Abstract
Weight loss interventions with probiotics have favourable effects on gut microbiota composition and derived metabolites. However, little is known about whether the consumption of natural probiotics, such as Bryndza cheeses, brings similar benefits. The purpose of the study was to find the effect of short-term weight loss programs and Bryndza cheese consumption on the structure of the gut microbiota, microbiota-derived metabolites and body composition in middle-aged women. We conducted a randomised controlled intervention study. Twenty-two female participants with a body fat percentage ≥25% underwent a short weight loss program (4 weeks). Subjects were randomised to either the control or intervention group according to diet. The intervention group comprised 13 participants, whose diet contained 30 g of “Bryndza” cheese daily (WLPB). The control group comprised nine participants without the regular consumption of Bryndza cheese (WLP) in their diet. Both interventions lead to a significant and favourable change of BMI, body fat, waist circumference and muscle mass. Moreover, the relative abundance of Erysipelotrichales significantly increased in both groups. However, the relative abundance of lactic acid bacteria (Lactobacillales, Streptococcaceae, Lactococcus and Streptococcus) significantly increased only in the WLPB group. Furthermore, short-chain fatty acid producers Phascolarctobacterium and Butyricimonas increased significantly in the WLPB group. A short-term weight loss program combined with Bryndza cheese consumption improves body composition and increases the abundance of lactic acid bacteria and short-chain fatty acid producers in middle-aged women.
1. Introduction
There is an abundance of literature on the positive effects of active lifestyle programs, diet, sport and exercise training on weight loss, metabolic health and prevention of chronic noncommunicable diseases [1,2,3,4]. However, it is difficult to find the optimal strategy; basically, weight loss programs differ in terms of diet, calorie intake, type, intensity and frequency of physical activity (if included).
Several studies have revealed that interventions including diet and moderate physical activity have a greater joint effect on body weight and composition than interventions featuring just one of these components [5,6]. Low caloric diet without physical activity is effective for short-term weight loss, but physical activity is vital for modifying body composition and improving physical fitness [7]. In addition, losing weight through an active lifestyle may modulate gut microbiota with a positive impact on metabolic regulation [8,9].
Active lifestyle programs are often associated with negative energy balance, diet composition or preferring some functional foods consumption [10,11,12,13]. Food containing probiotics is a functional food with a relative long history. Probiotics are defined by the World Health Organization (WHO) as “live microorganisms which when administered in adequate amounts, confer a beneficial health effect on the host” [14]. Traditional and efficient probiotics products are fermented dairy products [15]. Yogurt is one of the best-known fermented dairy foods [16,17] that has positive health effects [18,19]. Nevertheless, there are other natural fermented foods containing several probiotic strains [20]. Fermented sheep cheese from pasteurised milk (Bryndza) is a traditional dairy product from Slovakia [21]. Bryndza was recently granted the protected geographic indication (PGI), as it is produced in the defined Carpathian Mountains region of Slovakia [22,23]. Slovaks have been eating Bryndza cheese presumably since the 14th century. However, the word “Bryndza” comes from the Wallachian word, which Romanians used to name any type of salted cheese [24]. It is a natural, spreadable, white food made of fermented ripened ewe’s cheese [25]. Recent studies showed that the majority of Bryndza’s microbiota is formed by lactic acid bacteria such as Lactococcus, Streptococcus, Lactobacillus and Enterococcus [26,27,28]. The positive effects of lactic acid bacteria (LAB) on the structure and modulation of gut microbiota and metabolism are well known [29,30,31]. However, there is scarce evidence on the influence of regular Bryndza cheese consumption on gut microbiome and blood metabolites [32].
The purpose of this study was to find the effect of a short-term weight loss program on body composition and the structure of gut microbiota and selected blood metabolites. At the same time, we aimed at elucidating benefits of the consumption of Bryndza cheese on microbiome and metabolic variables.
2. Materials and Methods
2.1. Subjects
A total of 30 participants agreed to participate in the study, of which 22 met the inclusion criteria. The cohort included healthy females who signed up for a short-term weight loss program. The participants were allocated into 2 groups: (a) healthy females (average age 44 ± 13.0 years) who completed a 30-day weight loss program (WLP), and (b) healthy females (average age 51 ± 12.6 years) who completed a 30-day weight loss program that included the regular consumption of natural probiotics (Bryndza cheese).
Inclusive criteria were: (1) females aged 18–65 years and (2) BMI 20–40 kg/m2. Exclusive criteria included: (1) the use of antibiotics for a period of 2 weeks (wk) prior to the study, (2) supplementing probiotics 2 months prior to the study, (3) acute or chronic diseases or infections (including upper respiratory tract infections, fever, chronic inflammatory disorders, autoimmune disorders) 2 months prior to the study, (4), alcohol consumption, (5) smoking or drug abuse, (6) history of digestive diseases (such as inflammatory bowel disease, irritable bowel syndrome) and (7) previous gastrointestinal surgery.
