Exploring the Binding Interaction of Active Compound of Pineapple against Foodborne Bacteria and Novel Coronavirus (SARS-CoV-2) Based on Molecular Docking and Simulation Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Drug Compounds Preparation
2.2. Target Molecules Preparation
2.3. In Silico Interaction Analysis
2.4. Molecular Dynamics Simulation (MDS) Experimentation
3. Results and Discussion
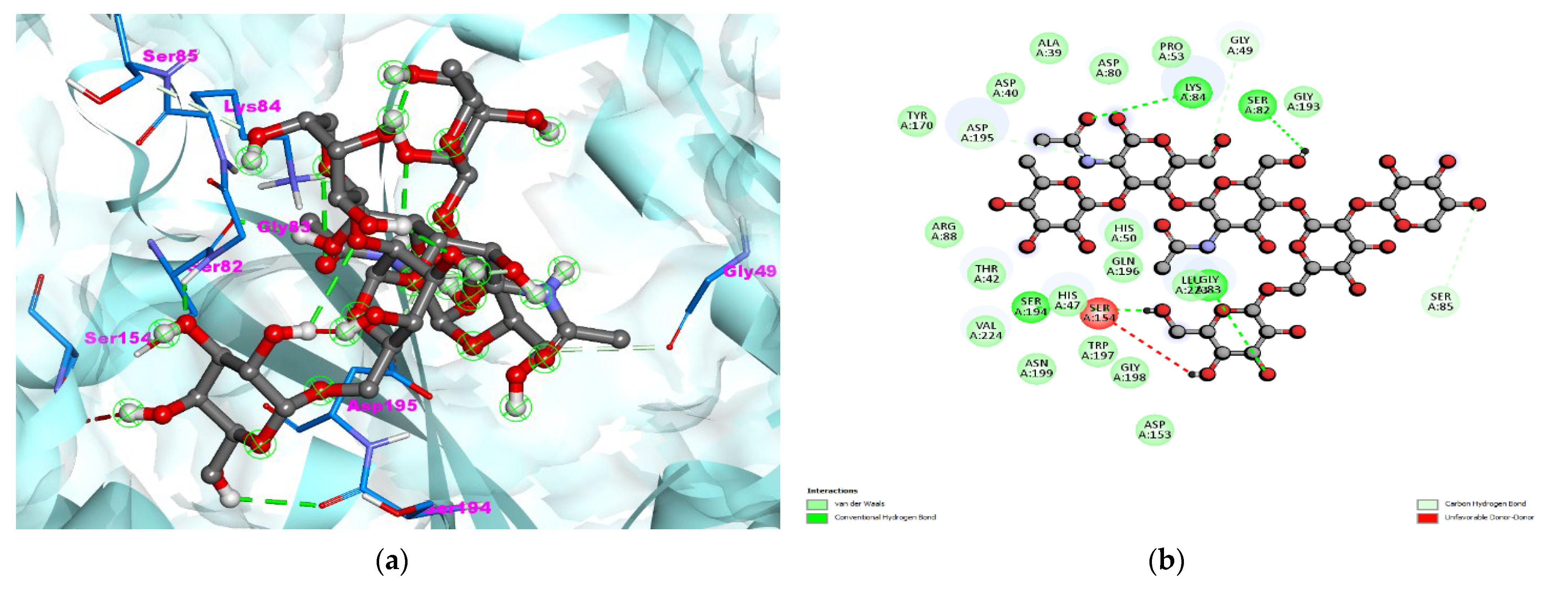
3.1. S. aureus TyrRS-Bromelain Docking Analysis
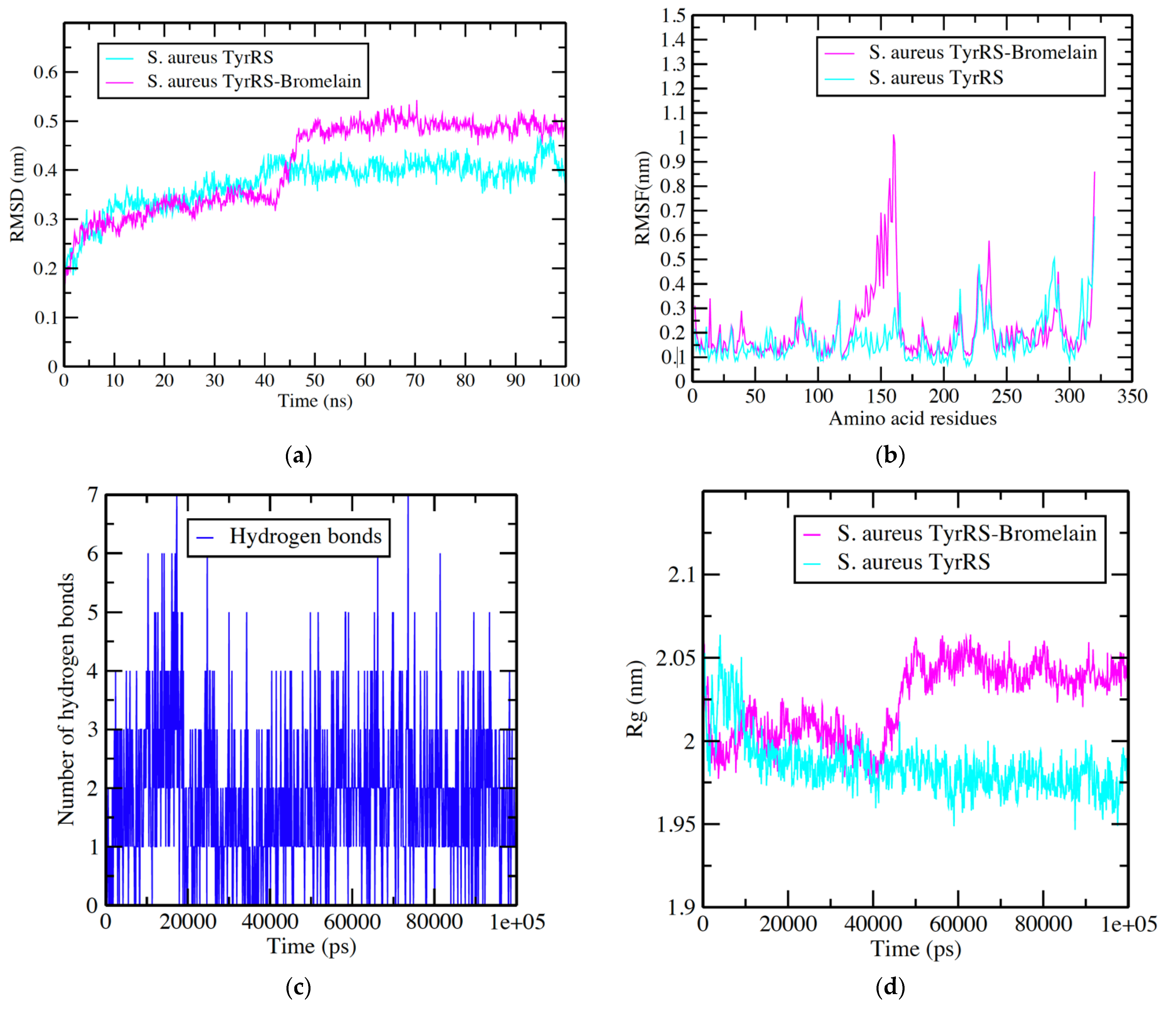
3.2. S. aureus TyrRS -Bromelain Simulation Analysis
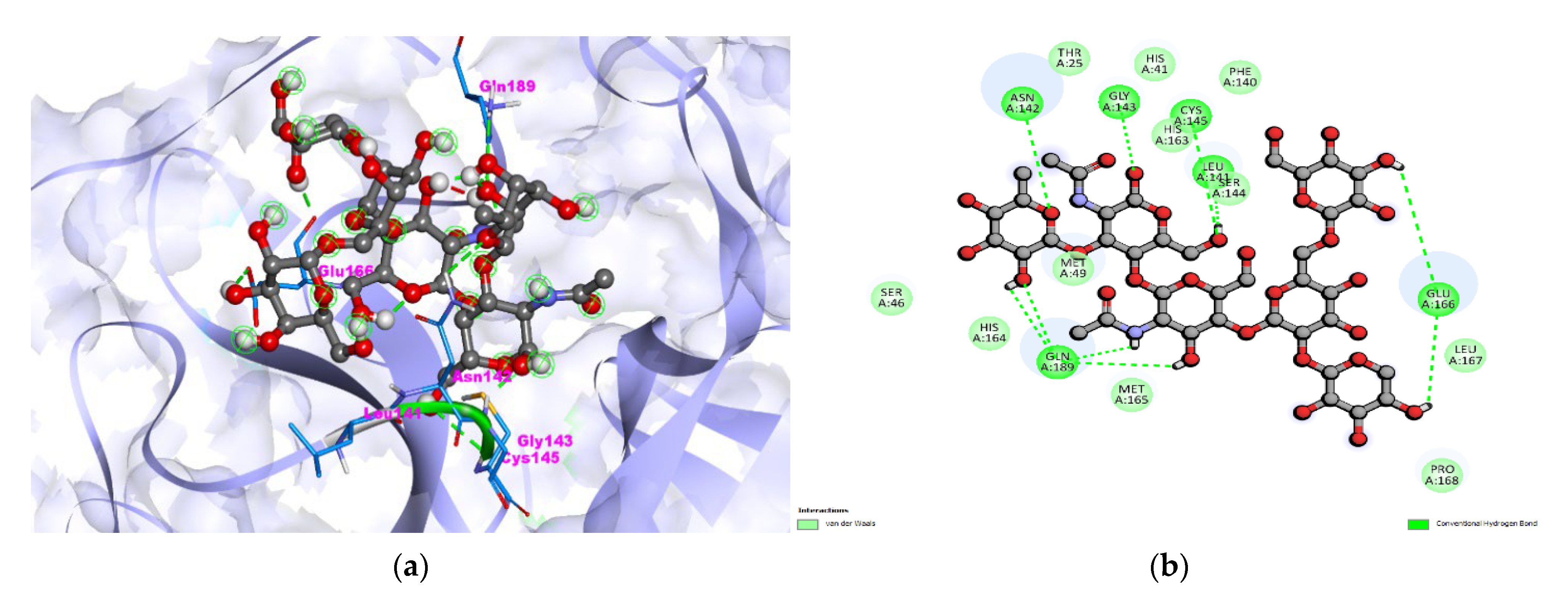
3.3. Protease-Bromelain Docking Analysis
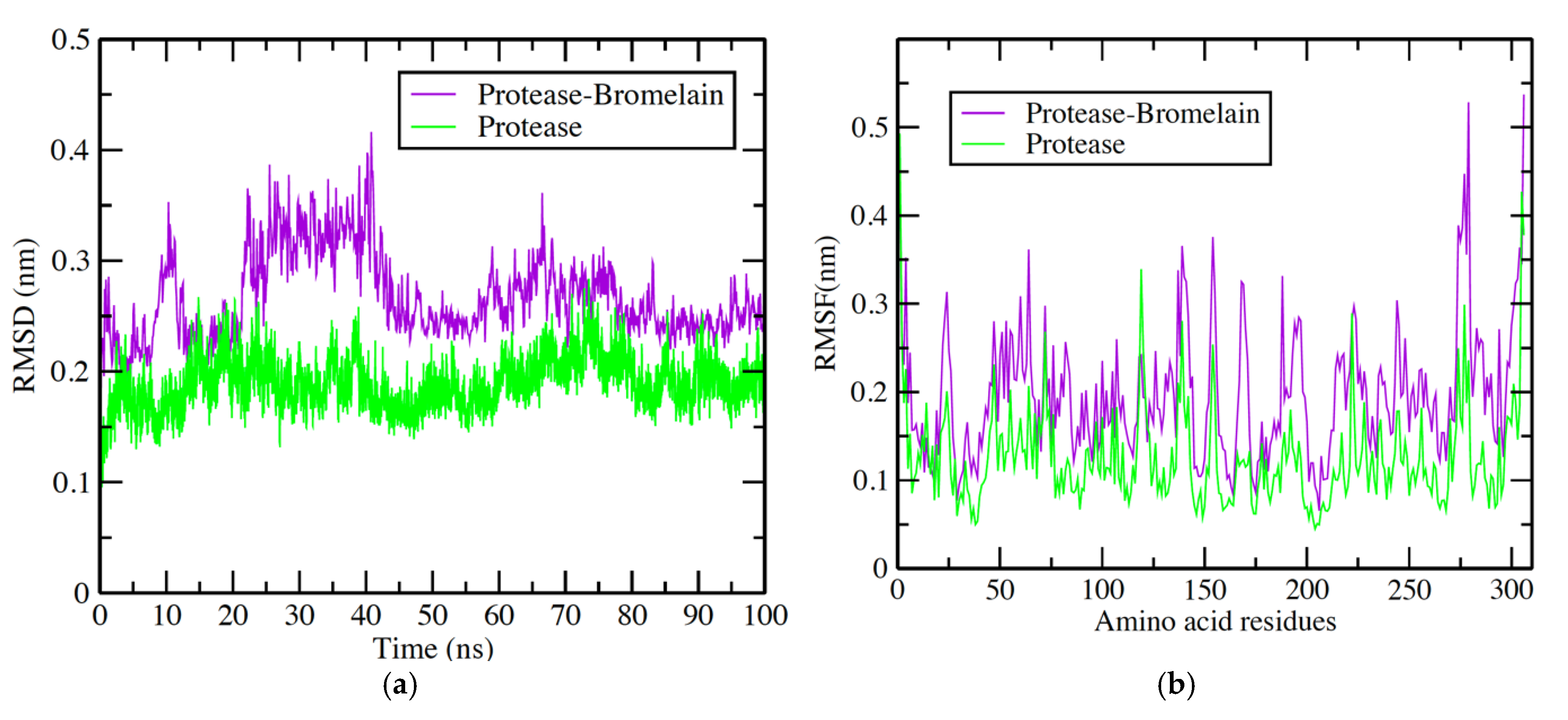
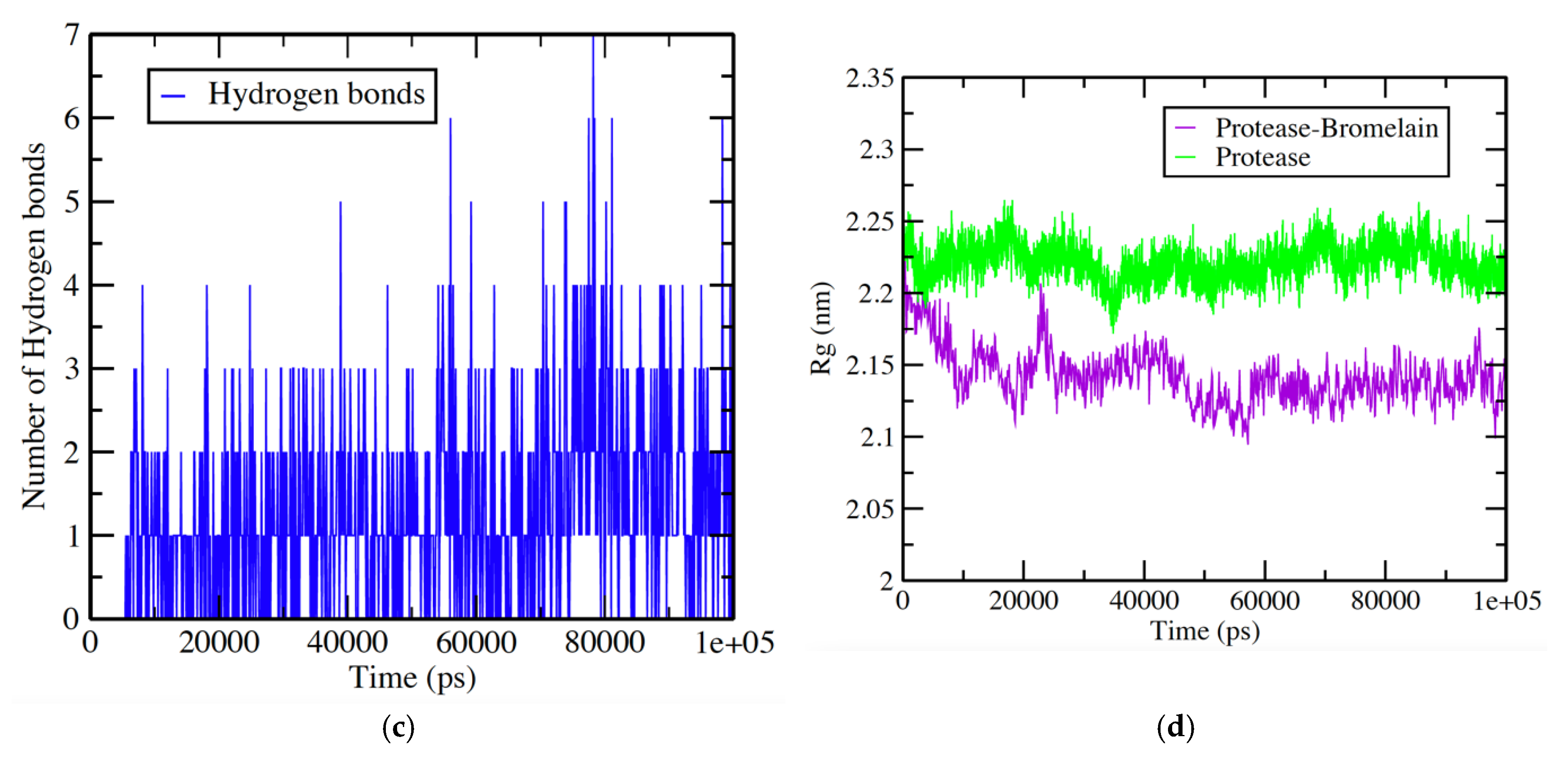
3.4. Protease-Bromelain Simulation Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pavan, R.; Jain, S.; Shraddha; Kumar, A. Properties and therapeutic application of bromelain: A review. Biotechnol. Res. Int. 2012, 2012, 976203. [Google Scholar] [CrossRef] [PubMed]
- Jančič, U.; Gorgieva, S. Bromelain and Nisin: The Natural Antimicrobials with High Potential in Biomedicine. Pharmaceutics 2021, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Khosa, S.; Frieg, B.; Mulnaes, D.; Kleinschrodt, D.; Hoeppner, A.; Gohlke, H.; Smits, S.H.J. Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 2016, 6, 18679. [Google Scholar] [CrossRef] [PubMed]
- Sulthana, R.; Archer, A. Bacteriocin nanoconjugates: Boon to medical and food industry. J. Appl. Microbiol. 2021, 131, 1056–1071. [Google Scholar] [CrossRef]
- Sharma, R. Viral Diseases and Antiviral Activity of Some Medicinal Plants with Special Reference to Ajmer. J. Antivir. Antiretrovir. 2019, 11, 183. [Google Scholar] [CrossRef][Green Version]
- Guzzi, P.H.; Mercatelli, D.; Ceraolo, C.; Giorgi, F.M. Master Regulator Analysis of the SARS-CoV-2/Human Interactome. J. Clin. Med. 2020, 9, 982. [Google Scholar] [CrossRef]
