Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Inclusion Criteria
- Cross-sectional studies or dietary interventions with MD, PD, or WD, or case-control studies of intestinal diseases such as colorectal cancer (CRC), colon adenoma (CA), colon polyposis (CP), ulcerative colitis (UC), and Crohn’s disease (CD). May include or not healthy subjects (HC), healthy familiars of IBD subjects (FC), or subjects at risk of developing CRC (RS).
- Available 16s rRNA gene sequences derived from human feces with associated quality scores and metadata.
- Samples obtained when subjects or patients were not under drug treatments.
- Data from at least 10 patients, subjects following a defined diet or healthy subjects with a minimum of 6000 reads each.
2.2. Study Groups
2.3. Data Analysis and Statistics
3. Results
3.1. Alpha and Beta Diversity
3.2. Microbiota Composition Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kopp, W. How western diet and lifestyle drive the pandemic of obesity and civilization diseases. Diabetes Metab. Syndr. Obes. Targets Ther. 2019, 12, 2221. [Google Scholar] [CrossRef] [Green Version]
- Ehlers, S.; Kaufmann, S.H. Infection, inflammation, and chronic diseases: Consequences of a modern lifestyle. Trends Immunol. 2010, 31, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z. Gut Microbiota: An Important Link between Western Diet and Chronic Diseases. Nutrients 2019, 11, 2287. [Google Scholar]
- Thorburn, A.N.; Macia, L.; Mackay, C.R. Diet, metabolites, and “western-lifestyle” inflammatory diseases. Immunity 2014, 40, 833–842. [Google Scholar] [CrossRef] [Green Version]
- Karunanithi, S.; Levi, L. High-fat diet and colorectal cancer: Myths and facts. Future Med. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Molendijk, I.; van der Marel, S.; Maljaars, P. Towards a Food Pharmacy: Immunologic Modulation through Diet. Nutrients 2019, 11, 1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucini, D.; Vigo, C.; Malacarne, M.; Gatzemeier, W.; Pagani, M. Lifestyle changes as internal medicine. Eur. J. Intern. Med. 2017, 43, 40–42. [Google Scholar] [CrossRef]
- Stephenson, J.; Heslehurst, N.; Hall, J.; Schoenaker, D.A.; Hutchinson, J.; Cade, J.E.; Poston, L.; Barrett, G.; Crozier, S.R.; Barker, M. Before the beginning: Nutrition and lifestyle in the preconception period and its importance for future health. Lancet 2018, 391, 1830–1841. [Google Scholar] [CrossRef]
- Willett, W.C.; Sacks, F.; Trichopoulou, A.; Drescher, G.; Ferro-Luzzi, A.; Helsing, E.; Trichopoulos, D. Mediterranean diet pyramid: A cultural model for healthy eating. Am. J. Clin. Nutr. 1995, 61, 1402S–1406S. [Google Scholar] [CrossRef] [PubMed]
- Sanches Machado d’Almeida, K.; Ronchi Spillere, S.; Zuchinali, P.; Corrêa Souza, G. Mediterranean diet and other dietary patterns in primary prevention of heart failure and changes in cardiac function markers: A systematic review. Nutrients 2018, 10, 58. [Google Scholar] [CrossRef] [Green Version]
- Di Daniele, N.; Noce, A.; Vidiri, M.F.; Moriconi, E.; Marrone, G.; Annicchiarico-Petruzzelli, M.; D’Urso, G.; Tesauro, M.; Rovella, V.; De Lorenzo, A. Impact of Mediterranean diet on metabolic syndrome, cancer and longevity. Oncotarget 2017, 8, 8947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddavide, R.; Rotolo, O.; Caruso, M.G.; Stasi, E.; Notarnicola, M.; Miraglia, C.; Nouvenne, A.; Meschi, T.; De’Angelis, G.L.; Di Mario, F. The role of diet in the prevention and treatment of Inflammatory Bowel Diseases. Acta Bio-Med. Atenei Parm. 2018, 89, 60. [Google Scholar]
- Farinetti, A.; Zurlo, V.; Manenti, A.; Coppi, F.; Mattioli, A.V. Mediterranean diet and colorectal cancer: A systematic review. Nutrition 2017, 43, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Zazpe, I.; Martínez, J.A.; Santiago, S.; Carlos, S.; Zulet, M.Á.; Ruiz-Canela, M. Scoping review of Paleolithic dietary patterns: A definition proposal. Nutr. Res. Rev. 2020, 34, 1–29. [Google Scholar]
- de Menezes, E.V.A.; de Carvalho Sampaio, H.A.; Carioca, A.A.F.; Parente, N.A.; Brito, F.O.; Moreira, T.M.M.; de Souza, A.C.C.; Arruda, S.P.M. Influence of Paleolithic diet on anthropometric markers in chronic diseases: Systematic review and meta-analysis. Nutr. J. 2019, 18, 41. [Google Scholar] [CrossRef]
- Otten, J.; Stomby, A.; Waling, M.; Isaksson, A.; Tellström, A.; Lundin-Olsson, L.; Brage, S.; Ryberg, M.; Svensson, M.; Olsson, T. Benefits of a Paleolithic diet with and without supervised exercise on fat mass, insulin sensitivity, and glycemic control: A randomized controlled trial in individuals with type 2 diabetes. Diabetes/Metab. Res. Rev. 2017, 33, e2828. [Google Scholar] [CrossRef]
- Ghaedi, E.; Mohammadi, M.; Mohammadi, H.; Ramezani-Jolfaie, N.; Malekzadeh, J.; Hosseinzadeh, M.; Salehi-Abargouei, A. Effects of a Paleolithic diet on cardiovascular disease risk factors: A systematic review and meta-analysis of randomized controlled trials. Adv. Nutr. 2019, 10, 634–646. [Google Scholar] [CrossRef]
- Otten, J.; Mellberg, C.; Ryberg, M.; Sandberg, S.; Kullberg, J.; Lindahl, B.; Larsson, C.; Hauksson, J.; Olsson, T. Strong and persistent effect on liver fat with a Paleolithic diet during a two-year intervention. Int. J. Obes. 2016, 40, 747–753. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Waliullah, S.; Godfrey, V.; Khan, M.A.W.; Ramachandran, R.A.; Cantarel, B.L.; Behrendt, C.; Peng, L.; Hooper, L.V.; Zaki, H. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 2020, 12, 567. [Google Scholar] [CrossRef]
- Chassaing, B.; Van de Wiele, T.; De Bodt, J.; Marzorati, M.; Gewirtz, A.T. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut 2017, 66, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Korem, T.; Zeevi, D.; Zilberman-Schapira, G.; Thaiss, C.A.; Maza, O.; Israeli, D.; Zmora, N.; Gilad, S.; Weinberger, A. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014, 514, 181–186. [Google Scholar] [CrossRef]
- Wan, Y.; Tong, W.; Zhou, R.; Li, J.; Yuan, J.; Wang, F.; Li, D. Habitual animal fat consumption in shaping gut microbiota and microbial metabolites. Food Funct. 2019, 10, 7973–7982. [Google Scholar] [CrossRef]
- Genoni, A.; Christophersen, C.T.; Lo, J.; Coghlan, M.; Boyce, M.C.; Bird, A.R.; Lyons-Wall, P.; Devine, A. Long-term Paleolithic diet is associated with lower resistant starch intake, different gut microbiota composition and increased serum TMAO concentrations. Eur. J. Nutr. 2020, 59, 1845–1858. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Lin, X.; Zhao, F.; Shi, X.; Li, H.; Li, Y.; Zhu, W.; Xu, X.; Li, C.; Zhou, G. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci. Rep. 2015, 5, 15220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koecher, K.J.; McKeown, N.M.; Sawicki, C.M.; Menon, R.S.; Slavin, J.L. Effect of whole-grain consumption on changes in fecal microbiota: A review of human intervention trials. Nutr. Rev. 2019, 77, 487–497. [Google Scholar] [CrossRef]