2.2. Intervention
2.2.1. Diet
The subjects had to undergo a 4-week weight loss intervention program, including reduced caloric intake and moderate to vigorous aerobic exercise. Each participant individually received detailed instructions and counselling about lifestyle changes, a personalised nutritional plan made using software PLANEAT (www.planeat.sk; accessed on 6 September 2014) and a plan of physical activity (File S1 in Supplementary Materials). Total daily calorie consumption consisted of 45% carbohydrates, 30% fat and 25% protein. In the WLPB group, their diet contained 30 g of the probiotic sheep cheese “Bryndza”. Altogether, 13 families, 24 genera and 44 species of microbiota were identified in Slovak cheese Bryndza [33]. The most abundant microorganisms in the Bryndza cheese are Lactococcus, Streptococcus, Lactobacillus and Enterococcus [21,26,34]. All the participants were asked to report their food consumption for feedback and control. Quantitative and qualitative data were analysed by the Planeat nutrition software (Planeat s.r.o, Bratislava, Slovakia). The food reports of all the participants were collected and analysed, and one random week was analysed and averaged.
2.2.2. Physical Activity
There were structured and guided exercise training sessions three times a week throughout the intervention period. Each session, including the warm-up, strength training, cardiovascular training, cool down and stretching, lasted 30 min. Exercise was performed on hydraulic strengthening machines in the form of circuit training. One circuit consisted of 9 hydraulic machines and 9 exercise mats for aerobic activity. The load interval was set to 30 s of vigorous-intensity physical activity. One training session comprised 3 entire circuits and lasted approximately 30 min. In addition, the participants were advised to engage in moderate-intensity activity for at least 150 min per week [35].
2.3. Body Composition
Body composition characteristics were measured before and after the intervention. BMI was calculated as weight in kilograms divided by height in metres squared. We measured the body weight, height, body fat percentage, amount of fat mass and muscle mass using the bioelectrical impedance (Omron 511BF, OMRON HEALTHCARE Co., Ltd. Kyoto, Japan). Waist circumference was measured with a flexible tape.
2.4. Resting and Total Energy Expenditure
Resting metabolic rate (RMR) was measured using indirect calorimetry. Respiratory variables were continuously measured using a Cosmed K4b2 breath-by-breath gas analyser. Before measurement, the subjects were advised to get 7 h–8 h of sleep, to refrain from intense physical activity in the previous 24 h and to fast overnight before arriving for the examination. A flow meter calibration was conducted, and before each use the metabolic cart was calibrated with reference gas. After achieving a steady state (after 5 min), the expired gases were collected for 20 min and the final 10 min of the data were averaged; the resting metabolic rate was calculated using the Weir formula [36].
2.5. Stool and Blood Sample Analysis
Faecal samples were collected from participants before and at the end of the intervention. Participants were instructed on how to prevent the contamination of samples during sample collection. They were provided with DNA/RNA Shield Fecal Collection Tubes for the collection and preservation of nucleic acids from stool specimens (ZymoResearch, Irvine, CA, USA). Samples were stored in the DNA/RNA Shield Fecal Collection Tubes at ambient temperature until delivered to the laboratory. Samples were aliquoted and stored at −80 °C.
2.6. DNA Extraction, High-Throughput Sequencing and Bioinformatics
Total DNA from the stool samples was extracted using the ZymoBiomics DNA/RNA mini kit (ZymoResearch Scientific, Irvine, CA, USA) in accordance with the manufacturer’s protocol. DNA from each sample was amplified using specific primers targeting the V1–V3 region of 16S rDNA [37]. PCR reaction contained 1 ng of DNA, 5xFIREPol MasterMix (Solis BioDyne, Tartu, Estonia) and 2 µM of each primer (10 µM). The reaction conditions for PCR amplification were 95 °C for 15 min; 25 cycles of 95 °C for 20 s, 56 °C for 30 s and 72 °C for 1 min; and final elongation at 72 °C for 5 min. The products of amplification were verified by agar electrophoresis. DNA libraries for Illumina sequencing were prepared using the index PCR reaction with input of 1 ng of DNA. The reaction conditions for the PCR were 95 °C for 15 min; 12 cycles of 95 °C for 10 s, 55 °C for 30 s and 72 °C for 90 s; and final elongation at 72 °C for 5 min. Index PCR amplification products were purified using 1.8x Agencourt AMPure XP magnetic beads (BeckmanCoulter, Brea, CA, USA). DNA libraries were validated by Agilent 2100 (Agilent Technologies, Santa Clara, CA, USA) and quantified by Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Mixed amplicons were pooled and sequenced using an Illumina MiSeq platform via a 300 bp paired-end reads Illumina sequencing system (Illumina, San Diego, CA, USA).
2.7. Illumina Data Processing
Samples were analysed using the QIIME2 Core 2020.8 pipeline [38]. Quality control and feature table construction were performed using the DADA2 QIIME2 plugin [39] with default parameters except --p-trunc-len-r 280 and --p-max-ee-r 3. For taxonomic classification, we used a pre-trained naive Bayes classifier version 2020.6.1 [40] in the q2-feature-classifier QIIME2 plugin [41]. This classifier was trained on the Silva 132 99% OTUs full-length sequences [42].
2.8. Plasma Metabolite Concentrations
The concentrations of the following 23 plasma metabolites were analysed: lactate, alanine, valine, leucine, isoleucine, glucose, creatinine, creatine, acetate, acetone, pyruvate, succinate, phenylalanine, tyrosine, glutamine, lysine, histidine, tryptophane, keto-leucine, keto-isoleucine, ketovaline and lipoprotein fractions Lipo1 (methyl [CH3] bands) and Lipo2 (long chain methylene [CH2] n bands) containing a fraction of VLDL, LDL, IDL and HDL.