- Detopoulou, P.; Tsouma, C.; Papamikos, V. COVID-19 and Nutrition: Summary of Official Recommendations. Top. Clin. Nutr. 2022, 37, 187–202. [Google Scholar] [CrossRef]
- Zhang, J.; Rao, X.; Li, Y.; Zhu, Y.; Liu, F.; Guo, G.; Luo, G.; Meng, Z.; De Backer, D.; Xiang, H.; et al. Pilot trial of high-dose vitamin C in critically ill COVID-19 patients. Ann. Intensive Care 2021, 11, 5. [Google Scholar] [CrossRef]
- Varilla, C.; Marcone, M.; Paiva, L.; Baptista, J. Bromelain, a Group of Pineapple Proteolytic Complex Enzymes (Ananas comosus) and Their Possible Therapeutic and Clinical Effects. A Summary. Foods 2021, 10, 2249. [Google Scholar] [CrossRef]
- Fadlalla, M.; Ahmed, M.; Ali, M.; Elshiekh, A.A.; Yousef, B.A. Molecular Docking as a Potential Approach in Repurposing Drugs Against COVID-19: A Systematic Review and Novel Pharmacophore Models. Curr. Pharmacol. Rep. 2022, 8, 212–226. [Google Scholar] [CrossRef] [PubMed]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T.; Mondal, A.H.; Haq, Q.M.R. Antibiotics, resistome and resistance mechanisms: A bacterial perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.R.; Brooks, C.L., 3rd; Mackerell, A.D., Jr.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The biomolecular simulation program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef] [PubMed]
- Dassault Systèmes BIOVIA. Discovery Studio Visualizer, Version 2020; Dassault Systèmes: San Diego, CA, USA, 2020. [Google Scholar]
- Qiu, X.; Janson, C.A.; Smith, W.W.; Green, S.M.; McDevitt, P.; Johanson, K.; Carter, P.; Hibbs, M.; Lewis, C.; Chalker, A.; et al. Crystal structure of Staphylococcus aureus tyrosyl-tRNA synthetase in complex with a class of potent and specific inhibitors. Protein Sci. 2009, 10, 2008–2016. [Google Scholar] [CrossRef] [PubMed]
