zhang, B.-P.; Zhao, L.-P.; Yang, Q.; Cui, C.; Chen, X.; et al. Neuroprotection of Fasting Mimicking Diet on MPTP-Induced Parkinson’s Disease Mice via Gut Microbiota and Metabolites. Neurotherapeutics 2019, 16, 741–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, M.; Wang, Y.; Han, R.; Xu, L.; Huang, M.; Zhao, J.; Lin, Y.; Song, S.; Chen, Y. Intermittent caloric restriction with a modified fasting-mimicking diet ameliorates autoimmunity and promotes recovery in a mouse model of multiple sclerosis. J. Nutr. Biochem. 2021, 87, 108493. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, Y.; Han, R.; Xu, L.; Huang, M.; Zhao, J.; Lin, Y.; Song, S.; Chen, Y. Ketogenic diet for human diseases: The underlying mechanisms and potential for clinical implementations. Signal Transduct. Target. Ther. 2022, 7, 11. [Google Scholar] [CrossRef]
- Weber, D.D.; Aminzadeh-Gohari, S.; Tulipan, J.; Catalano, L.; Feichtinger, R.G.; Kofler, B. Ketogenic diet in the treatment of cancer—Where do we stand? Mol. Metab. 2020, 33, 102–121. [Google Scholar] [CrossRef] [PubMed]
- Gregor, A.; Fragner, L.; Trajanoski, S.; Li, W.; Sun, X.; Weckwerth, W.; König, J.; Duszka, K. Cage bedding modifies metabolic and gut microbiota profiles in mouse studies applying dietary restriction. Sci. Rep. 2020, 10, 20835. [Google Scholar] [CrossRef] [PubMed]
- Gregor, A.P.; Fahrngruber, C.M.; Bayer, S.; Somoza, V.; König, K.; Duszka, K. Caloric restriction increases levels of taurine in the intestine and stimulates taurine uptake by conjugation to glutathione. JNB 2021, 96, 108781. [Google Scholar] [CrossRef]
- Locker, F.; Szarzynski, A.; Beikbaghban, T.; Gregor, A.; Duszka, K.; Walter, I.; Kofler, B.; Pohl, E.E. Triglyceride mediated upregulation of UCP3 in murine heart. Biophys. J. 2022, 121, 513a. [Google Scholar] [CrossRef]
- Lilja, S.; Stoll, C.; Krammer, U.; Hippe, B.; Duszka, K.; Debebe, T.; Höfinger, I.; König, J.; Pointner, A.; Haslberger, A. Five Days Periodic Fasting Elevates Levels of Longevity Related Christensenella and Sirtuin Expression in Humans. Int. J. Mol. Sci. 2021, 22, 2331. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Bottcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [Green Version]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Reiter, S.; Dunkel, A.; Metwaly, A.; Panes, J.; Salas, A.; Haller, D.; Hofmann, T. Development of a Highly Sensitive Ultra-High-Performance Liquid Chromatography Coupled to Electrospray Ionization Tandem Mass Spectrometry Quantitation Method for Fecal Bile Acids and Application on Crohn’s Disease Studies. J. Agric. Food Chem. 2021, 69, 5238–5251. [Google Scholar] [CrossRef]
- Reiter, S.; Dunkel, A.; Dawid, C.; Hofmann, T. Targeted LC-MS/MS Profiling of Bile Acids in Various Animal Tissues. J. Agric. Food Chem. 2021, 69, 10572–10580. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Gaynor, R.B. IkappaB kinases: Key regulators of the NF-kappaB pathway. Trends Biochem. Sci. 2004, 29, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Foley, M.H.; O’Flaherty, S.; Barrangou, R.; Theriot, C.M. Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 2019, 15, e1007581. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duszka, K.; Gregor, A.; Guillou, H.; Konig, J.; Wahli, W. Peroxisome Proliferator-Activated Receptors and Caloric Restriction-Common Pathways Affecting Metabolism, Health, and Longevity. Cells 2020, 9, 1708. [Google Scholar] [CrossRef]
- Bagherniya, M.; Butler, A.E.; Barreto, G.E.; Sahebkar, A. The effect of fasting or calorie restriction on autophagy induction: A review of the literature. Ageing Res. Rev. 2018, 47, 183–197. [Google Scholar] [CrossRef]
- Liu, W.; Hendren, J.; Qin, X.J.; Shen, J.; Liu, K.J. Normobaric hyperoxia attenuates early blood-brain barrier disruption by inhibiting MMP-9-mediated occludin degradation in focal cerebral ischemia. J. Neurochem 2009, 108, 811–820. [Google Scholar] [CrossRef] [Green Version]