2.9. Blood Plasma Metabolites; NMR Data Acquisition
The plasma fraction was deproteinated by adding 600 µL of methanol to 300 µL of plasma. The mixture was vortexed for a few seconds and stored at −20 °C for 20 min. Subsequently, the mixture was centrifuged for 30 min at 14,000 rpm. Finally, 700 µL of supernatant was dried out and mixed with 100 µL of stock solution (150 mM phosphate buffer and 0.3 mM TSP-d4 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt as a chemical shift reference in deuterated water) and 500 µL of deuterated water. The final mixture (550 µL) was transferred into a 5 mm NMR tube.
NMR data were obtained by an Avance III 600 MHz NMR spectrometer equipped with cryoprobe (Brukker, Ettlingen, Germany). Initial settings were made from an independent sample and adopted for measurements. Before measurement, samples were stored in Sample Jet at 6 °C for no longer than 3 h and randomly ordered for acquisition. Measurements were carried out at 310 K. An exponential noise filter was used to introduce 0.3 Hz line broadening before the Fourier transform. We used standard Bruker profiling protocols with the following modifications: profiling: 1D NOESY with pre-saturation (noesygppr1d): FID size: 64 k, dummy scans: 4, number of scans: 128, spectral width: 20.4750 ppm; COSY with pre-saturation (cosygpprqf): FID size: 4k, dummy scans: 8, number of scans: 1, spectral width: 16.0125 ppm; homonuclear: J-resolved (jresgpprqf): FID size: 8 k, dummy scans: 16, number of scans: 4; profiling: CPMG with pre-saturation (cpmgpr1d, L4 = 126, d20 = 3 ms): FID size: 64k, dummy scans: 4, number of scans: 128, spectral width: 20.0156 ppm. All experiments were conducted with a relaxation delay of 4 s; all data were once zero filled.
2.10. Statistical Analysis
The data were explored and analysed by R ver. 4.0.3 [43], through the use of the emmeans [44], randomForestSRC [45] and pROC [46]. Exploratory data analysis involved data visualising by swarmplots overlaid with boxplots. Data were subjected to a two-way mixed ANOVA model followed by post hoc comparisons. A Random Forest (RF) machine learning algorithm was used to obtain the out-of-bag ROC curve for predicting pre/post status in the WLPB group and thus to estimate the discriminative ability of the selected physical, microbiome and metabolite markers. Machine learning techniques have several advantages over classical statistical models [47]. The correlations between the characteristics of the gut microbiota and metabolism were analysed using the Spearman correlation coefficient. The significance level of all statistical analyses was set at 0.05. Power calculations for this study were based on previous studies of effect of the probiotic intake on gut microbiota [48]. Assuming a mean of Bifidobacterium abundance 5.2(±4.6) in the placebo group and a standard mean of difference 4.6, our crossover design with n = 11 had a power of 0.80 at an alpha of 0.05 to detect at least 10% increase in the LB group following the diet with probiotics.
ClustVis was used to visualise multidimensional data using principal component analysis (PCA) [49,50].
3. Results
3.1. Body Composition Analysis
Twenty-two participants completed the study. After completing the short-term weight loss program, significant differences were observed in the body composition characteristics in both groups (Table 1). There was significant decrease in body weight, BMI and body fat (%) in both WLPB and WLP groups. Furthermore, there was an increase in muscle mass (%) in both WLPB and WLP groups. There were no significant differences between weight, BMI, fat loss and muscle gain between the groups after intervention.

Table 1.
Physical characteristics at baseline and after completing the short-term weight loss program.
3.2. Nutrition Analysis
All the participants received a nutrition plan with food recipes for the duration of the intervention (File S1 in Supplementary Materials). The plan consisted of four meals per day. Based on the measured RMR, prescribed daily calorie consumption was 1526 ± 240 kcal, containing 45% carbohydrates, 30% fat and 25% protein. The food reports of all the participants were collected and analysed, and one random week was analysed and averaged. Total real calorie consumption and macronutrient intake was similar and did not differ between groups. It included 55–60% carbohydrates, 25–30% proteins and 18–20% lipids. Moreover, we did not find significant differences between the prescribed nutritional plan and self-reported food intake (Table 2).

Table 2.
Calorie and nutrient intake calculated from self-reported food intake.
3.3. Microbial Analysis
Gut microbiota composition was influenced after both interventions. Table 3 shows the significant differences in the abundance of certain microbial taxa after interventions. We identified 12 different phyla in the WLPB groups and 11 phylum taxa in the WLP groups. There were no significant changes in the relative abundance of bacteria at the phylum level within or between the groups. The microbiota alpha diversity of 18–65 year-old women defined by the Shannon, Simpson and Chao1 index did not differ within and between the groups.

Table 3.
Microbial taxa (order, family, genus) differentially present after interventions.
The higher abundance of order Erysipelotrichales was measured within the groups (WLPB-pre vs. WLPB-post; p = 0.001; WLP-pre vs. WLP-post; p = 0.027). Moreover, family Lachnospiraceae decreased in the WLPB group (p = 0.006). It was also numerically decreased in the WLP group; however, the difference was not significant (p = 0.13).