- Lafitte, D.; Lamour, V.; Tsvetkov, P.O.; Makarov, A.A.; Klich, M.; Deprez, P.; Moras, D.; Briand, C.; Gilli, R. DNA gyrase interaction with coumarin-based inhibitors: The role of the hydroxybenzoate isopentenyl moiety and the 5′-methyl group of the noviose. Biochemistry 2002, 41, 7217–7223. [Google Scholar] [CrossRef]
- Oefner, C.; Parisi, S.; Schulz, H.; Lociuro, S.; Dale, G.E. Inhibitory properties and X-ray crystallographic study of the binding of AR-101, AR-102 and iclaprim in ternary complexes with NADPH and dihydrofolate reductase from Staphylococcus aureus. Acta Crystallogr. D Biol. Crystallogr. 2009, 65 Pt 8, 751–757. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–422. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AUTODOCK4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Goodsell, D.S.; Morris, G.M.; Olson, A.J. Automated docking of flexible ligands: Applications of AutoDock. J. Mol. Recognit. 1996, 9, 1–5. [Google Scholar] [CrossRef]
- Tsai, C.W.; Chen, J.L.; Yang, C.S. An improved LGA for protein-ligand docking prediction. In Proceedings of the 2012 IEEE Congress on Evolutionary Computation, Brisbane, QLD, Astralia, 10–15 June 2012. [Google Scholar]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Tiwari, N.; Verma, J.; Waseem, M.; Subbarao, N.; Munde, M. Estimation of a stronger heparin binding locus in fibronectin domain III14 using thermodynamics and molecular dynamics. RSC Adv. 2020, 10, 20288–20301. [Google Scholar] [CrossRef] [PubMed]
- Kufareva, I.; Abagyan, R. Methods of protein structure comparison. Methods Mol. Biol. 2011, 857, 231–257. [Google Scholar]
- Kuzmanic, A.; Zagrovic, B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010, 98, 861–871. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Mirzaei, R.; Goodarzi, P.; Asadi, M.; Soltani, A.; Aljanabi, H.A.; Jeda, A.S.; Dashtbin, S.; Jalalifar, S.; Mohammadzadeh, R.; Teimoori, A.; et al. Bacterial co-infections with SARS-CoV-2. IUBMB Life 2020, 72, 2097–2111. [Google Scholar] [CrossRef]