- Wachtel, M.; Wachtel, M.; Frei, K.; Ehler, E.; Fontana, A.; Winterhalter, K.; Gloor, S.M. Occludin proteolysis and increased permeability in endothelial cells through tyrosine phosphatase inhibition. J. Cell Sci. 1999, 112, 4347–4356. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Zeng, R.; Gorodeski, G.I. Changes in tight junctional resistance of the cervical epithelium are associated with modulation of content and phosphorylation of occludin 65-kilodalton and 50-kilodalton forms. Endocrinology 2006, 147, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Corfield, A.P. Mucins: A biologically relevant glycan barrier in mucosal protection. Biochim. Biophys. Acta 2015, 1850, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Szentkuti, L.; Lorenz, K. The thickness of the mucus layer in different segments of the rat intestine. Histochem. J. 1995, 27, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Andrews, C.; McLean, M.H.; Durum, S.K. Cytokine Tuning of Intestinal Epithelial Function. Front. Immunol. 2018, 9, 1270. [Google Scholar] [CrossRef] [PubMed]
- Sakai, H.; Jinawath, A.; Yamaoka, S.; Yuasa, Y. Upregulation of MUC6 mucin gene expression by NFkappaB and Sp factors. Biochem. Biophys. Res. Commun. 2005, 333, 1254–1260. [Google Scholar] [CrossRef]
- Lewin, M.R.; El Masri, S.H.; Clark, C.G. Effects of bile acids on mucus secretion in the dog colon. Eur. Surg. Res. 1979, 11, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Shimotoyodome, A.; Meguro, S.; Hase, T.; Tokimitsu, I.; Sakata, T. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2000, 125, 525–531. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Chamignon, C.; Mhedbi-Hajri, N.; Chain, F.; Derrien, M.; Escribano-Vázquez, U.; Garault, P.; Cotillard, A.; Pham, H.P.; Chervaux, C.; et al. The potential probiotic Lactobacillus rhamnosus CNCM I-3690 strain protects the intestinal barrier by stimulating both mucus production and cytoprotective response. Sci. Rep. 2019, 9, 5398. [Google Scholar] [CrossRef] [Green Version]
- Cheng, C.W.; Biton, M.; Haber, A.L.; Gunduz, N.; Eng, G.; Gaynor, L.T.; Tripathi, S.; Calibasi-Kocal, G.; Rickelt, S.; Butty, V.L.; et al. Ketone Body Signaling Mediates Intestinal Stem Cell Homeostasis and Adaptation to Diet. Cell 2019, 178, 1115–1131.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, T.Y.; Liu, H.W.; Hung, T.M. The Ketogenic Effect of Medium-Chain Triacylglycerides. Front. Nutr. 2021, 8, 747284. [Google Scholar] [CrossRef] [PubMed]
- Chwen, L.T.; Foo, H.L.; Thanh, N.T.; Choe, D.W. Growth performance, plasma Fatty acids, villous height and crypt depth of preweaning piglets fed with medium chain triacylglycerol. Asian-Australas J. Anim. Sci. 2013, 26, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Randall-Demllo, S.; Chieppa, M.; Eri, R. Intestinal epithelium and autophagy: Partners in gut homeostasis. Front. Immunol. 2013, 4, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagy, P.; Szatmári, Z.; Sándor, G.O.; Lippai, M.; Hegedűs, K.; Juhász, G. Drosophila Atg16 promotes enteroendocrine cell differentiation via regulation of intestinal Slit/Robo signaling. Development 2017, 144, 3990–4001. [Google Scholar] [CrossRef] [Green Version]
- Patel, K.K.; Miyoshi, H.; Beatty, W.L.; Head, R.D.; Malvin, N.P.; Cadwell, K.; Guan, J.-L.; Saitoh, T.; Akira, S.; Seglen, P.; et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013, 32, 3130–3144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picca, A.; Riezzo, G.; Lezza, A.M.S.; Clemente, C.; Pesce, V.; Orlando, A.; Chimienti, G.; Russo, F. Mitochondria and redox balance in coeliac disease: A case-control study. Eur. J. Clin. Investig. 2018, 48, e12877. [Google Scholar] [CrossRef]
- Jackson, D.N.; Theiss, A.L. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes 2020, 11, 285–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mottawea, W.; Chiang, C.-K.; Mühlbauer, M.; Starr, A.E.; Butcher, J.; Abujamel, T.; Deeke, S.A.; Brandel, A.; Zhou, H.; Shokralla, S.; et al. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn’s disease. Nat. Commun. 2016, 7, 13419. [Google Scholar] [CrossRef] [PubMed]