- Fernando, W.; Hill, J.; Zello, G.; Tyler, R.; Dahl, W.; Van Kessel, A. Diets supplemented with chickpea or its main oligosaccharide component raffinose modify faecal microbial composition in healthy adults. Benef. Microbes 2010, 1, 197–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Romano, K.A.; Vivas, E.I.; Amador-Noguez, D.; Rey, F.E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio 2015, 6, e02481-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnenburg, J.L.; Bäckhed, F. Diet–microbiota interactions as moderators of human metabolism. Nature 2016, 535, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Yap, Y.A.; Mariño, E. An insight into the intestinal web of mucosal immunity, microbiota, and diet in inflammation. Front. Immunol. 2018, 9, 2617. [Google Scholar] [CrossRef]
- Telle-Hansen, V.H.; Holven, K.B.; Ulven, S.M. Impact of a healthy dietary pattern on gut microbiota and systemic inflammation in humans. Nutrients 2018, 10, 1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belizário, J.E.; Faintuch, J.; Garay-Malpartida, M. Gut microbiome dysbiosis and immunometabolism: New frontiers for treatment of metabolic diseases. Mediat. Inflamm. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Grochowska, M.; Wojnar, M.; Radkowski, M. The gut microbiota in neuropsychiatric disorders. Acta Neurobiol. Exp. 2018, 78, 69–81. [Google Scholar] [CrossRef] [Green Version]
- Zeng, M.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral bacteria and intestinal dysbiosis in colorectal cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef] [Green Version]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [Green Version]
- PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov (accessed on 15 August 2020).
- Sequence Read Archive. Available online: https://www.ncbi.nlm.nih.gov/sra (accessed on 15 August 2020).
- Vitaglione, P.; Mennella, I.; Ferracane, R.; Rivellese, A.A.; Giacco, R.; Ercolini, D.; Gibbons, S.M.; La Storia, A.; Gilbert, J.A.; Jonnalagadda, S. Whole-grain wheat consumption reduces inflammation in a randomized controlled trial on overweight and obese subjects with unhealthy dietary and lifestyle behaviors: Role of polyphenols bound to cereal dietary fiber. Am. J. Clin. Nutr. 2015, 101, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Barone, M.; Turroni, S.; Rampelli, S.; Soverini, M.; D’Amico, F.; Biagi, E.; Brigidi, P.; Troiani, E.; Candela, M. Gut microbiome response to a modern Paleolithic diet in a Western lifestyle context. PLoS ONE 2019, 14, e0220619. [Google Scholar] [CrossRef] [Green Version]
- Sofi, F.; Dinu, M.; Pagliai, G.; Marcucci, R.; Casini, A. Validation of a literature-based adherence score to Mediterranean diet: The MEDI-LITE score. Int. J. Food Sci. Nutr. 2017, 68, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Agnoli, C.; Krogh, V.; Grioni, S.; Sieri, S.; Palli, D.; Masala, G.; Sacerdote, C.; Vineis, P.; Tumino, R.; Frasca, G. A priori–defined dietary patterns are associated with reduced risk of stroke in a large Italian cohort. J. Nutr. 2011, 141, 1552–1558. [Google Scholar] [CrossRef] [Green Version]
- Hess, A.L.; Benítez-Páez, A.; Blædel, T.; Larsen, L.H.; Iglesias, J.R.; Madera, C.; Sanz, Y.; Larsen, T.M.; Consortium, M. The effect of inulin and resistant maltodextrin on weight loss during energy restriction: A randomised, placebo-controlled, double-blinded intervention. Eur. J. Nutr. 2020, 59, 2507–2524. [Google Scholar] [CrossRef]
- Tap, J.; Furet, J.P.; Bensaada, M.; Philippe, C.; Roth, H.; Rabot, S.; Lakhdari, O.; Lombard, V.; Henrissat, B.; Corthier, G. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ. Microbiol. 2015, 17, 4954–4964. [Google Scholar] [CrossRef]
- Hiel, S.; Bindels, L.B.; Pachikian, B.D.; Kalala, G.; Broers, V.; Zamariola, G.; Chang, B.P.; Kambashi, B.; Rodriguez, J.; Cani, P.D. Effects of a diet based on inulin-rich vegetables on gut health and nutritional behavior in healthy humans. Am. J. Clin. Nutr. 2019, 109, 1683–1695. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Baxter, N.T.; Koumpouras, C.C.; Rogers, M.A.; Ruffin, M.T.; Schloss, P.D. DNA from fecal immunochemical test can replace stool for detection of colonic lesions using a microbiota-based model. Microbiome 2016, 4, 59. [Google Scholar] [CrossRef] [Green Version]
- Jacob, V.; Crawford, C.; Cohen-Mekelburg, S.; Viladomiu, M.; Putzel, G.G.; Schneider, Y.; Chabouni, F.; O’Neil, S.; Bosworth, B.; Woo, V. Single delivery of high-diversity fecal microbiota preparation by colonoscopy is safe and effective in increasing microbial diversity in active ulcerative colitis. Inflamm. Bowel Dis. 2017, 23, 903–911. [Google Scholar] [CrossRef] [Green Version]
- Forbes, J.D.; Chen, C.-Y.; Knox, N.C.; Marrie, R.-A.; El-Gabalawy, H.; de Kievit, T.; Alfa, M.; Bernstein, C.N.; Van Domselaar, G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases—Does a common dysbiosis exist? Microbiome 2018, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.P.; Goudarzi, M.; Singh, N.; Tong, M.; McHardy, I.H.; Ruegger, P.; Asadourian, M.; Moon, B.-H.; Ayson, A.; Borneman, J. A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 750–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pisanu, S.; Palmas, V.; Madau, V.; Casula, E.; Deledda, A.; Cusano, R.; Uva, P.; Vascellari, S.; Boi, F.; Loviselli, A. Impact of a Moderately Hypocaloric Mediterranean Diet on the Gut Microbiota Composition of Italian Obese Patients. Nutrients 2020, 12, 2707. [Google Scholar] [CrossRef]
- Pagliai, G.; Russo, E.; Niccolai, E.; Dinu, M.; Di Pilato, V.; Magrini, A.; Bartolucci, G.; Baldi, S.; Menicatti, M.; Giusti, B. Influence of a 3-month low-calorie Mediterranean diet compared to the vegetarian diet on human gut microbiota and SCFA: The CARDIVEG Study. Eur. J. Nutr. 2020, 59, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Dadkhah, E.; Sikaroodi, M.; Korman, L.; Hardi, R.; Baybick, J.; Hanzel, D.; Kuehn, G.; Kuehn, T.; Gillevet, P.M. Gut microbiome identifies risk for colorectal polyps. BMJ Open Gastroenterol. 2019, 6, e000297. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef]