After an analysis of the relative abundance of LAB bacteria, we found some significant changes. In the WLPB group, we observed an increase in Lactobacillales and Streptococcaceae. Further at a lower taxonomic level, WLPB intervention increased the abundance of the genera Lactococcus and Streptococcus (Figure 1). However, no significant changes in the abundance of any LAB bacteria were observed after WLP intervention. We also found some bacterial shifts in SCFA-producing bacteria (Figure 1). Phascolarctobacterium and Butyricimonas increased at the genus level in the WLPB group compared to baseline (p = 0.019, p = 0.023 respectively).
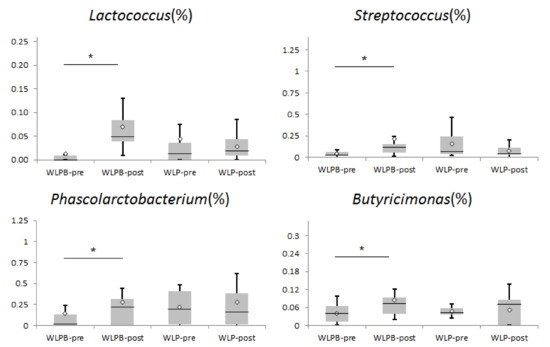
Figure 1.
Relative abundance of gut bacteria taxa in the WLP and WLPB groups compared to baseline. WLPB—short-term weight loss program + Bryndza consumption; WLP—short-term weight loss program. * p < 0.05.
Selected bacterial genera enabled the discrimination between the groups using principal component analysis (PCA) (Figure 2).
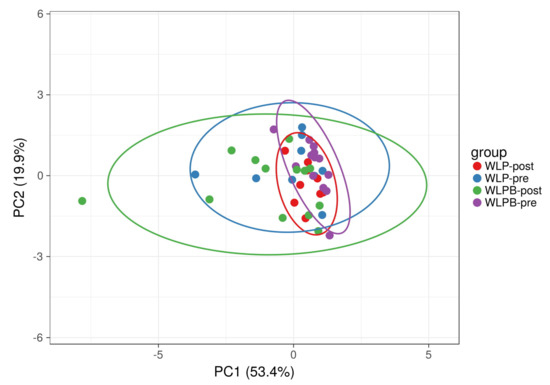
Figure 2.
Beta diversity of analysed samples represented by significantly altered (p < 0.05) bacterial taxa, selected by a Random Forest machine learning analysis, before and after a short-term weight loss program in Bryndza and control groups visualised by PCA. SVD with imputation is used to calculate principal components. The X and Y axis show principal component 1 and principal component 2 that explain 53.4% and 19.9% of the total variance, respectively. Prediction ellipses are such that with a probability of 0.95, a new observation from the same group will fall inside the ellipse (n = 44 data points).
3.4. Plasma Metabolite Analysis
File S2 in Supplementary Materials shows the significant differences in the concentration of certain metabolites after the interventions. In the intra-group comparison, we identified a significant decrease in the lipoprotein fraction Lipo1 between WLPB-pre and WLPB-post (p = 0.036) and WLP-pre and WLP-post (p = 0.026). After the WLPB treatment there was a decrease in the Lipo2 fraction, although it did not reach significance (p = 0.055). Moreover, there was a significant decrease in isoleucine but only in the intra-group WLP. The decrease in isoleucine in the intra-group WLPB was not significant (p = 0.13).
3.5. Machine Learning Analysis of Selected Variables/Predictors
Additionally, we have ranked selected physical, microbiome and metabolite markers used in the Random Forest (RF) machine learning (ML) analyses. The machine learning analysis identified Lactococcus, Lactobacillales, Erysipelotrichales, Streptococcus, Lipo1, BMI, Lachnospiraceae and muscle mass as the important predictors, and it led to an ROC curve with an AUC of 0.83, indicating a good ability to discriminate probands from WLPB-pre and WLPB-post groups (Figure 3).
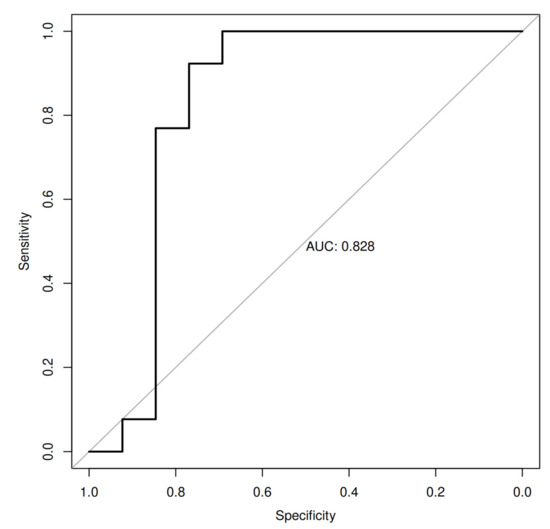
Figure 3.
ROC (receiver operating characteristic) curves with the area under the ROC curve (AUC) for the RF-ML algorithm with Lactococcus, Lactobacillales, Erysipelotrichales, Streptococcus, Lipo1, BMI, Lachnospiraceae and muscle mass identified as the important predictors for discriminating between WLPB-pre and WLPB-post groups. Abbreviations: FPR, false positive rate; RFM-L, Random Forest machine learning; TPR, true positive rate.