- Maurer, H.R. Bromelain: Biochemistry, pharmacology and medical use. Cell. Mol. Life Sci. 2001, 58, 1234–1245. [Google Scholar] [CrossRef]
- Taussig, S.J.; Batkin, S. Bromelain, the enzyme complex of pineapple (Ananas comosus) and its clinical application. An update. J. Ethnopharmacol. 1998, 22, 191–203. [Google Scholar] [CrossRef]
- Praveen, N.C.; Rajesh, A.; Madan, M.; Chaurasia, V.R.; Hiremath, N.V.; Sharma, A.M. In vitro Evaluation of Antibacterial Efficacy of Pineapple Extract (Bromelain) on Periodontal Pathogens. J. Int. Oral Health 2014, 6, 96–98. [Google Scholar]
- Srikanth, S.; Chen, Z. Plant Protease Inhibitors in Therapeutics-Focus on Cancer Therapy. Front. Pharmacol. 2016, 7, 470. [Google Scholar] [CrossRef] [PubMed]
- Ávalos-Flores, E.; López-Castillo, L.M.; Wielsch, N.; Hupfer, Y.; Winkler, R.; Magaña-Ortiz, D. Protein extract of Bromelia Karatas L. Rich in cysteine proteases (ananain- and bromelain-like) has antibacterial activity against foodborne pathogens listeria monocytogenes and salmonella typhimurium. Folia Microbiol. 2021, 67, 1–13. [Google Scholar] [CrossRef]
- Narkhede, R.R.; Cheke, R.S.; Ambhore, J.P.; Shinde, S.D. The Molecular Docking Study of Potential Drug Candidates Showing Anti-COVID-19 Activity by Exploring of Therapeutic Targets of SARS-CoV-2. Eurasian J. Med. Oncol. 2020, 4, 185–195. [Google Scholar]
- Aman, F.; Masood, S. How Nutrition can help to fight against COVID-19. Pandemic. Pak. J. Med. Sci. 2020, 36, S121–S123. [Google Scholar] [CrossRef]
- Jamal, Q.M.S.; Ahmad, V.; Alharbi, A.H.; Ansari, M.A.; Alzohairy, M.A.; Almatroudi, A.; Alghamdi, S.; Alomary, M.N.; AlYahya, S.; Shesha, N.T.; et al. Therapeutic development by repurposing drugs targeting SARS-CoV-2 spike protein interactions by simulation studies. Saudi J. Biol. Sci. 2021, 28, 4560–4568. [Google Scholar] [CrossRef]
