Microbiota and Metabolite Profiling as Markers of Mood Disorders: A Cross-Sectional Study in Obese Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Anthropometric Characteristics
2.3. Dietary Anamnesis
2.4. Gut Microbiota Composition
2.5. Non-Targeted Metabolomics
2.6. Psychological Measures
2.7. Statistical Analyses
3. Results
3.1. Mood Status Characterization and Related Psychological and Behavioural Profiles
3.2. The Low Mood Score Group Did Not Display Specific Clinical Features
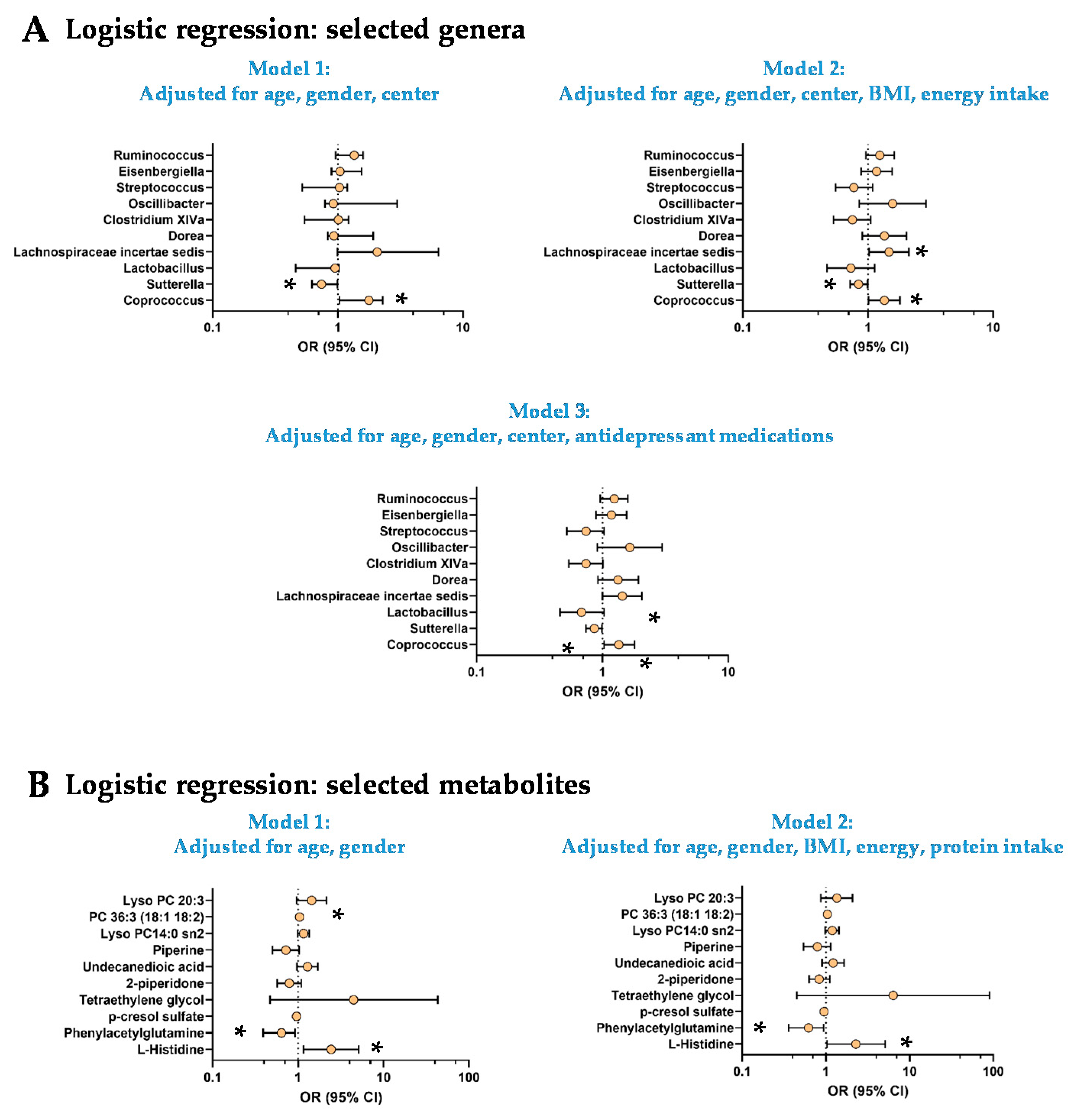
3.3. The Low Mood Score Group Are Characterized by Specific Gut Microbiome Composition