- Zackular, J.P.; Rogers, M.A.; Ruffin, M.T.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Pearson, T.; Caporaso, J.G.; Yellowhair, M.; Bokulich, N.A.; Padi, M.; Roe, D.J.; Wertheim, B.C.; Linhart, M.; Martinez, J.A.; Bilagody, C. Effects of ursodeoxycholic acid on the gut microbiome and colorectal adenoma development. Cancer Med. 2019, 8, 617–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Elabd, E.M.Y.; Morsy, S.M.; Elmalt, H.A. Investigating of Moringa oleifera role on Gut microbiota composition and inflammation associated with obesity following high fat diet feeding. Open Access Maced. J. Med. Sci. 2018, 6, 1359. [Google Scholar] [CrossRef] [Green Version]
- Prieto, I.; Hidalgo, M.; Segarra, A.B.; Martínez-Rodríguez, A.M.; Cobo, A.; Ramírez, M.; Abriouel, H.; Gálvez, A.; Martínez-Cañamero, M. Influence of a diet enriched with virgin olive oil or butter on mouse gut microbiota and its correlation to physiological and biochemical parameters related to metabolic syndrome. PLoS ONE 2018, 13, e0190368. [Google Scholar] [CrossRef] [Green Version]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J. Gastroenterol. 2015, 50, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chávez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Dinakaran, V.; Mandape, S.N.; Shuba, K.; Pratap, S.; Sakhare, S.S.; Tabatabai, M.A.; Smoot, D.T.; Farmer-Dixon, C.M.; Kesavalu, L.N.; Adunyah, S.E. Identification of specific oral and gut pathogens in full thickness colon of colitis patients: Implications for colon motility. Front. Microbiol. 2019, 9, 3220. [Google Scholar] [CrossRef] [PubMed]
- De Cruz, P.; Kang, S.; Wagner, J.; Buckley, M.; Sim, W.H.; Prideaux, L.; Lockett, T.; McSweeney, C.; Morrison, M.; Kirkwood, C.D. Association between specific mucosa-associated microbiota in Crohn’s disease at the time of resection and subsequent disease recurrence: A pilot study. J. Gastroenterol. Hepatol. 2015, 30, 268–278. [Google Scholar] [CrossRef]
- Richard, M.L.; Liguori, G.; Lamas, B.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Pierluigi Di Simone, M.; Calabrese, C.; Poggioli, G.; Langella, P. Mucosa-associated microbiota dysbiosis in colitis associated cancer. Gut Microbes 2018, 9, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Group, E.S. An international association between Helicobacter pylori infection and gastric cancer. Lancet 1993, 341, 1359–1363. [Google Scholar] [CrossRef]
- Baj, J.; Korona-Głowniak, I.; Forma, A.; Maani, A.; Sitarz, E.; Rahnama-Hezavah, M.; Radzikowska, E.; Portincasa, P. Mechanisms of the Epithelial–Mesenchymal Transition and Tumor Microenvironment in Helicobacter pylori-Induced Gastric Cancer. Cells 2020, 9, 1055. [Google Scholar] [CrossRef] [Green Version]
- Maeda, M.; Moro, H.; Ushijima, T. Mechanisms for the induction of gastric cancer by Helicobacter pylori infection: Aberrant DNA methylation pathway. Gastric Cancer 2017, 20, 8–15. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Marusawa, H.; Kinoshita, K.; Endo, Y.; Kou, T.; Morisawa, T.; Azuma, T.; Okazaki, I.-M.; Honjo, T.; Chiba, T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007, 13, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Meucci, G.; Tatarella, M.; Vecchi, M.; Ranzi, M.; Biguzzi, E.; Beccari, G.; Clerici, E.; De Franchis, R. High prevalence of Helicobacter pylori infection in patients with colonic adenomas and carcinomas. J. Clin. Gastroenterol. 1997, 25, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Strofilas, A.; Lagoudianakis, E.E.; Seretis, C.; Pappas, A.; Koronakis, N.; Keramidaris, D.; Koukoutsis, I.; Chrysikos, I.; Manouras, I.; Manouras, A. Association of helicobacter pylori infection and colon cancer. J. Clin. Med. Res. 2012, 4, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maggio-Price, L.; Treuting, P.; Zeng, W.; Tsang, M.; Bielefeldt-Ohmann, H.; Iritani, B.M. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006, 66, 828–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum—Symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; DeVinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.; Chen, Y.; Chen, Y.; Su, W.; Zhan, N.; Dong, W. Fusobacterium nucleatum activates endoplasmic reticulum stress to promote crohn’s disease development via the upregulation of CARD3 expression. Front. Pharmacol. 2020, 11, 106. [Google Scholar] [CrossRef] [Green Version]
- Tahara, T.; Hirata, I.; Nakano, N.; Tahara, S.; Horiguchi, N.; Kawamura, T.; Okubo, M.; Ishizuka, T.; Yamada, H.; Yoshida, D. Potential link between Fusobacterium enrichment and DNA methylation accumulation in the inflammatory colonic mucosa in ulcerative colitis. Oncotarget 2017, 8, 61917. [Google Scholar] [CrossRef] [Green Version]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y. Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jangi, S.; Gandhi, R.; Cox, L.M.; Li, N.; Von Glehn, F.; Yan, R.; Patel, B.; Mazzola, M.A.; Liu, S.; Glanz, B.L. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgård, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef] [Green Version]
- Million, M.; Maraninchi, M.; Henry, M.; Armougom, F.; Richet, H.; Carrieri, P.; Valero, R.; Raccah, D.; Vialettes, B.; Raoult, D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2012, 36, 817–825. [Google Scholar] [CrossRef] [Green Version]
- Picchianti Diamanti, A.; Panebianco, C.; Salerno, G.; Di Rosa, R.; Salemi, S.; Sorgi, M.L.; Meneguzzi, G.; Mariani, M.B.; Rai, A.; Iacono, D. Impact of Mediterranean Diet on Disease Activity and Gut Microbiota Composition of Rheumatoid Arthritis Patients. Microorganisms 2020, 8, 1989. [Google Scholar] [CrossRef]
- Garcia-Mantrana, I.; Selma-Royo, M.; Alcantara, C.; Collado, M.C. Shifts on gut microbiota associated to mediterranean diet adherence and specific dietary intakes on general adult population. Front. Microbiol. 2018, 9, 890. [Google Scholar] [CrossRef]
- van de Pol, J.A.; van Best, N.; Mbakwa, C.A.; Thijs, C.; Savelkoul, P.H.; Arts, I.C.; Hornef, M.W.; Mommers, M.; Penders, J. Gut colonization by methanogenic archaea is associated with organic dairy consumption in children. Front. Microbiol. 2017, 8, 355. [Google Scholar] [CrossRef]
- Hustoft, T.; Hausken, T.; Ystad, S.; Valeur, J.; Brokstad, K.; Hatlebakk, J.; Lied, G. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol. Motil. 2017, 29, e12969. [Google Scholar] [CrossRef]