- Jamal, Q.M.S.; Alharbi, A.H.; Ahmad, V. Identification of doxorubicin as a potential therapeutic against SARS-CoV-2 (COVID-19) protease: A molecular docking and dynamics simulation studies. J. Biomol. Struct. Dyn. 2021, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ataide, J.A.; de Carvalho, N.M.; Rebelo, M.A.; Chaud, M.V.; Grotto, D.; Gerenutti, M.; Rai, M.; Mazzola, P.G.; Jozala, A.F. Bacterial Nanocellulose Loaded with Bromelain: Assessment of Antimicrobial, Antioxidant and Physical-Chemical Properties. Sci. Rep. 2017, 7, 18031. [Google Scholar] [CrossRef]
- Secor, E.R., Jr.; Carson, W.F.; Cloutier, M.M.; Guernsey, L.A.; Schramm, C.M.; Wu, C.A.; Thrall, R.S. Bromelain exerts anti-inflammatory effects in an ovalbumin-induced murine model of allergic airway disease. Cell. Immunol. 2005, 237, 68–75. [Google Scholar] [CrossRef]
- Rathnavelu, V.; Alitheen, N.B.; Sohila, S.; Kanagesan, S.; Ramesh, R. Potential role of bromelain in clinical and therapeutic applications. Biomed. Rep. 2016, 5, 283–288. [Google Scholar] [CrossRef]

| S.No | Compound Name | Molecular Formula | Molecular Weight | Structure | SMILES ID | PubChem ID |
|---|---|---|---|---|---|---|
| 1. | Bromelain | C39H66N2O29 | 1026.9 g/mol | 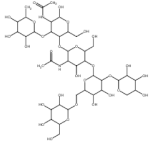 | CC1C(C(C(C(O1)OC2C(C(OC(C2OC3C(C(C(C(O3)CO)OC4C(C(C(C(O4)COC5C(C(C(C(O5)CO)O)O)O)O)O)OC6C(C(C(CO6)O)O)O)O)NC(=O)C)CO)O)NC(=O)C)O)O)O | CID: 44263865 |
| 2. | Artemisinin | C15H22O5 | 282.33 g/mol | 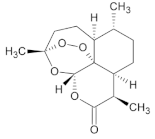 | CC1CCC2C(C(=O)OC3C24C1CCC(O3)(OO4)C)C | CID:68827 |
| PDB IDs | Binding Affinity (Kcal/mol) | Hydrogen Bond Details | Hydrogen Bond Length Angstrom | Hydrophobic Residues/Van der Waals | Other Interaction |
|---|---|---|---|---|---|
| 1JIJ (Crystal structure of S. aureus TyrRS in complex with SB-239629) | −7.8 | A:GLY83:HN–:UNK1:O42 | 2.90 | TYR170,ARG88,THR42,HIS47,ASN199,TRP197,GLY198, ASP153,GLN186,GLY193,PRP53, ASP80,ALA39,ASP40 | UNFAVORABLE DONOR = SER154 |
| A:LYS84:HZ1–:UNK1:O66 | 2.66 | ||||
| :UNK1:H6–A:SER194:O | 2.03 | ||||