3.4. The Low Mood Score Group Exhibited Selective Profile of Plasma Metabolites
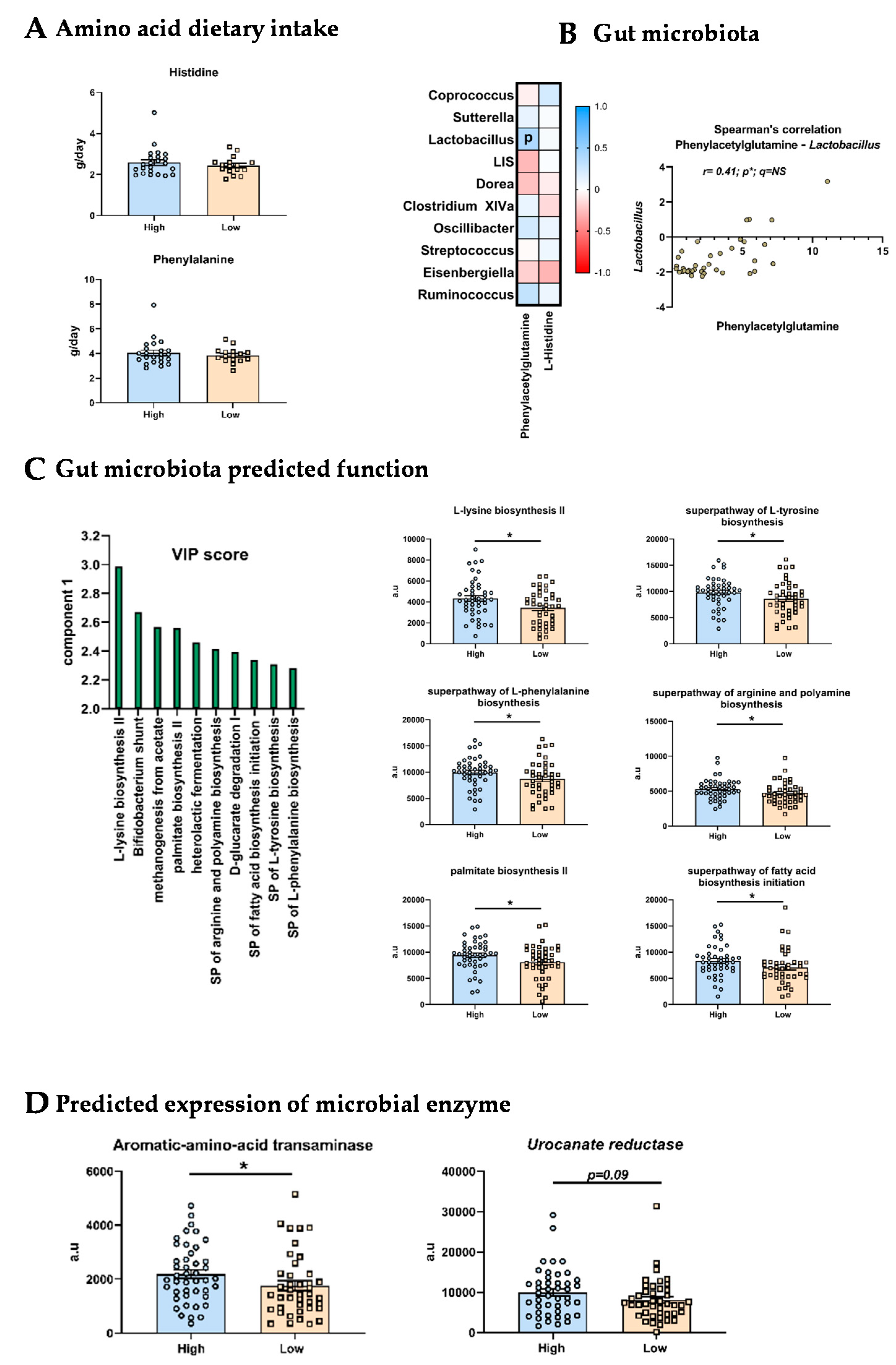
3.5. Origins of the Differences in L-Histidine and Phenylacetylglutamine Levels
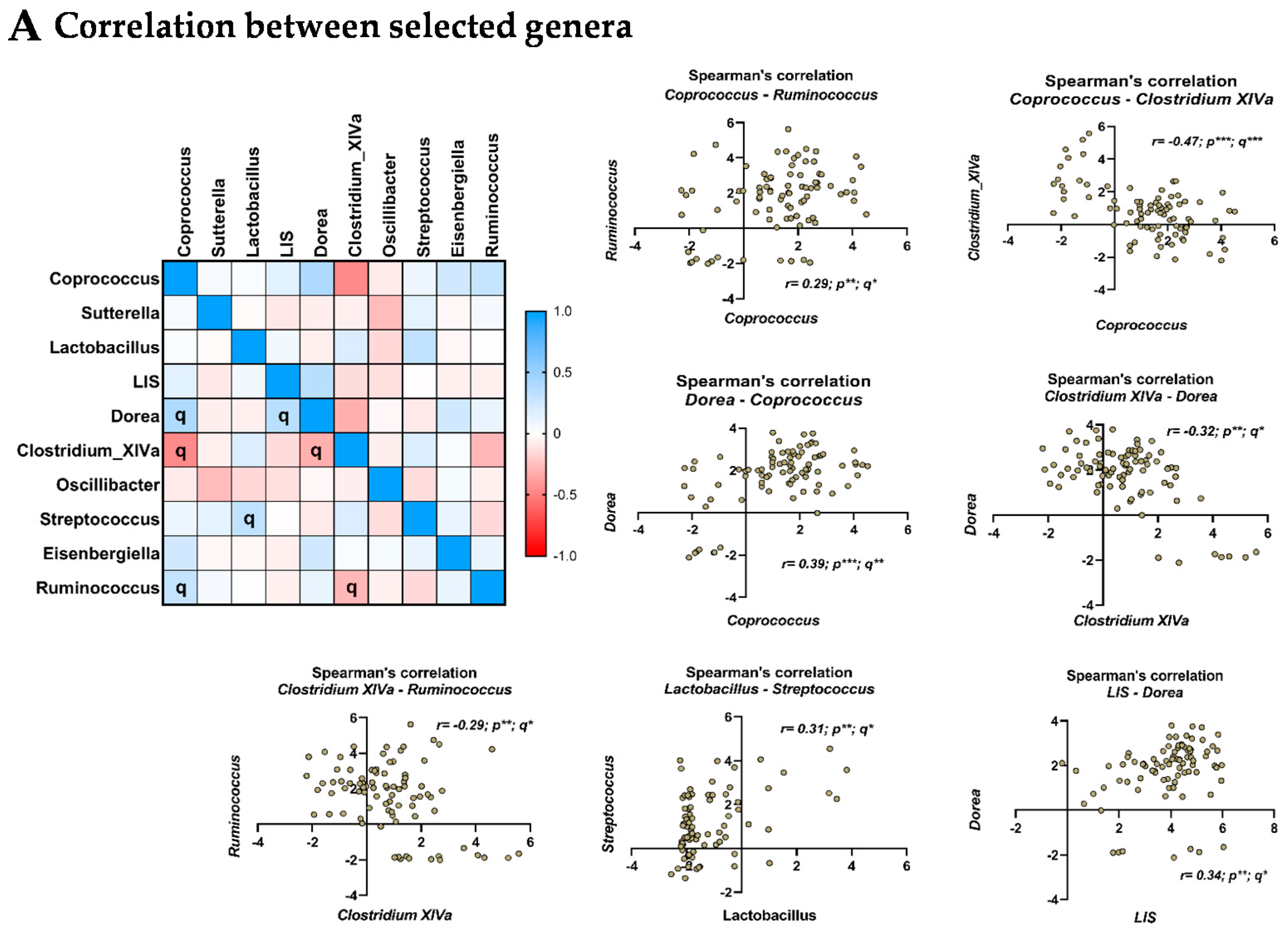