- Miyamoto, J.; Watanabe, K.; Taira, S.; Kasubuchi, M.; Li, X.; Irie, J.; Itoh, H.; Kimura, I. Barley β-glucan improves metabolic condition via short-chain fatty acids produced by gut microbial fermentation in high fat diet fed mice. PLoS ONE 2018, 13, e0196579. [Google Scholar] [CrossRef]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leahy, S.; Higgins, D.; Fitzgerald, G.; Van Sinderen, D. Getting better with bifidobacteria. J. Appl. Microbiol. 2005, 98, 1303–1315. [Google Scholar] [CrossRef]
- Mayo, B.; Vázquez, L.; Flórez, A.B. Equol: A bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients 2019, 11, 2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, C.; Frankenfeld, C.L.; Lampe, J.W. Gut bacterial metabolism of the soy isoflavone daidzein: Exploring the relevance to human health. Exp. Biol. Med. 2005, 230, 155–170. [Google Scholar] [CrossRef]
- Setchell, K.D.; Cole, S.J. Method of defining equol-producer status and its frequency among vegetarians. J. Nutr. 2006, 136, 2188–2193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caesar, R.; Tremaroli, V.; Kovatcheva-Datchary, P.; Cani, P.D.; Bäckhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015, 22, 658–668. [Google Scholar] [CrossRef] [Green Version]
- Sartor, R.B.; Mazmanian, S.K. Intestinal microbes in inflammatory bowel diseases. Am. J. Gastroenterol. Suppl. 2012, 1, 15. [Google Scholar] [CrossRef]
- Gomez-Arango, L.F.; Barrett, H.L.; Wilkinson, S.A.; Callaway, L.K.; McIntyre, H.D.; Morrison, M.; Dekker Nitert, M. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes 2018, 9, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Astbury, S.; Atallah, E.; Vijay, A.; Aithal, G.P.; Grove, J.I.; Valdes, A.M. Lower gut microbiome diversity and higher abundance of proinflammatory genus Collinsella are associated with biopsy-proven nonalcoholic steatohepatitis. Gut Microbes 2020, 11, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wright, K.; Davis, J.M.; Jeraldo, P.; Marietta, E.V.; Murray, J.; Nelson, H.; Matteson, E.L.; Taneja, V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nagpal, R.; Neth, B.J.; Wang, S.; Craft, S.; Yadav, H. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment. EBioMedicine 2019, 47, 529–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.-S.; Kitahara, M.; Sakamoto, M.; Hattori, M.; Benno, Y. Slackia equolifaciens sp. nov., a human intestinal bacterium capable of producing equol. Int. J. Syst. Evol. Microbiol. 2010, 60, 1721–1724. [Google Scholar] [CrossRef] [Green Version]
- Matthies, A.; Loh, G.; Blaut, M.; Braune, A. Daidzein and genistein are converted to equol and 5-hydroxy-equol by human intestinal Slackia isoflavoniconvertens in gnotobiotic rats. J. Nutr. 2012, 142, 40–46. [Google Scholar] [CrossRef]
- Kim, K.-S.; Rowlinson, M.-C.; Bennion, R.; Liu, C.; Talan, D.; Summanen, P.; Finegold, S.M. Characterization of Slackia exigua isolated from human wound infections, including abscesses of intestinal origin. J. Clin. Microbiol. 2010, 48, 1070–1075. [Google Scholar] [CrossRef] [Green Version]
- Albertsen, M.; Hugenholtz, P.; Skarshewski, A.; Nielsen, K.L.; Tyson, G.W.; Nielsen, P.H. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 2013, 31, 533–538. [Google Scholar] [CrossRef]
- Kuehbacher, T.; Rehman, A.; Lepage, P.; Hellmig, S.; Fölsch, U.R.; Schreiber, S.; Ott, S.J. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J. Med Microbiol. 2008, 57, 1569–1576. [Google Scholar] [CrossRef] [Green Version]
- He, X.; McLean, J.S.; Edlund, A.; Yooseph, S.; Hall, A.P.; Liu, S.-Y.; Dorrestein, P.C.; Esquenazi, E.; Hunter, R.C.; Cheng, G. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2015, 112, 244–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic Bacteria With Increased Prevalence in IBD Mucosa AugmentIn VitroUtilization of Mucin by Other Bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Song, K.; Zhang, Y.; Zhu, C.; Shen, Z.; Wu, S.; Luo, W.; Tan, B.; Yang, Z.; Wang, X. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem. Biophys. Res. Commun. 2018, 501, 791–799. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menni, C.; Hernandez, M.M.; Vital, M.; Mohney, R.P.; Spector, T.D.; Valdes, A.M. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes 2019, 10, 688–695. [Google Scholar] [CrossRef] [Green Version]
- Negatu, D.A.; Gengenbacher, M.; Dartois, V.A.; Dick, T. Indole propionic acid, an unusual antibiotic produced by the gut microbiota, with anti-inflammatory and antioxidant properties. Front. Microbiol. 2020, 11, 2654. [Google Scholar] [CrossRef]
- Bang, S.-J.; Kim, G.; Lim, M.Y.; Song, E.-J.; Jung, D.-H.; Kum, J.-S.; Nam, Y.-D.; Park, C.-S.; Seo, D.-H. The influence of in vitro pectin fermentation on the human fecal microbiome. Amb Express 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.; Ferrocino, I.; Calabrese, F.M.; De Filippis, F.; Cavallo, N.; Siragusa, S.; Rampelli, S.; Di Cagno, R.; Rantsiou, K.; Vannini, L. Diet influences the functions of the human intestinal microbiome. Sci. Rep. 2020, 10, 4247. [Google Scholar] [CrossRef] [Green Version]
- Schreiner, P.; Yilmaz, B.; Rossel, J.-B.; Franc, Y.; Misselwitz, B.; Scharl, M.; Zeitz, J.; Frei, P.; Greuter, T.; Vavricka, S.R. Vegetarian or gluten-free diets in patients with inflammatory bowel disease are associated with lower psychological well-being and a different gut microbiota, but no beneficial effects on the course of the disease. United Eur. Gastroenterol. J. 2019, 7, 767–781. [Google Scholar] [CrossRef] [Green Version]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Hong, J.; Xu, X.; Feng, Q.; Zhang, D.; Gu, Y.; Shi, J.; Zhao, S.; Liu, W.; Wang, X. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017, 23, 859. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Del Pugar, E.M.G.; López-Almela, I.; Moya-Pérez, Á.; Codoñer-Franch, P.; Sanz, Y. Depletion of Blautia species in the microbiota of obese children relates to intestinal inflammation and metabolic phenotype worsening. Msystems 2020, 5, e00857-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolenbrander, P. The genus Veillonella. Prokaryotes 2006, 4, 1022–1040. [Google Scholar]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Del Dot, T.; Osawa, R.; Stackebrandt, E. Phascolarctobacterium faecium gen. nov, spec. nov., a novel taxon of the Sporomusa group of bacteria. Syst. Appl. Microbiol. 1993, 16, 380–384. [Google Scholar] [CrossRef]
- Watanabe, Y.; Nagai, F.; Morotomi, M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl. Environ. Microbiol. 2012, 78, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Kes, M.M.; Van den Bossche, J.; Griffioen, A.W.; Huijbers, E.J. Oncometabolites lactate and succinate drive pro-angiogenic macrophage response in tumors. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188427. [Google Scholar] [CrossRef]