4. Discussion
We performed an intervention study to find the effect of a short-term weight loss program with vs. without Bryndza cheese consumption on the gut microbiota composition of healthy women. This is the first gut microbiome study to examine the combination of natural probiotics in the form of Bryndza and exercise training applied in a weight loss program. We hypothesised that a short-term weight loss program would have a positive influence on body composition and could favourably modulate gut microbiota. We expected that regular consumption of Bryndza cheese would have an additional effect on gut microbiota and metabolic parameters. Our main findings demonstrated similar effects of both intervention modalities, which led to decreased body weight, BMI index and body fat in both groups after completing a 4-week weight loss program. Furthermore, we found a higher abundance of the LAB bacteria after Bryndza consumption (Lactobacillales, Streptococcaceae, Lactococcus and Streptococcus). We also noticed an increase in SCFA-producing bacteria Phascolacrtobacterium and Butyricimonas after Bryndza consumption.
These data are in accordance with previous studies, where it was suggested that similar programs have a positive impact on the reduction of BMI and body fat [7,51,52]. Recent studies demonstrated that the intake of dairy products, such as milk and kefir, promoted higher body fat loss and lean mass gain in weight loss [53,54]. We recorded no significant differences between BMI, body fat and waist circumference loss between the WLPB and WLP groups since both recorded equal calorie consumption, which was confirmed by the calorie intake checks (Table 3). Interestingly, the positive influence of probiotics on weight loss is associated with decreased appetite due to the SCFA production of butyrate producing bacteria [55,56]. However, as mentioned above, the diet in our study was isocaloric for both groups.
The main purpose of the study was to find out if a short-term weight loss program combined with Bryndza sheep cheese consumption would change the relative abundance of LAB. Šaková et al. used an independent method to identify some of the LAB bacteria in the Bryndza cheese that were mainly Lactococcus spp. followed by Streptococcus spp. and Leuconostoc spp. [22]. Based on this evidence, we expected a higher relative abundance of the LAB bacteria in stool samples after the probiotic intervention by Bryndza. We detected a significant increase in the relative abundance of the Lactobacillales population. Furthermore, the Random Forest machine learning analysis identified Lactococcus as the most important predictor in a small group of predictors with a good ability to discriminate between the subjects from WLPB-pre and WLPB-post groups. This taxa is associated with improved gut integrity, reduced gut permeability and induced anti-inflammatory response [57,58]. In another study, Lactobacillales were increased after kefir administration in patients with metabolic syndrome [59]. After a short-term weight loss program with Bryndza cheese consumption, we found a significant increase in additional LAB bacteria, specifically Lactococcus and Streptococcus. These genera are commonly used as starter cultures in fermented dairy foods [60]. This is in accordance with previous studies reporting an increased abundance of Lactococcus and Streptococcus as a result of the high consumption of fermented milk products [30,61]. Lactococcus is a homolactic fermentative bacteria that converts its carbon source to lactate from pyruvate with lactate dehydrogenase enzyme utilisation [62]. This genus does not gain too much interest for its probiotic activity because the main attention is focused on the other LAB genera as Lactobacillus and Bifidobacterium [63]. However, some studies have shown the beneficial effects of products fermented with Lactococcus in the gut [64]. A previous study on subjects with slightly elevated blood lipid or blood sugar levels demonstrated that the administration of yogurt fermented with the Lactococcus lactis 11/19-B1 strain for 8 weeks significantly reduced total cholesterol, LDL and LDL/HDL ratio [64]. Moreover, 8 weeks of drinking skim milk fermented with Lactococcus genera has shown a positive impact on the systolic blood pressure level in prehypertensive individuals [65]. There is also some evidence to suggest that the consumption of fermented dairy foods influences the abundance of Streptococcus genera in the gut. Burton et al. showed a significant increase in Streptococcus after a 2-week intervention with probiotic yogurt in young males [66]. Similar to Lactococcus, it has been reported that a higher abundance of Streptococcus, induced by fermented milk intake, improved the lipid profile in cholesterolaemic adults [67]. These results agree with our findings of increased Lactococcus population in the WLPB group. Although our participants were non-obese, we found a significant decrease in lipoprotein fraction 1 after the weight loss program in both WLPB and WLP groups. However, we also found decreased lipoprotein fraction 2 that almost reached statistical significance (p = 0.055) in the WLPB group. In previous studies it was demonstrated that probiotic bacteria had a favourable impact on lipid profile [68,69,70]. In this regard, we believe that Bryndza consumption could represent an added benefit in reducing the lipoprotein fraction.
Furthermore, we detected a decrease in the relative abundance of metabolite isoleucine in the WLP intra-group. We suggest that it could be attributed to the negative energy balance of the short-term weight loss program. These results are in agreement with a previous study on Japanese adults with metabolic syndrome, where they observed the significant correlation between weight loss and reduction of isoleucine concentration [71]. However, in the WLPB group, who consumed the Bryndza cheese, the decrease in isoleucine did not reach a significant value (p = 0.13). Amino acid isoleucine is abundant in whey proteins and ewe’s milk [72]. Rigamonti et al. demonstrated that the administration of whey protein significantly increased the concentration of isoleucine and other amino acids in young, obese women [73]. Therefore, we believe that the protein content in Bryndza may compensate for the catabolic effect due to the weight loss program.