3.6. Relationship between the Selected Microbial Genera
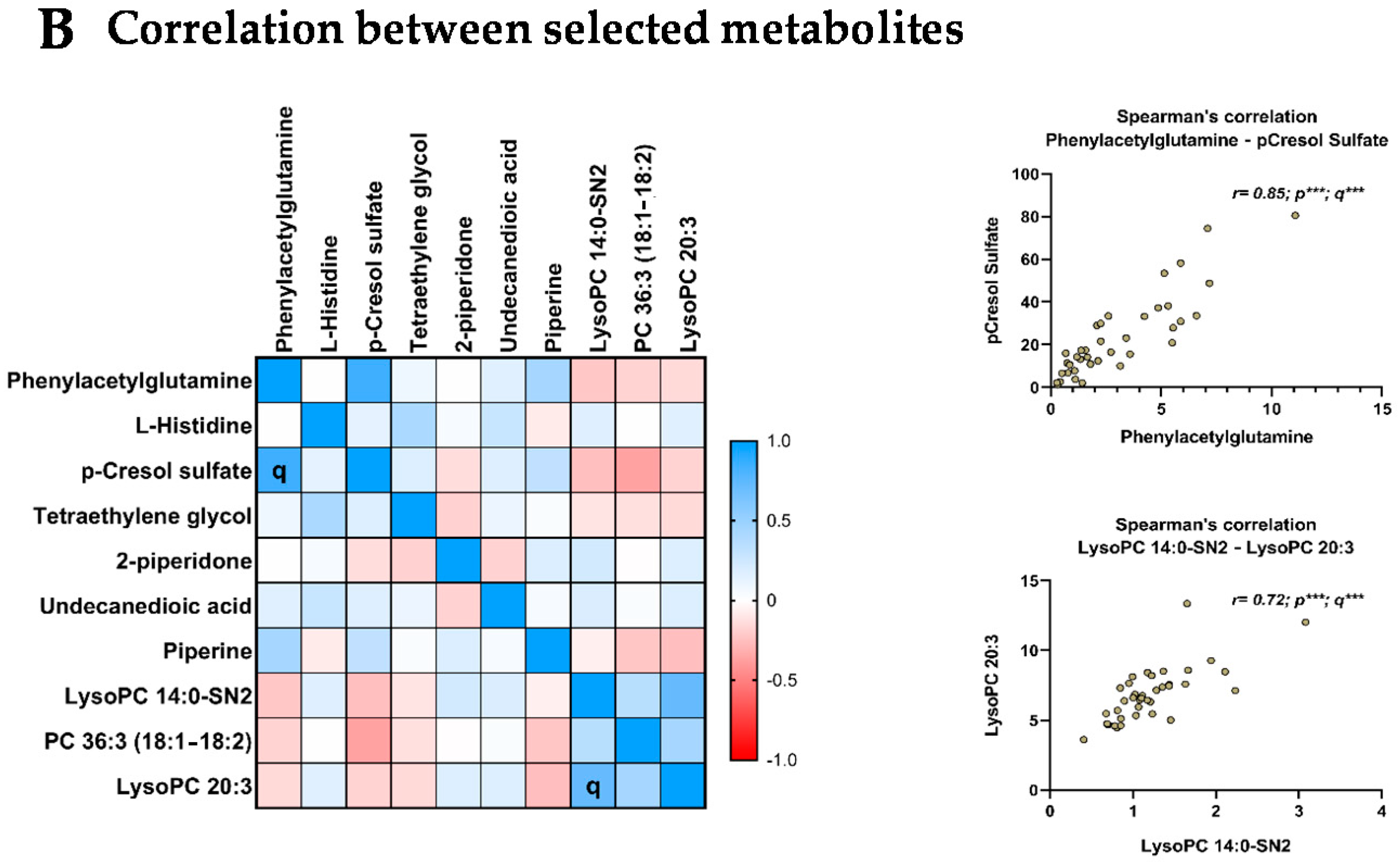
3.7. Relationship between the Selected Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Obesity and Overweight. 2016. Available online: http://www.who.int/mediacentre/factsheets/fs311/en/ (accessed on 16 December 2019).