- Tannahill, G.; Curtis, A.; Adamik, J.; Palsson-McDermott, E.; McGettrick, A.; Goel, G.; Frezza, C.; Bernard, N.; Kelly, B.; Foley, N. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Vernia, P.; Caprilli, R.; Latella, G.; Barbetti, F.; Magliocca, F.M.; Cittadini, M. Fecal lactate and ulcerative colitis. Gastroenterology 1988, 95, 1564–1568. [Google Scholar] [CrossRef]
- Barrett, H.L.; Gomez-Arango, L.F.; Wilkinson, S.A.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Dekker Nitert, M. A vegetarian diet is a major determinant of gut microbiota composition in early pregnancy. Nutrients 2018, 10, 890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whisner, C.M.; Martin, B.R.; Nakatsu, C.H.; Story, J.A.; MacDonald-Clarke, C.J.; McCabe, L.D.; McCabe, G.P.; Weaver, C.M. Soluble corn fiber increases calcium absorption associated with shifts in the gut microbiome: A randomized dose-response trial in free-living pubertal females. J. Nutr. 2016, 146, 1298–1306. [Google Scholar] [CrossRef] [Green Version]
- Martínez, I.; Lattimer, J.M.; Hubach, K.L.; Case, J.A.; Yang, J.; Weber, C.G.; Louk, J.A.; Rose, D.J.; Kyureghian, G.; Peterson, D.A. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013, 7, 269–280. [Google Scholar] [CrossRef] [Green Version]
- Citronberg, J.S.; Curtis, K.R.; White, E.; Newcomb, P.A.; Newton, K.; Atkinson, C.; Song, X.; Lampe, J.W.; Hullar, M.A. Association of gut microbial communities with plasma lipopolysaccharide-binding protein (LBP) in premenopausal women. ISME J. 2018, 12, 1631–1641. [Google Scholar] [CrossRef] [Green Version]
- Bajer, L.; Kverka, M.; Kostovcik, M.; Macinga, P.; Dvorak, J.; Stehlikova, Z.; Brezina, J.; Wohl, P.; Spicak, J.; Drastich, P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J. Gastroenterol. 2017, 23, 4548. [Google Scholar] [CrossRef]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [Green Version]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Matera, G.; Muto, V.; Vinci, M.; Zicca, E.; Abdollahi-Roodsaz, S.; van de Veerdonk, F.L.; Kullberg, B.-J.; Liberto, M.C.; van der Meer, J.W.; Foca, A. Receptor recognition of and immune intracellular pathways for Veillonella parvula lipopolysaccharide. Clin. Vaccine Immunol. 2009, 16, 1804–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van den Bogert, B.; Meijerink, M.; Zoetendal, E.G.; Wells, J.M.; Kleerebezem, M. Immunomodulatory properties of Streptococcus and Veillonella isolates from the human small intestine microbiota. PLoS ONE 2014, 9, e114277. [Google Scholar] [CrossRef] [PubMed]
- Santoru, M.L.; Piras, C.; Murgia, A.; Palmas, V.; Camboni, T.; Liggi, S.; Ibba, I.; Lai, M.A.; Orrù, S.; Blois, S. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 2017, 7, 9523. [Google Scholar] [CrossRef] [PubMed]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Russell, J.B. Acidaminococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–3. [Google Scholar]
- Marchandin, H.; Juvonen, R.; Haikara, A. Megasphaera. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–16. [Google Scholar]
- Willems, A.; Collins, M.D. Mitsuokella. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–9. [Google Scholar]
- Dai, Z.-L.; Zhang, J.; Wu, G.; Zhu, W.-Y. Utilization of amino acids by bacteria from the pig small intestine. Amino Acids 2010, 39, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-W.; Chee, K.-M.; Cho, S.-J. Nutritional quality of rice bran protein in comparison to animal and vegetable protein. Food Chem. 2015, 172, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; De Vos, W.M. Microbes inside—From diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, Á.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019, 9, 15683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.S.; Wang, J.; Yannie, P.J.; Sandhu, Y.K.; Korzun, W.J.; Ghosh, S. Dietary supplementation with galactooligosaccharides attenuates high-fat, high-cholesterol diet–induced glucose intolerance and disruption of colonic mucin layer in C57BL/6 mice and reduces atherosclerosis in Ldlr–/–mice. J. Nutr. 2020, 150, 285–293. [Google Scholar] [CrossRef]
- Hashemi, Z.; Fouhse, J.; Im, H.S.; Chan, C.B.; Willing, B.P. Dietary pea fiber supplementation improves glycemia and induces changes in the composition of gut microbiota, serum short chain fatty acid profile and expression of mucins in glucose intolerant rats. Nutrients 2017, 9, 1236. [Google Scholar] [CrossRef] [Green Version]
- Sanapareddy, N.; Legge, R.M.; Jovov, B.; McCoy, A.; Burcal, L.; Araujo-Perez, F.; Randall, T.A.; Galanko, J.; Benson, A.; Sandler, R.S. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012, 6, 1858–1868. [Google Scholar] [CrossRef] [Green Version]
- Byrd, J.C.; Bresalier, R.S. Mucins and mucin binding proteins in colorectal cancer. Cancer Metastasis Rev. 2004, 23, 77–99. [Google Scholar] [CrossRef]
- Betge, J.; Schneider, N.I.; Harbaum, L.; Pollheimer, M.J.; Lindtner, R.A.; Kornprat, P.; Ebert, M.P.; Langner, C. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: Expression profiles and clinical significance. Virchows Arch. 2016, 469, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Wu, Y.; Li, J.; Bao, Y.; Guo, Y.; Yang, W. The dynamic changes of gut microbiota in Muc2 deficient mice. Int. J. Mol. Sci. 2018, 19, 2809. [Google Scholar] [CrossRef] [Green Version]
- Liso, M.; De Santis, S.; Verna, G.; Dicarlo, M.; Calasso, M.; Santino, A.; Gigante, I.; Eri, R.; Raveenthiraraj, S.; Sobolewski, A. A specific mutation in Muc2 determines early dysbiosis in colitis-prone Winnie mice. Inflamm. Bowel Dis. 2020, 26, 546–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepage, P.; Häsler, R.; Spehlmann, M.E.; Rehman, A.; Zvirbliene, A.; Begun, A.; Ott, S.; Kupcinskas, L.; Doré, J.; Raedler, A. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 2011, 141, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the immune relevance of Prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. 2017, 69, 964–975. [Google Scholar] [CrossRef] [Green Version]
- Marietta, E.V.; Murray, J.A.; Luckey, D.H.; Jeraldo, P.R.; Lamba, A.; Patel, R.; Luthra, H.S.; Mangalam, A.; Taneja, V. Suppression of inflammatory arthritis by human gut-derived Prevotella histicola in humanized mice. Arthritis Rheumatol. 2016, 68, 2878–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Microbiome CRC Biomarker Study. Available online: http://mothur.org/MicrobiomeBiomarkerCRC (accessed on 10 June 2020).