| :UNK1:H91–A:SER82:O | 3.70 | ||||
| A:SER85:CB–:UNK1:O53 | 3.47 | ||||
| :UNK1:C10–A:ASP195:OD1 | 3.42 | ||||
| :UNK1:C61–A:GLY49:O | 3.42 | ||||
| 1KZN (Crystal Structure of E. coli 24 kDa Domain in Complex with Clorobiocin) | −6.0 | A:GLN135:–:UNK1:O7 | 2.39 | VAL133,MET166,THR165,GLY164, THR160,ILE140,CYS56,GLY75,ARG76,LYS162,ASP74,GLU58,ILE60, ARG206 | UNFAVORABLE = LYS57 |
| :UNK1:H6–A:THR163:OG1 | 2.89 | ||||
| :UNK1:H8–A:HIS55:O | 2.30 | ||||
| :UNK1:H35–A:GLN72:OE1 | 2.73 | ||||
| :UNK1:C2–A:GLN72:OE1 | 3.49 | ||||
| 3FYV (Staph. aureus DHFR complexed with NADPH and AR-102) | −5.8 | X:THR46:HG1–:UNK1:O62 | 2.00 | TRP22,VAL6,ALA7,VAL31,LEU28, LEU5,ILE50,GLN19,LEU62,GLU17, ARG44,LEU97,LYS45,GLY15, PHE98,LEU20, | Pi-sigma = PHE92 |
| X:GLN95:HN–:UNK1:O56 | 2.04 | ||||
| X:THR96:HG1–:UNK1:O32 | 2.54 | ||||
| X:THR121:HG1–:UNK1:O23 | 3.01 | ||||
| :UNK1:H91–X:ASN18:OD1 | 2.76 | ||||
| :UNK1:H22–X:ILE14:O | 2.52 | ||||
| :UNK1:H29–X:THR46:O | 2.87 | ||||
| X:SER49:CB–:UNK1:O12 | 2.75 | ||||
| X:GLY93:CA–:UNK1:O59 | 3.08 | ||||
| X:GLY94:CA–:UNK1:O59 | 3.11 | ||||
| :UNK1:C16–X:ILE14:O | 3.65 | ||||
| :UNK1:C35–X:THR96:OG1 | 2.57 | ||||
| :UNK1:C61–X:ASN18:O | 3.56 | ||||
| :UNK1:C61–X:SER49:OG | 2.98 |
| S. No | Compound Name | Final Intermolecular Energy (kcal/mol) | vdW + Hbond + Desolv Energy (kcal/mol) | Electrostatic Energy (kcal/mol) | Inhibition Constant | Hydrogen Bond Details | Hydrogen Bonds Length (Angstrom) | Residues Involved in Hydrophobic Interaction |
|---|---|---|---|---|---|---|---|---|
| 1. | Bromelain | −9.37 | −8.85 | −0.51 | 15.46 uM | A:ASN142:HD22–:UNK1:O7 | 3.01 | Thr25, His41, Ser46, Met49, Phe140, Leu141, Asn142, Gly143, Ser144, Cys145, His163, His164,Met165, Glu166, Leu167, Pro168, Gln189 |
| A:GLY143:HN–:UNK1:O63 | 1.84 | |||||||
| A:CYS145:HN–:UNK1:O62 | 2.87 | |||||||
| A:GLN189:HE22–:UNK1:O68 | 3.03 | |||||||
| :UNK1:H–A:GLU166:OE1 | 2.32 | |||||||
| :UNK1:H–A:GLU166:O | 1.91 | |||||||