- Guh, D.P.; Zhang, W.; Bansback, N.; Amarsi, Z.; Birmingham, C.L.; Anis, A.H. The incidence of co-morbidities related to obesity and overweight: A systematic review and meta-analysis. BMC Public Health 2009, 9, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uranga, R.M.; Keller, J.N. The Complex Interactions between Obesity, Metabolism and the Brain. Front. Neurosci. 2019, 13, 513. [Google Scholar] [CrossRef] [Green Version]
- Capuron, L.; Lasselin, J.; Castanon, N. Role of Adiposity-Driven Inflammation in Depressive Morbidity. Neuropsychopharmacology 2016, 42, 115–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Torres-Fuentes, C.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. The microbiota–gut–brain axis in obesity. Lancet Gastroenterol. Hepatol. 2017, 2, 747–756. [Google Scholar] [CrossRef]
- Leclercq, S.; Le Roy, T.; Furgiuele, S.; Coste, V.; Bindels, L.B.; Leyrolle, Q.; Neyrinck, A.M.; Quoilin, C.; Amadieu, C.; Petit, G.; et al. Gut Microbiota-Induced Changes in β-Hydroxybutyrate Metabolism Are Linked to Altered Sociability and Depression in Alcohol Use Disorder. Cell Rep. 2020, 33, 108238. [Google Scholar] [CrossRef]
- Kelly, J.; Borre, Y.; Brien, C.O.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef]
- Crovesy, L.; Masterson, D.; Rosado, E.L. Profile of the gut microbiota of adults with obesity: A systematic review. Eur. J. Clin. Nutr. 2020, 74, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2020, 70, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Genet. 2020, 19, 77–94. [Google Scholar] [CrossRef]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-Cardoso, V.F.; Corlianò, M.; Singaraja, R.R. Bile Acids: A Communication Channel in the Gut-Brain Axis. NeuroMolecular Med. 2020, 23, 99–117. [Google Scholar] [CrossRef]
- Arnoriaga-Rodríguez, M.; Mayneris-Perxachs, J.; Burokas, A.; Contreras-Rodríguez, O.; Blasco, G.; Coll, C.; Biarnés, C.; Miranda-Olivos, R.; Latorre, J.; Moreno-Navarrete, J.-M.; et al. Obesity Impairs Short-Term and Working Memory through Gut Microbial Metabolism of Aromatic Amino Acids. Cell Metab. 2020, 32, 548–560.e7. [Google Scholar] [CrossRef] [PubMed]
- Arnoriaga-Rodríguez, M.; Mayneris-Perxachs, J.; Contreras-Rodríguez, O.; Burokas, A.; Ortega-Sanchez, J.-A.; Blasco, G.; Coll, C.; Biarnés, C.; Castells-Nobau, A.; Puig, J.; et al. Obesity-associated deficits in inhibitory control are phenocopied to mice through gut microbiota changes in one-carbon and aromatic amino acids metabolic pathways. Gut 2021. [Google Scholar] [CrossRef] [PubMed]
- Bar, N.; The IMI DIRECT Consortium; Korem, T.; Weissbrod, O.; Zeevi, D.; Rothschild, D.; Leviatan, S.; Kosower, N.; Lotan-Pompan, M.; Weinberger, A.; et al. A reference map of potential determinants for the human serum metabolome. Nature 2020, 588, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Hiel, S.; Gianfrancesco, M.A.; Rodriguez, J.; Portheault, D.; Leyrolle, Q.; Bindels, L.B.; Cauduro, C.G.D.S.; Mulders, M.D.; Zamariola, G.; Azzi, A.-S.; et al. Link between gut microbiota and health outcomes in inulin -treated obese patients: Lessons from the Food4Gut multicenter randomized placebo-controlled trial. Clin. Nutr. 2020, 39, 3618–3628. [Google Scholar] [CrossRef]