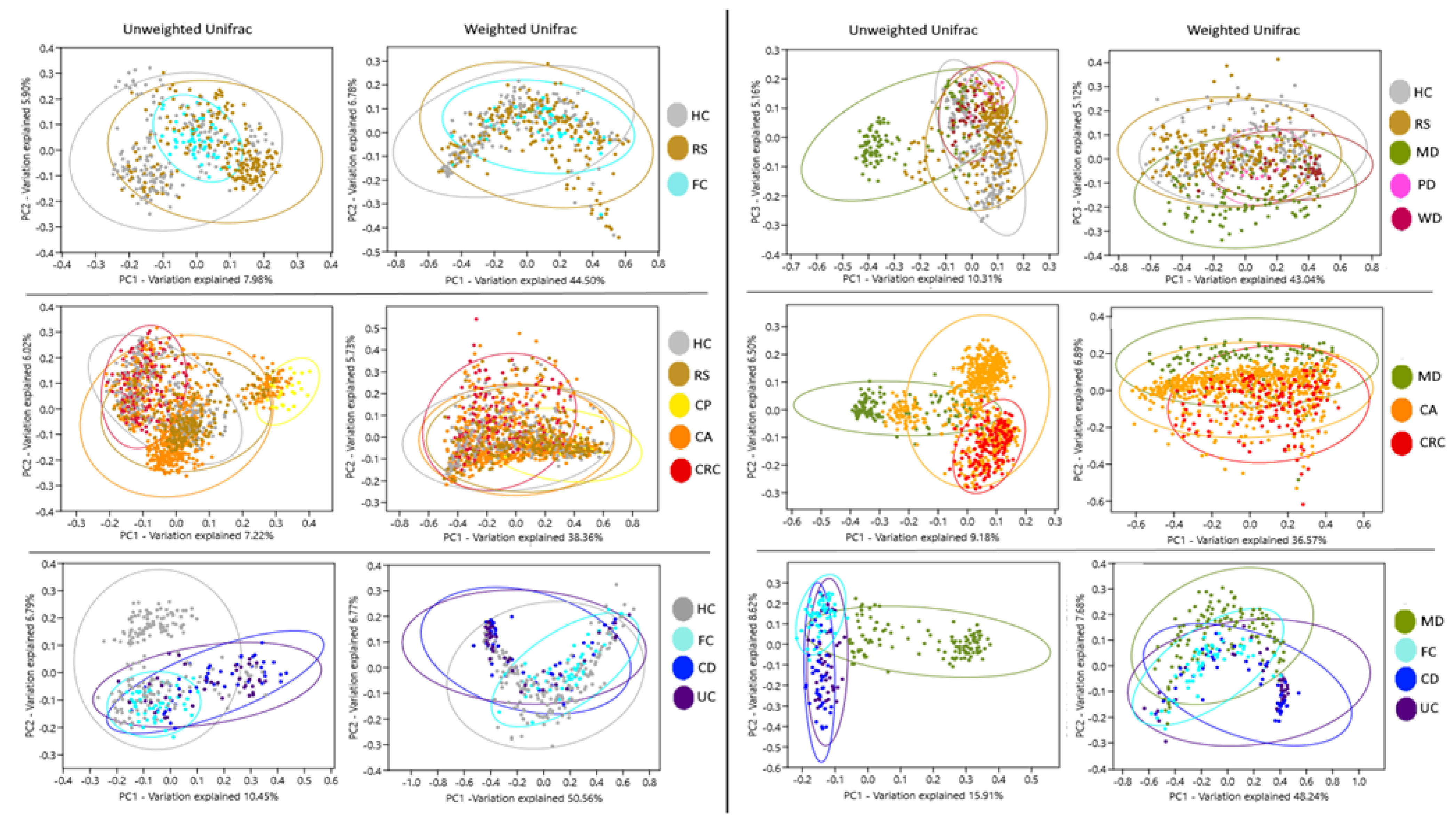


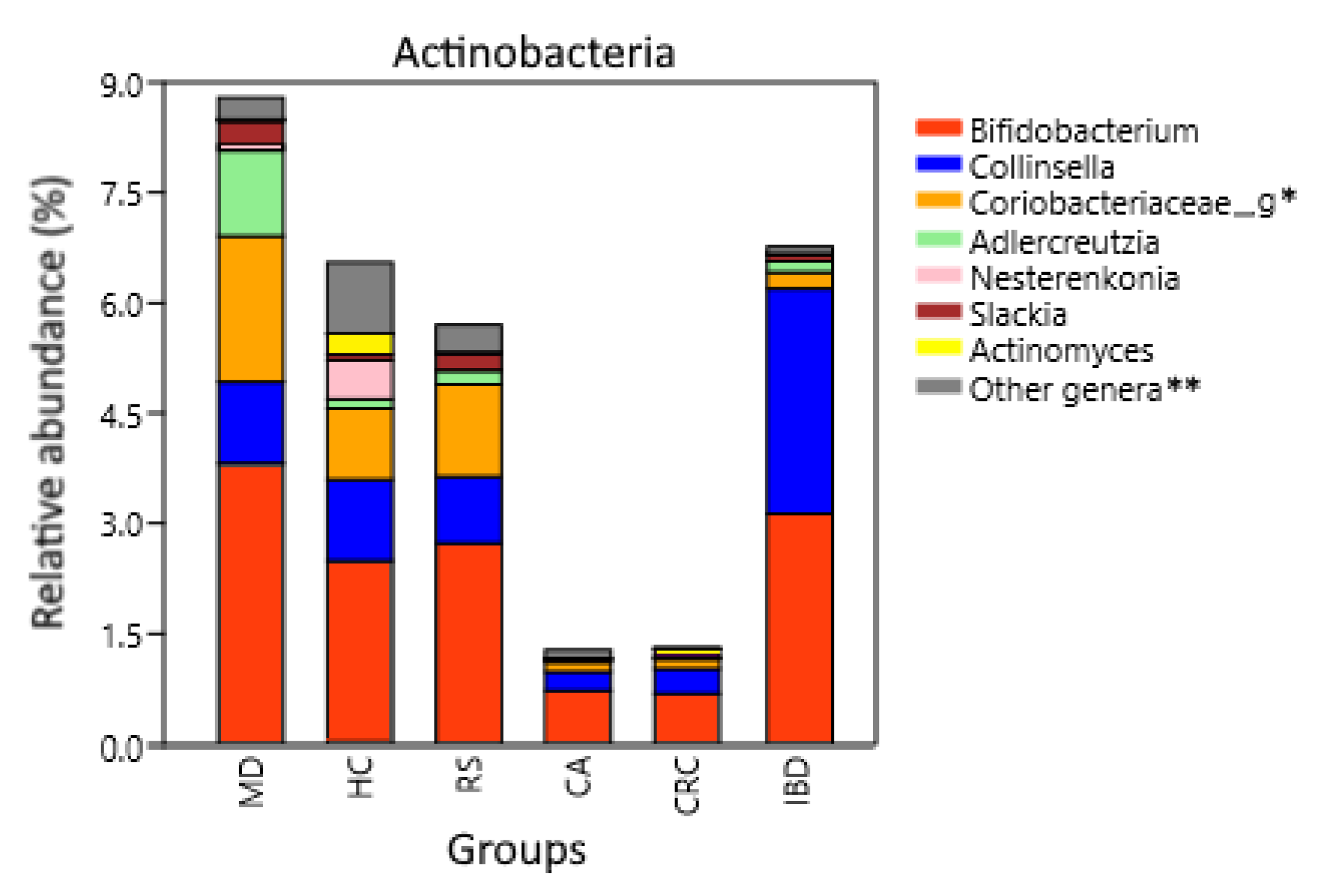
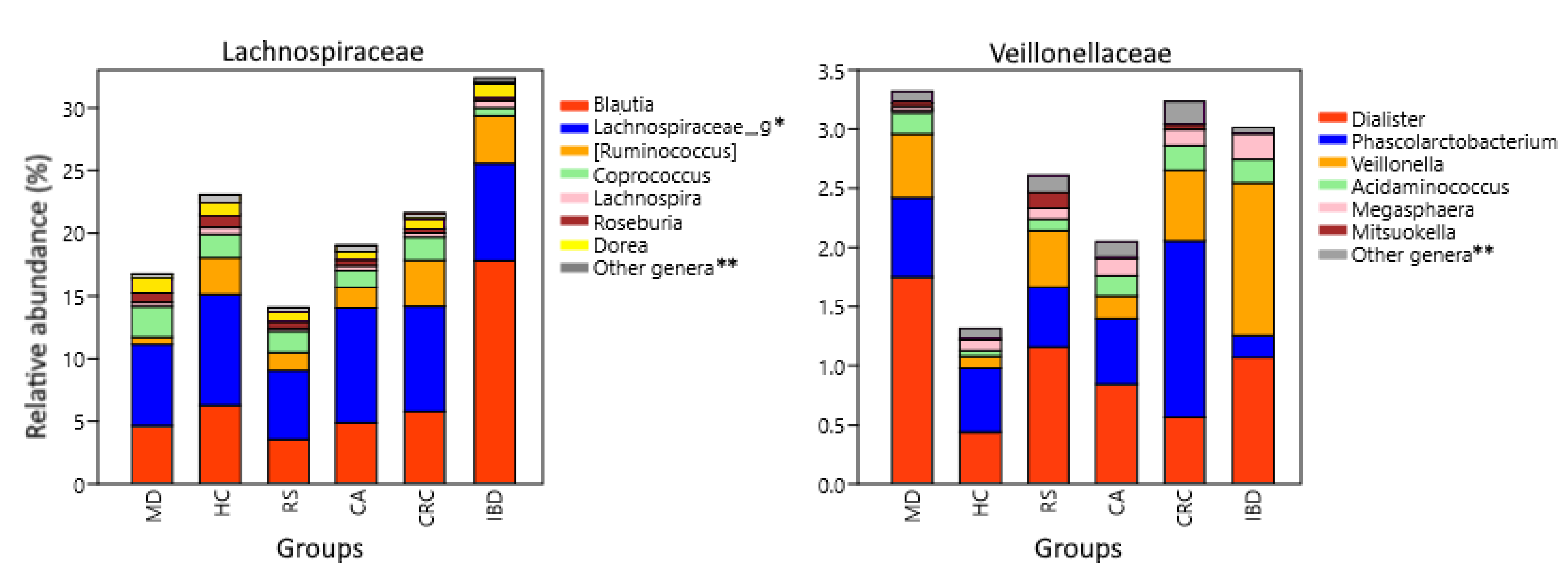
| Group 1 | Diet | Disease Status | Subjects | Description | Studies |
|---|---|---|---|---|---|
| HC | Undefined | No disease | 196 | Healthy subjects non-related to patients | [27,47,48,57,58,59] |
| FC | Undefined | No disease | 54 | Healthy familiars of IBD patients | [60] |
| RS | Undefined | No disease | 214 | Subjects at risk of CRC 2 | [44,57,61,62,63] |
| WD | Western-type | No disease | 38 | Diet subjects. Healthy | [42] |
| PD | Modern Paleolithic | No disease | 15 | Diet subjects. Healthy | [43] |
| MD | Mediterranean | No disease | 123 | Diet subjects. Healthy | [61,62,64] |
| CP | Undefined | Colon polyposis | 23 | Patients | [63] |
| CA | Undefined | Colorectal adenoma | 662 | Patients | [57,63,65,66,67] |
| CRC | Undefined | Colorectal cancer | 155 | Patients | [57,65,66] |
| UC | Undefined | Ulcerative colitis | 38 | Patients | [59,60] |
| CD | Undefined | Crohn’s disease | 45 | Patients | [58,59,60] |
| Phylum 1 | HC | RS | FC | MD | PD | WD | CP | CA | CRC | IBD |
|---|---|---|---|---|---|---|---|---|---|---|
| Firmicutes | 58.9506 | 52.9856 | 50.5467 | 60.7094 | 62.6186 | 68.4219 | 43.9497 | 60.7098 | 57.1596 | 57.2953 |
| Bacteroidetes | 27.2735 | 33.3556 | 44.5428 | 23.4625 | 27.3412 | 15.0817 | 50.2231 | 27.0028 | 23.5202 | 30.5508 |
| Proteobacteria | 3.4721 | 4.7896 | 1.1461 | 3.3301 | 4.4188 | 1.6484 | 3.8428 | 6.5936 | 9.7906 | 3.8073 |
| Actinobacteria | 6.5486 | 5.6996 | 1.7042 | 8.7902 | 3.6509 | 11.9093 | 0.6523 | 1.2800 | 1.1112 | 6.7657 |
| Verrucomicrobia | 2.6702 | 2.5748 | 1.2910 | 3.1041 | 1.4947 | 0.9764 | 0.0091 | 2.5214 | 3.9575 | 0.8956 |
| Euryarchaeota | 0.5848 | 0.2252 | 0.2990 | 0.0847 | 0.0536 | 0.4117 | 0 | 1.0877 | 1.7885 | 0.0168 |
| Fusobacteria | 0.0146 | 0.0197 | 0.0029 | 0.0025 | 0.0042 | 0.0848 | 0.0117 | 0.2145 | 2.0454 | 0.2526 |
| Tenericutes | 0.3459 | 0.1043 | 0.3512 | 0.0482 | 0.2094 | 0.7785 | 0.0971 | 0.3692 | 0.2732 | 0.0329 |
| Cyanobacteria | 0.0581 | 0.1052 | 0.1002 | 0.2465 | 0.1488 | 0.2541 | 0.0536 | 0.0625 | 0.0579 | 0.0284 |