| :UNK1:H–A:GLN189:OE1 | 1.95 | |||||||
| :UNK1:H–A:GLN189:OE1 | 2.35 | |||||||
| :UNK1:H–A:GLN189:OE1 | 1.82 | |||||||
| :UNK1:H–A:LEU141:O | 1.68 | |||||||
| 2. | Artemisinin | −6.94 | −6.80 | −0.14 | 8.19 uM | A:HIS163:HE2–:UNK1:O17 | 1.93 | His41,Phe140,Leu141, Asn142,Gly143,Ser144,Cys145,His163,His164,Met165,Glu166,His172,Gln189 |
| A:GLU166:HN–:UNK1:O9 | 1.79 | |||||||
| A:MET165:CA–:UNK1:O18 | 3.32 |
| Property | Model Name | Predicted Value | Unit |
|---|---|---|---|
| Absorption | Water solubility | −2.87 | Numeric (log mol/L) |
| Absorption | Caco2 permeability | −1.262 | Numeric (log Papp in 10−6 cm/s) |
| Absorption | Intestinal absorption (human) | 0 | Numeric (% Absorbed) |
| Absorption | Skin Permeability | −2.735 | Numeric (log Kp) |
| Absorption | P-glycoprotein substrate | Yes | Categorical (Yes/No) |
| Absorption | P-glycoprotein I inhibitor | No | Categorical (Yes/No) |
| Absorption | P-glycoprotein II inhibitor | No | Categorical (Yes/No) |
| Distribution | VDss (human) | −0.327 | Numeric (log L/kg) |
| Distribution | Fraction unbound (human) | 0.392 | Numeric (Fu) |
| Distribution | BBB permeability | −2.689 | Numeric (log BB) |
| Distribution | CNS permeability | −5.75 | Numeric (log PS) |
| Metabolism | CYP2D6 substrate | No | Categorical (Yes/No) |
| Metabolism | CYP3A4 substrate | No | Categorical (Yes/No) |
| Metabolism | CYP1A2 inhibitior | No | Categorical (Yes/No) |
| Metabolism | CYP2C19 inhibitior | No | Categorical (Yes/No) |
| Metabolism | CYP2C9 inhibitior | No | Categorical (Yes/No) |
| Metabolism | CYP2D6 inhibitior | No | Categorical (Yes/No) |
| Metabolism | CYP3A4 inhibitior | No | Categorical (Yes/No) |
| Excretion | Total Clearance | 1.686 | Numeric (log ml/min/kg) |
| Excretion | Renal OCT2 substrate | No | Categorical (Yes/No) |
| Toxicity | AMES toxicity | No | Categorical (Yes/No) |
| Toxicity | Max. tolerated dose (human) | 0.377 | Numeric (log mg/kg/day) |