- Chimienti, G.; Orlando, A.; Lezza, A.; D’Attoma, B.; Notarnicola, M.; Gigante, I.; Pesce, V.; Russo, F. The Ketogenic Diet Reduces the Harmful Effects of Stress on Gut Mitochondrial Biogenesis in a Rat Model of Irritable Bowel Syndrome. Int. J. Mol. Sci. 2021, 22, 3498. [Google Scholar] [CrossRef]
- Newell, C.; Shutt, T.E.; Ahn, Y.; Hittel, D.S.; Khan, A.; Rho, J.M.; Shearer, J. Tissue Specific Impacts of a Ketogenic Diet on Mitochondrial Dynamics in the BTBR(T+tf/j) Mouse. Front. Physiol. 2016, 7, 654. [Google Scholar] [CrossRef] [Green Version]
- Hutfles, L.J.; Wilkins, H.; Koppel, S.J.; Weidling, I.W.; Selfridge, J.E.; Tan, E.; Thyfault, J.P.; Slawson, C.; Fenton, A.W.; Zhu, H.; et al. A bioenergetics systems evaluation of ketogenic diet liver effects. Appl. Physiol. Nutr. Metab. 2017, 42, 955–962. [Google Scholar] [CrossRef] [Green Version]
- Gabande-Rodriguez, E.; Gomez de Las Heras, M.M.; Mittelbrunn, M. Control of Inflammation by Calorie Restriction Mimetics: On the Crossroad of Autophagy and Mitochondria. Cells 2019, 9, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Brocker, C.N.; Yan, T.; Xie, C.; Krausz, K.W.; Xiang, R.; Gonzalez, F.J. Metabolic adaptation to intermittent fasting is independent of peroxisome proliferator-activated receptor alpha. Mol. Metab. 2018, 7, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Lanza, I.R.; Zabielski, P.; Klaus, K.A.; Morse, D.M.; Heppelmann, C.J.; Bergen, H.R., 3rd; Dasari, S.; Walrand, S.; Short, K.R.; Johnson, M.L.; et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012, 16, 777–788. [Google Scholar] [CrossRef] [Green Version]
- Martin-Montalvo, A.; de Cabo, R. Mitochondrial metabolic reprogramming induced by calorie restriction. Antioxid. Redox. Signal 2013, 19, 310–320. [Google Scholar] [CrossRef] [Green Version]
- Northcutt, R.C.; Mella, G.W.; Dickson, L.G. Analysis of human chromosomes. Postgrad. Med. 1966, 39, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Papada, E.; Kaliora, A.C.; Gioxari, A.; Papalois, A.; Forbes, A. Anti-inflammatory effect of elemental diets with different fat composition in experimental colitis. Br. J. Nutr. 2014, 111, 1213–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locker, F.; Leitner, J.; Aminzadeh-Gohari, S.; Weber, D.D.; Sanio, P.; Koller, A.; Feichtinger, R.G.; Weiss, R.; Kofler, B.; Lang, R. The Influence of Ketogenic Diets on Psoriasiform-Like Skin Inflammation. J. Investig. Dermatol. 2020, 140, 707–710.e7. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregor, A.; Huber, L.; Auernigg-Haselmaier, S.; Sternberg, F.; Billerhart, M.; Dunkel, A.; Somoza, V.; Ogris, M.; Kofler, B.; Longo, V.D.; et al. A Comparison of the Impact of Restrictive Diets on the Gastrointestinal Tract of Mice. Nutrients 2022, 14, 3120. https://doi.org/10.3390/nu14153120
Gregor A, Huber L, Auernigg-Haselmaier S, Sternberg F, Billerhart M, Dunkel A, Somoza V, Ogris M, Kofler B, Longo VD, et al. A Comparison of the Impact of Restrictive Diets on the Gastrointestinal Tract of Mice. Nutrients. 2022; 14(15):3120. https://doi.org/10.3390/nu14153120
Chicago/Turabian StyleGregor, András, Laura Huber, Sandra Auernigg-Haselmaier, Felix Sternberg, Magdalena Billerhart, Andreas Dunkel, Veronika Somoza, Manfred Ogris, Barbara Kofler, Valter D. Longo, and et al. 2022. "A Comparison of the Impact of Restrictive Diets on the Gastrointestinal Tract of Mice" Nutrients 14, no. 15: 3120. https://doi.org/10.3390/nu14153120
APA StyleGregor, A., Huber, L., Auernigg-Haselmaier, S., Sternberg, F., Billerhart, M., Dunkel, A., Somoza, V., Ogris, M., Kofler, B., Longo, V. D., König, J., & Duszka, K. (2022). A Comparison of the Impact of Restrictive Diets on the Gastrointestinal Tract of Mice. Nutrients, 14(15), 3120. https://doi.org/10.3390/nu14153120