| Synergistetes | 0.0098 | 0.0513 | 0.0014 | 0.0441 | 0.007 | 0.0041 | 0.4129 | 0.0848 | 0.1576 | 0.0005 |
| Chloroflexi | 0.0024 | 0.0342 | 0.0008 | 0.0047 | 0 | 0.0179 | 0.4694 | 0.03 | 0.0011 | 0.0007 |
| TM7 | 0.0083 | 0.0191 | 0.0055 | 0.0653 | 0.007 | 0.0039 | 0.0013 | 0.0033 | 0.0014 | 0.0210 |
| Family 1 | HC | RS | FC | MD | PD | WD | CP | CA | CRC | IBD |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacteroidaceae | 20.0833 | 21.4943 | 34.1572 | 15.4143 | 15.5215 | 10.1769 | 28.9815 | 23.0839 | 15.5119 | 17.7059 |
| Ruminococcaceae | 19.1884 | 19.3032 | 17.7934 | 23.7961 | 26.1617 | 15.8293 | 17.3777 | 21.0300 | 17.0954 | 12.9261 |
| Lachnospiraceae | 23.0466 | 13.8709 | 19.8088 | 16.7415 | 13.7672 | 25.2004 | 14.8706 | 19.0354 | 21.5969 | 32.3826 |
| Prevotellaceae | 3.4126 | 6.4874 | 2.9435 | 3.3205 | 7.9948 | 0.9122 | 7.1719 | 2.1619 | 2.4454 | 0.5454 |
| Verrucomicrobiaceae | 2.5702 | 2.5748 | 1.3390 | 3.1041 | 1.4059 | 1.0218 | 0.0091 | 2.6860 | 4.7985 | 1.4723 |
| Enterobacteriaceae | 1.8800 | 2.3051 | 0.1756 | 1.4505 | 1.5145 | 0.9643 | 2.8031 | 3.1807 | 4.6496 | 2.8912 |
| Rikenellaceae | 1.2078 | 2.3913 | 3.3995 | 0.9956 | 0.6892 | 1.3703 | 1.5607 | 3.7645 | 1.3509 | 1.4844 |
| Veillonellaceae | 1.3149 | 2.6069 | 1.5675 | 3.3201 | 3.1355 | 0.9408 | 1.4446 | 2.0480 | 3.2348 | 3.0156 |
| Erysipelotrichaceae | 1.3873 | 2.9033 | 1.6074 | 3.5306 | 2.7406 | 3.0238 | 2.0810 | 2.1707 | 1.2688 | 1.9607 |
| Clostridiaceae | 2.1895 | 1.5247 | 1.5840 | 1.8002 | 2.9558 | 10.5602 | 2.4377 | 1.8124 | 2.0468 | 3.6841 |
| Bifidobacteriaceae | 2.4882 | 3.0881 | 1.4775 | 3.8193 | 0.9905 | 8.5908 | 0.1076 | 0.7255 | 0.6851 | 4.1287 |
| Porphyromonadaceae | 1.4686 | 1.9259 | 2.1814 | 1.3922 | 0.6344 | 0.5258 | 1.9845 | 2.1017 | 2.6539 | 0.7964 |
| Coriobacteriaceae | 2.2984 | 2.8772 | 0.1868 | 4.8781 | 2.4784 | 3.3779 | 0.3076 | 0.4466 | 0.5358 | 3.5795 |
| Streptococcaceae | 0.7604 | 1.1587 | 0.1391 | 1.1514 | 1.6407 | 1.5708 | 0.8787 | 0.8000 | 1.3647 | 2.4764 |
| [Barnesiellaceae] | 0.8793 | 1.0808 | 0.7642 | 0.4972 | 0.5040 | 0.4677 | 0.4404 | 0.8730 | 0.4621 | 0.1233 |