Fermented dairy foods affect the abundance of bacteria producing short chain fatty acids (SCFA) [74]. In our study, Butyricimonas and Phascolarctobacterium, SCFA producers, were elevated in the WLPB intra-group. A recent clinical study revealed a higher abundance of Phascolarctobacterium after the intervention of Kimchi, which is another lactic acid bacteria-fermented food [75]. Previous studies demonstrated the positive correlation between these taxa and an amelioration of the glucose homeostasis [76,77]. Moreno-indias et al. demonstrated a negative correlation between Butyricimonas and plasma glucose. They also found an elevated level of Phascolarctobacterium in appendix samples with insulin sensitivity compared to insulin-resistant obese individuals [77]. These data agree with our results, where we found a negative correlation between the relative abundance of Butyricimonas and fasting insulin level after the WLPB intervention. Moreover, Naderpoor et al. [76] showed a positive correlation between the higher abundance of Phascolarctobacterium and insulin sensitivity in overweight adults [76]. We did not find any significant differences in fasting glucose levels, probably due to the normal glucose levels of the subjects.
We assumed that the short-term weight loss program without Bryndza sheep cheese would, to some extent, affect the composition of gut microbiota too, as there is evidence to suggest that weight loss programs may affect the composition of gut microbiota in humans [78,79,80]. Our microbial analysis did not reveal many significant bacterial shifts after the short-term weight loss program. This may be due to the fact that the participants in our study were not obese and lost an average 3.65 ± 1.70 kg (WLPB) and 3.66 ± 2.50 kg (WLP) of body weight. However, the interesting finding is that the population of Erysipelotrichales increased significantly in both WLPB and WLP groups after intervention, which may be explained by a response to the weight loss program rather than the probiotic intervention. Interestingly the Random Forest machine learning analysis identified Erysipelotrichales as the third most important discrimination factor for subjects from WLPB-pre and WLPB-post groups. The increased number of Erysipelotrichaceae was previously measured in healthy subjects with higher cardiorespiratory fitness levels [81]. Even though we did not examine the physical fitness of our responders, we suppose that 4 weeks of exercise training might increase the physical fitness in both groups. Furthermore, the Erysipelotrichaceae family is considered the key SCFA producer [81]. The gut bacteria-derived SCFA play a role in maintaining intestinal and immune homeostasis and anti-inflammatory response [82,83].
We also identified a decrease in the Lachnospiraceae family in the WLPB and WLP intra-groups. There is strong evidence to suggest that Lachnospiraceae were positively associated with certain metabolic disturbances [84,85,86,87]. In an observational study, Chávez-Carbajal et al. confirmed that Lachnospiraceae was significantly more abundant in obese and extremely obese women [85]. In addition, Lachnospiraceae count is positively associated with diet-related obesity in British men [86]. These results are consistent with our data, where we report decreased weight, body fat and BMI.
Our findings substantiate the benefits of a physically active lifestyle and diet. We strongly recommend further research into the combination of other natural probiotics and physical activity for health promotion and disease prevention.
The main limitation of our study is the small number of participants. We observed elevated drop out from the study mainly due to seasonal flu or non-compliance. However, the majority of participants who completed the study were in the Bryndza group. Further long-term clinical trials with the diet containing a natural probiotic as a weight loss intervention are necessary to investigate the promising protective effect of this intervention on serum/plasma metabolites, body composition and health. The second limitation of our study was age variance. However, all of the subjects were highly motivated to improve their diet and lose body weight. Third, we did not measure the exact amount of physical activity during the intervention.
5. Conclusions
Short-term weight loss intervention both with and without Bryndza cheese consumption improves body composition and increases the count of SCFA producers, although Bryndza consumption results further the health benefits by increasing the relative abundance of lactic acid bacteria. From the broader perspective, our findings suggest health benefits of consuming natural probiotics, such as Bryndza cheese, and emphasise their role in the diet of modern Man.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/nu13061753/s1, File S1: Nutrition plan. File S2: Selected metabolites differentially present after interventions.
Author Contributions
Conceptualisation, K.Š. and V.B.; Methodology V.B., K.Š. and M.K.; Software, M.G.; Validation, M.G.; Formal analysis, A.P., Ľ.K. and G.B.; Investigation, L.K., A.P., Ž.R. and I.H.; Data curation, S.U., S.Š., Ľ.K., G.B. and E.B.; Writing—original draft preparation, I.H.; Writing—review and editing, V.B.; Visualisation, K.Š. and M.G.; Supervision, V.B., K.Š. and M.K.; Project administration, K.Š. and A.P.; Funding acquisition, V.B. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the following granting schemes: APVV-17-0099, VEGA 1/0129/20, VEGA 1/0260/21 and Comenius University Science Park II phase (ITMS 313021D075).