- Leyrolle, Q.; Cserjesi, R.; Mulders, M.D.; Zamariola, G.; Hiel, S.; Gianfrancesco, M.A.; Portheault, D.; Amadieu, C.; Bindels, L.B.; Leclercq, S.; et al. Prebiotic effect on mood in obese patients is determined by the initial gut microbiota composition: A randomized, controlled trial. Brain Behav. Immun. 2021, 94, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Vineis, J.H.; Morrison, H.; Sogin, M.L. A Filtering Method to Generate High Quality Short Reads Using Illumina Paired-End Technology. PLoS ONE 2013, 8, e66643. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klåvus, A.; Kokla, M.; Noerman, S.; Koistinen, V.M.; Tuomainen, M.; Zarei, I.; Meuronen, T.; Häkkinen, M.R.; Rummukainen, S.; Babu, A.F.; et al. “Notame”: Workflow for Non-Targeted LC–MS Metabolic Profiling. Metabolites 2020, 10, 135. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Brasseur, S.; Grégoire, J.; Bourdu, R.; Mikolajczak, M. The Profile of Emotional Competence (PEC): Development and Validation of a Self-Reported Measure that Fits Dimensions of Emotional Competence Theory. PLoS ONE 2013, 8, e62635. [Google Scholar] [CrossRef] [Green Version]
- Watson, D.; Clark, L.A.; Tellegen, A. Development and validation of brief measures of positive and negative affect: The PANAS scales. J. Pers. Soc. Psychol. 1988, 54, 1063–1070. [Google Scholar] [CrossRef]
- Diener, E.; Wirtz, D.; Tov, W.; Kim-Prieto, C.; Choi, D.-W.; Oishi, S.; Biswas-Diener, R. New Well-being Measures: Short Scales to Assess Flourishing and Positive and Negative Feelings. Soc. Indic. Res. 2009, 97, 143–156. [Google Scholar] [CrossRef]
- Poesen, R.; Claes, K.; Evenepoel, P.; de Loor, H.; Augustijns, P.; Kuypers, D.R.; Meijers, B. Microbiota-Derived Phenylacetylglutamine Associates with Overall Mortality and Cardiovascular Disease in Patients with CKD. J. Am. Soc. Nephrol. 2016, 27, 3479–3487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmanski, T.; Diener, C.; Rappaport, N.; Patwardhan, S.; Wiedrick, J.; Lapidus, J.; Earls, J.C.; Zimmer, A.; Glusman, G.; Robinson, M.; et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat. Metab. 2021, 3, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Collino, S.; Montoliu, I.; Martin, F.-P.; Scherer, M.; Mari, D.; Salvioli, S.; Bucci, L.; Ostan, R.; Monti, D.; Biagi, E.; et al. Metabolic Signatures of Extreme Longevity in Northern Italian Centenarians Reveal a Complex Remodeling of Lipids, Amino Acids, and Gut Microbiota Metabolism. PLoS ONE 2013, 8, e56564. [Google Scholar] [CrossRef]
- Ottosson, F.; Brunkwall, L.; Smith, E.; Orho-Melander, M.; Nilsson, P.M.; Fernandez, C.; Melander, O. The gut microbiota-related metabolite phenylacetylglutamine associates with increased risk of incident coronary artery disease. J. Hypertens. 2020, 38, 2427–2434. [Google Scholar] [CrossRef] [PubMed]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Gui, X.; Wu, L.; Tian, S.; Wang, H.; Xie, L.; Wu, W. Amino acid metabolism, lipid metabolism, and oxidative stress are associated with post-stroke depression: A metabonomics study. BMC Neurol. 2020, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Miyata, N.; Hata, T.; Takakura, S.; Yoshihara, K.; Morita, C.; Mikami, K.; Nomoto, K.; Miyazaki, K.; Tsuji, H.; Sudo, N. Metabolomics profile of Japanese female patients with restricting-type anorexia nervosa. Physiol. Behav. 2020, 228, 113204. [Google Scholar] [CrossRef]
- HoleČek, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; Molinaro, A.; Ståhlman, M.; Khan, M.T.; Schmidt, C.; Mannerås-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC. Cell 2018, 175, 947–961.e17. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, T.; Nakamura, T.; Shibakusa, T.; Sugita, M.; Naganuma, F.; Iida, T.; Miura, Y.; Mohsen, A.; Harada, R.; Yanai, K. Insufficient Intake of L-Histidine Reduces Brain Histamine and Causes Anxiety-Like Behaviors in Male Mice. J. Nutr. 2014, 144, 1637–1641. [Google Scholar] [CrossRef] [PubMed]