| [Paraprevotellaceae] | 0.4943 | 0.8159 | 0.3284 | 0.8542 | 0.8998 | 0.6662 | 3.1720 | 0.7737 | 0.6244 | 0.0257 |
| S24–7 | 0.6452 | 0.8367 | 0.4641 | 0.5315 | 0.2762 | 0.3106 | 0.6800 | 0.5968 | 0.9590 | 0.1841 |
| Methanobacteriaceae | 0.6146 | 0.2509 | 0.2882 | 0.0847 | 0.0512 | 0.4403 | 0 | 0.6862 | 1.5798 | 0.0292 |
| Alcaligenaceae | 0.6979 | 0.8799 | 0.5232 | 0.2415 | 0.6768 | 0.2323 | 0.1827 | 0.2702 | 0.2863 | 0.3770 |
| [Odoribacteraceae] | 0.5183 | 0.2700 | 0.1939 | 0.2048 | 0.3140 | 0.2704 | 0.1536 | 0.2848 | 0.3808 | 0.0449 |
| Desulfovibrionaceae | 0.2213 | 0.4774 | 0.1547 | 0.2684 | 0.2958 | 0.1048 | 0.1304 | 0.2288 | 0.3753 | 0.1004 |
| Peptostreptococcaceae | 0.2905 | 0.3369 | 0.0330 | 0.3157 | 0.2815 | 0.2195 | 0.5588 | 0.1831 | 0.4749 | 0.5393 |
| Christensenellaceae | 0.2410 | 0.1657 | 0.2954 | 0.1624 | 0.5880 | 0.0704 | 0.1304 | 0.2189 | 0.7917 | 0.0163 |
| Sphingomonadaceae | 0.0051 | 0.0001 | 0.0001 | 0.0007 | 0 | 0.1645 | 0.0001 | 0.3026 | 1.3384 | 0.0001 |
| Lactobacillaceae | 0.2327 | 0.0474 | 0.0032 | 0.3848 | 0.0170 | 0.1321 | 0.3623 | 0.1945 | 0.3568 | 0.4582 |
| Fusobacteriaceae | 0.0173 | 0.0191 | 0.0035 | 0.0020 | 0.0040 | 0.0519 | 0.0030 | 0.2023 | 1.1520 | 0.2526 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Illescas, O.; Rodríguez-Sosa, M.; Gariboldi, M. Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies. Nutrients 2021, 13, 2234. https://doi.org/10.3390/nu13072234
Illescas O, Rodríguez-Sosa M, Gariboldi M. Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies. Nutrients. 2021; 13(7):2234. https://doi.org/10.3390/nu13072234
Chicago/Turabian StyleIllescas, Oscar, Miriam Rodríguez-Sosa, and Manuela Gariboldi. 2021. "Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies" Nutrients 13, no. 7: 2234. https://doi.org/10.3390/nu13072234
APA StyleIllescas, O., Rodríguez-Sosa, M., & Gariboldi, M. (2021). Mediterranean Diet to Prevent the Development of Colon Diseases: A Meta-Analysis of Gut Microbiota Studies. Nutrients, 13(7), 2234. https://doi.org/10.3390/nu13072234