| Toxicity | hERG I inhibitor | No | Categorical (Yes/No) |
| Toxicity | hERG II inhibitor | Yes | Categorical (Yes/No) |
| Toxicity | Oral Rat Acute Toxicity (LD50) | 2.467 | Numeric (mol/kg) |
| Toxicity | Oral Rat Chronic Toxicity (LOAEL) | 2.368 | Numeric (log mg/kg_bw/day) |
| Toxicity | Hepatotoxicity | No | Categorical (Yes/No) |
| Toxicity | Skin Sensitisation | No | Categorical (Yes/No) |
| Toxicity | T.Pyriformis toxicity | 0.285 | Numeric (log ug/L) |
| Toxicity | Minnow toxicity | 28.03 | Numeric (log mM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abuzinadah, M.F.; Ahmad, V.; Al-Thawdi, S.; Zakai, S.A.; Jamal, Q.M.S. Exploring the Binding Interaction of Active Compound of Pineapple against Foodborne Bacteria and Novel Coronavirus (SARS-CoV-2) Based on Molecular Docking and Simulation Studies. Nutrients 2022, 14, 3045. https://doi.org/10.3390/nu14153045
Abuzinadah MF, Ahmad V, Al-Thawdi S, Zakai SA, Jamal QMS. Exploring the Binding Interaction of Active Compound of Pineapple against Foodborne Bacteria and Novel Coronavirus (SARS-CoV-2) Based on Molecular Docking and Simulation Studies. Nutrients. 2022; 14(15):3045. https://doi.org/10.3390/nu14153045
Chicago/Turabian StyleAbuzinadah, Mohammed F., Varish Ahmad, Salwa Al-Thawdi, Shadi Ahmed Zakai, and Qazi Mohammad Sajid Jamal. 2022. "Exploring the Binding Interaction of Active Compound of Pineapple against Foodborne Bacteria and Novel Coronavirus (SARS-CoV-2) Based on Molecular Docking and Simulation Studies" Nutrients 14, no. 15: 3045. https://doi.org/10.3390/nu14153045
APA StyleAbuzinadah, M. F., Ahmad, V., Al-Thawdi, S., Zakai, S. A., & Jamal, Q. M. S. (2022). Exploring the Binding Interaction of Active Compound of Pineapple against Foodborne Bacteria and Novel Coronavirus (SARS-CoV-2) Based on Molecular Docking and Simulation Studies. Nutrients, 14(15), 3045. https://doi.org/10.3390/nu14153045