Institutional Review Board Statement
The purpose, procedures and risks of the study were explained to each subject prior to their inclusion, and all of the subjects were enrolled in the study after providing their written informed consent. All of the procedures were reviewed and approved by the ethics committee of the Faculty of Physical Education and Sport of Comenius University (FTVS UK-6/19).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
The authors are grateful to all of the subjects for their participation in this study.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Imayama, I.; Alfano, C.M.; Kong, A.; Foster-Schubert, K.E.; Bain, C.E.; Xiao, L.; Duggan, C.; Wang, C.Y.; Campbell, K.L.; Blackburn, G.L.; et al. Dietary weight loss and exercise interventions effects on quality of life in overweight/obese postmenopausal women: A randomized controlled trial. Int. J. Behav. Nutr. Phys. Act. 2011, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Van Gemert, W.A.M.; van der Palen, J.; Monninkhof, E.M.; Rozeboom, A.; Peters, R.; Wittink, H.; Schuit, A.J.; Peeters, P.H. Quality of life after diet or exercise-induced weight loss in overweight to obese postmenopausal women: The SHAPE-2 randomised controlled trial. PLoS ONE 2015, 10, e0127520. [Google Scholar] [CrossRef] [PubMed]
- Van Gemert, W.A.M.; Schuit, A.J.; van der Palen, J.; May, A.M.; Iestra, J.A.; Wittink, H.; Peeters, P.H.; Monninkhof, E.M. Effect of weight loss, with or without exercise, on body composition and sex hormones in postmenopausal women: The SHAPE-2 trial. Breast Cancer Res. 2015, 17, 120. [Google Scholar] [CrossRef] [PubMed]
- Salas-Salvadó, J.; Díaz-López, A.; Ruiz-Canela, M.; Basora, J.; Fitó, M.; Corella, D.; Serra-Majem, L.; Wärnberg, J.; Romaguera, D.; Estruch, R.; et al. Effect of a Lifestyle Intervention Program With Energy-Restricted Mediterranean Diet and Exercise on Weight Loss and Cardiovascular Risk Factors: One-Year Results of the PREDIMED-Plus Trial. Diabetes Care 2019, 42, 777–788. [Google Scholar] [CrossRef]
- Foster-Schubert, K.E.; Alfano, C.M.; Duggan, C.R.; Xiao, L.; Campbell, K.L.; Kong, A.; Bain, C.E.; Wang, C.Y.; Blackburn, G.L.; Mctiernan, A. Effect of diet and exercise, alone or combined, on weight and body composition in overweight-to-obese postmenopausal women. Obesity 2012, 20, 1628–1638. [Google Scholar] [CrossRef]
- Villareal, D.T.; Chode, S.; Parimi, N.; Sinacore, D.R.; Hilton, T.; Armamento-Villareal, R.; Napoli, N.; Qualls, C.; Shah, K. Weight loss, exercise, or both and physical function in obese older adults. Obstet. Gynecol. Surv. 2011, 66, 488–489. [Google Scholar] [CrossRef]
- Hernández-Reyes, A.; Cámara-Martos, F.; Molina-Luque, R.; Romero-Saldaña, M.; Molina-Recio, G.; Moreno-Rojas, R. Changes in body composition with a hypocaloric diet combined with sedentary, moderate and high-intense physical activity: A randomized controlled trial. BMC Women’s Health 2019, 19, 167. [Google Scholar] [CrossRef]
- Heinsen, F.A.; Fangmann, D.; Müller, N.; Schulte, D.M.; Rühlemann, M.C.; Türk, K.; Settgast, U.; Lieb, W.; Baines, J.F.; Schreiber, S.; et al. Beneficial Effects of a Dietary Weight Loss Intervention on Human Gut Microbiome Diversity and Metabolism Are Not Sustained during Weight Maintenance. Obes. Facts 2017, 9, 379–391. [Google Scholar] [CrossRef]
- Louis, S.; Tappu, R.M.; Damms-Machado, A.; Huson, D.H.; Bischoff, S.C. Characterization of the gut microbial community of obese patients following a weight-loss intervention using whole metagenome shotgun sequencing. PLoS ONE 2016, 11, e0149564. [Google Scholar] [CrossRef]
- Winkler, J.K.; Schultz, J.H.; Woehning, A.; Piel, D.; Gartner, L.; Hildebrand, M.; Roeder, E.; Nawroth, P.P.; Wolfrum, C.; Rudofsky, G. Effectiveness of a low-calorie weight loss program in moderately and severely obese patients. Obes. Facts 2013, 6, 469–480. [Google Scholar] [CrossRef]
- Moreira Carvalho Alves, A.P.; Duarte, F. Effects of food intervention in adults with overweight or obesity. Adv. Obes. Weight Manag. Control 2019, 9, 122–125. [Google Scholar] [CrossRef]
- Medina-Vera, I.; Sanchez-Tapia, M.; Noriega-López, L.; Granados-Portillo, O.; Guevara-Cruz, M.; Flores-López, A.; Avila-Nava, A.; Fernández, M.L.; Tovar, A.R.; Torres, N. A dietary intervention with functional foods reduces metabolic endotoxaemia and attenuates biochemical abnormalities by modifying faecal microbiota in people with type 2 diabetes. Diabetes Metab. 2019, 45, 122–131. [Google Scholar] [CrossRef]