- Moro, J.; Tomé, D.; Schmidely, P.; Demersay, T.-C.; Azzout-Marniche, D. Histidine: A Systematic Review on Metabolism and Physiological Effects in Human and Different Animal Species. Nutrients 2020, 12, 1414. [Google Scholar] [CrossRef]
- Sasahara, I.; Fujimura, N.; Nozawa, Y.; Furuhata, Y.; Sato, H. The effect of histidine on mental fatigue and cognitive performance in subjects with high fatigue and sleep disruption scores. Physiol. Behav. 2015, 147, 238–244. [Google Scholar] [CrossRef] [Green Version]
- Geliebter, A.A.; Hashim, S.A.; Van Itallie, T.B. Oral L-histidine fails to reduce taste and smell acuity but induces anorexia and urinary zinc excretion. Am. J. Clin. Nutr. 1981, 34, 119–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valles-Colomer, M.; Falony, G.; Darzi, Y.; Tigchelaar, E.F.; Wang, J.; Tito, R.Y.; Schiweck, C.; Kurilshikov, A.; Joossens, M.; Wijmenga, C.; et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 2019, 4, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids from Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Barandouzi, Z.A.; Starkweather, A.R.; Henderson, W.; Gyamfi, A.; Cong, X.S. Altered Composition of Gut Microbiota in Depression: A Systematic Review. Front. Psychiatry 2020, 11, 541. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Liu, H.; Zhang, H.; Bao, Y.; Di, J.; Hu, C. The genus Sutterella is a potential contributor to glucose metabolism improvement after Roux-en-Y gastric bypass surgery in T2D. Diabetes Res. Clin. Pract. 2020, 162, 108116. [Google Scholar] [CrossRef]
- Kaakoush, N.O. Sutterella Species, IgA-degrading Bacteria in Ulcerative Colitis. Trends Microbiol. 2020, 28, 519–522. [Google Scholar] [CrossRef]
- Morgan, X.C.; Kabakchiev, B.; Waldron, L.; Tyler, A.D.; Tickle, T.; Milgrom, R.; Stempak, J.M.; Gevers, D.; Xavier, R.J.; Silverberg, M.S.; et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol. 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butera, A.; Di Paola, M.; Vitali, F.; De Nitto, D.; Covotta, F.; Borrini, F.; Pica, R.; De Filippo, C.; Cavalieri, D.; Giuliani, A.; et al. IL-13 mRNA Tissue Content Identifies Two Subsets of Adult Ulcerative Colitis Patients With Different Clinical and Mucosa-Associated Microbiota Profiles. J. Crohn’s Coliti 2019, 14, 369–380. [Google Scholar] [CrossRef]
- Amaretti, A.; Gozzoli, C.; Simone, M.; Raimondi, S.; Righini, L.; Pérez-Brocal, V.; Lopez, R.G.; Moya, A.; Rossi, M. Profiling of Protein Degraders in Cultures of Human Gut Microbiota. Front. Microbiol. 2019, 10, 2614. [Google Scholar] [CrossRef]
- Takada, M.; Nishida, K.; Kataoka-Kato, A.; Gondo, Y.; Ishikawa, H.; Suda, K.; Kawai, M.; Hoshi, R.; Watanabe, O.; Igarashi, T.; et al. Probiotic Lactobacillus casei strain Shirota relieves stress-associated symptoms by modulating the gut–brain interaction in human and animal models. Neurogastroenterol. Motil. 2016, 28, 1027–1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, C.; Wu, R.Y.; Wong, A.; Stanisz, A.M.; Yan, R.; Min, K.K.; Pasyk, M.; Neufeld, K.-A.M.; Karamat, M.I.; Foster, J.A.; et al. Lactobacillus rhamnosusstrain JB-1 reverses restraint stress-induced gut dysmotility. Neurogastroenterol. Motil. 2016, 29, e12903. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fuskevåg, O.M.; Broster, A.; Davies, N.; Cudmore, T.; Hodges, T.; Griffin, M. Lactobacillus Casei Shirota Modulation of Ammonia Metabolism in Physical Exercise. Int. J. Probiotics Prebiotics 2012, 7, 13–16. [Google Scholar]
- Zheng, P.; Zeng, B.; Zhou, C.; Liu, M.; Fang, Z.; Xu, X.; Zeng, L.; Chen, J.-J.; Fan, S.-H.; Du, X.; et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol. Psychiatry 2016, 21, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhao, N.; Zhang, C.; Lam, Y.Y.; Zhao, L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 2021, 13, 1–12. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H.; et al. Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef]
- Neis, E.P.J.G.; DeJong, C.H.C.; Rensen, S.S. The Role of Microbial Amino Acid Metabolism in Host Metabolism. Nutrients 2015, 7, 2930–2946. [Google Scholar] [CrossRef] [Green Version]
- Crawford, J.R.; Henry, J.D. The Positive and Negative Affect Schedule (PANAS): Construct validity, measurement properties and normative data in a large non-clinical sample. Br. J. Clin. Psychol. 2004, 43, 245–265. [Google Scholar] [CrossRef]
| High | Low | p | Model 1 | Model 2 | |||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Mean ± SD | OR | p | OR | p | ||
| PANAS PA | 36.4 ± 4.90 | 27.2 ± 6.36 | <0.0001 | 0.75 | <0.0001 | 0.75 | <0.0001 |
| PANAS NA | 11.9 ± 3.26 | 19.5 ± 8.06 | <0.0001 | 1.36 | <0.0001 | 1.42 | <0.0001 |
| PANAS PA-NA | 24.5 ± 5.17 | 7.77 ± 8.36 | <0.0001 | 1.92 × 10−13 | <0.0001 | 1.06 × 10−6 | <0.0001 |
| PEC TOT | 3.42 ± 0.44 | 3.18 ± 0.47 | 0.012 | 0.26 | 0.006 | 0.26 | 0.016 |
| PEC INTRA | 3.37 ± 0.50 | 3.11 ± 0.55 | 0.027 | 0.33 | 0.016 | 0.36 | 0.034 |
| PEC INTER | 3.40 ± 0.49 | 3.20 ± 0.51 | 0.041 | 0.36 | 0.032 | 0.34 | 0.029 |
| PEC Reg Self | 3.26 ± 0.80 | 2.85 ± 0.86 | 0.013 | 0.54 | 0.025 | 0.58 | 0.048 |
| SPANE.PE | 20.5 ± 2.62 | 17.2 ± 8.77 | <0.0001 | 0.86 | 0.024 | 0.89 | 0.078 |
| SPANE.NE | 10.9 ± 3.07 | 13.7 ± 4.03 | 0.0003 | 1.25 | 0.001 | 1.24 | 0.003 |
| SPANE.BE | 9.65 ± 5.09 | 3.53 ± 10.6 | <0.0001 | 0.89 | 0.003 | 0.90 | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leyrolle, Q.; Cserjesi, R.; Demeure, R.; Neyrinck, A.M.; Amadieu, C.; Rodriguez, J.; Kärkkäinen, O.; Hanhineva, K.; Paquot, N.; Cnop, M.; et al. Microbiota and Metabolite Profiling as Markers of Mood Disorders: A Cross-Sectional Study in Obese Patients. Nutrients 2022, 14, 147. https://doi.org/10.3390/nu14010147
Leyrolle Q, Cserjesi R, Demeure R, Neyrinck AM, Amadieu C, Rodriguez J, Kärkkäinen O, Hanhineva K, Paquot N, Cnop M, et al. Microbiota and Metabolite Profiling as Markers of Mood Disorders: A Cross-Sectional Study in Obese Patients. Nutrients. 2022; 14(1):147. https://doi.org/10.3390/nu14010147
Chicago/Turabian StyleLeyrolle, Quentin, Renata Cserjesi, Romane Demeure, Audrey M. Neyrinck, Camille Amadieu, Julie Rodriguez, Olli Kärkkäinen, Kati Hanhineva, Nicolas Paquot, Miriam Cnop, and et al. 2022. "Microbiota and Metabolite Profiling as Markers of Mood Disorders: A Cross-Sectional Study in Obese Patients" Nutrients 14, no. 1: 147. https://doi.org/10.3390/nu14010147
APA StyleLeyrolle, Q., Cserjesi, R., Demeure, R., Neyrinck, A. M., Amadieu, C., Rodriguez, J., Kärkkäinen, O., Hanhineva, K., Paquot, N., Cnop, M., Cani, P. D., Thissen, J.-P., Bindels, L. B., Klein, O., Luminet, O., & Delzenne, N. M. (2022). Microbiota and Metabolite Profiling as Markers of Mood Disorders: A Cross-Sectional Study in Obese Patients. Nutrients, 14(1), 147. https://doi.org/10.3390/nu14010147