- Storck, L.J.; Meffert, P.J.; Rausch, J.; Gärtner, S.; Aghdassi, A.A.; Kühn, J.P.; Kraft, M.; Pietzner, M.; Lerch, M.M.; Steveling, A. Efficiency of a 15-Week Weight-Loss Program, including a Low-Calorie Formula Diet, on Glycemic Control in Patients with Type 2 Diabetes Mellitus and Overweight or Obesity. Obes. Facts 2021, 14, 45–55. [Google Scholar] [CrossRef]
- FAO; WHO. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. In Food and Agriculture Organization of the United Nations and World Health Organization Expert Consultation Report; WHO: Geneva, Switzerland, 2001; pp. 413–426. [Google Scholar]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented foods as a dietary source of live organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef]
- González, S.; Fernández-Navarro, T.; Arboleya, S.; De Los Reyes-Gavilán, C.G.; Salazar, N.; Gueimonde, M. Fermented dairy foods: Impact on intestinal microbiota and health-linked biomarkers. Front. Microbiol. 2019, 10, 1046. [Google Scholar] [CrossRef]
- Pei, R.; Martin, D.A.; DiMarco, D.M.; Bolling, B.W. Evidence for the effects of yogurt on gut health and obesity. Crit. Rev. Food Sci. Nutr. 2017, 57, 1569–1583. [Google Scholar] [CrossRef]
- Makino, S.; Ikegami, S.; Kume, A.; Horiuchi, H.; Sasaki, H.; Orii, N. Reducing the risk of infection in the elderly by dietary intake of yoghurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. Br. J. Nutr. 2010, 104, 998–1006. [Google Scholar] [CrossRef]
- Ataie-Jafari, A.; Larijani, B.; Alavi Majd, H.; Tahbaz, F. Cholesterol-lowering effect of probiotic yogurt in comparison with ordinary yogurt in mildly to moderately hypercholesterolemic subjects. Ann. Nutr. Metab. 2009, 54, 22–27. [Google Scholar] [CrossRef]
- Salque, M.; Bogucki, P.I.; Pyzel, J.; Sobkowiak-Tabaka, I.; Grygiel, R.; Szmyt, M.; Evershed, R.P. Earliest evidence for cheese making in the sixth millennium bc in northern Europe. Nature 2013, 493, 522–525. [Google Scholar] [CrossRef]
- Jurkovič, D.; Križková, L.; Dušinský, R.; Belicová, A.; Sojka, M.; Krajčovič, J.; Ebringer, L. Identification and characterization of enterococci from bryndza cheese. Lett. Appl. Microbiol. 2006, 42, 553–559. [Google Scholar] [CrossRef]
- Šaková, N.; Sádecká, J.; Lejková, J.; Puškárová, A.; Korenová, J.; Kolek, E.; Valík, L.; Kuchta, T.; Pangallo, D. Characterization of May bryndza cheese from various regions in Slovakia based on microbiological, molecular and principal volatile odorants examination. J. Food Nutr. Res. 2015, 54, 239–251. [Google Scholar]
- Commission Regulation (Ec) No 676/2008 of 16 July 2008 Registering certain names in the register of protected designations of origin and protected geographical indications (Ail De La Drôme (Pgi), Všestarská Cibule (Pdo), Slovenská Bryndza (Pgi). Ajo. Morad. J. Eur. Union 2008, L189, 19–20.
- Podolák, J. Tradičné Ovčiarstvo na Slovensku; Veda: Bratislava, Slovakia, 1982. [Google Scholar]
- Kačániová, M.; Kunová, S.; Štefániková, J.; Felšöciová, S.; Godočíková, L.; Horská, E.; Nagyová, Ľ.; Haščík, P.; Terentjeva, M. Microbiota of the traditional slovak sheep cheese “bryndza”. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 482–486. [Google Scholar] [CrossRef]
- Pangallo, D.; Šaková, N.; Koreňová, J.; Puškárová, A.; Kraková, L.; Valík, L.; Kuchta, T. Microbial diversity and dynamics during the production of May bryndza cheese. Int. J. Food Microbiol. 2014, 170, 38–43. [Google Scholar] [CrossRef]
- Chebeňová-Turcovská, V.; Ženišová, K.; Kuchta, T.; Pangallo, D.; Brežná, B. Culture-independent detection of microorganisms in traditional Slovakian bryndza cheese. Int. J. Food Microbiol. 2011, 150, 73–78. [Google Scholar] [CrossRef]
- Markusková, B.; Lichvariková, A.; Szemes, T.; Koreňová, J.; Kuchta, T.; Drahovská, H. Genome analysis of lactic acid bacterial strains selected as potential starters for traditional Slovakian bryndza cheese. FEMS Microbiol. Lett. 2018, 365, fny257. [Google Scholar] [CrossRef]
- Mathur, H.; Beresford, T.P.; Cotter, P.D. Health benefits of lactic acid bacteria (Lab) fermentates. Nutrients 2020, 12, 1679. [Google Scholar] [CrossRef]
- Veiga, P.; Pons, N.; Agrawal, A.; Oozeer, R.; Guyonnet, D.; Brazeilles, R.; Faurie, J.M.; Van Hylckama Vlieg, J.E.T.; Houghton, L.A.; Whorwell, P.J.; et al. Changes of the human gut microbiome induced by a fermented milk product. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Kamodyová, N.; Minárik, G.; Hodosy, J.; Celec, P. Single Consumption of Bryndza Cheese Temporarily Affects Oral Microbiota and Salivary Markers of Oxidative Stress. Curr. Microbiol. 2014, 69, 716–724. [Google Scholar] [CrossRef]
- Kačániová, M.; Terentjeva, M.; Kunová, S.; Haščík, P.; Kowalczewski, P.Ł.; Štefániková, J. Diversity of microbiota in Slovak summer ewes’ cheese “bryndza”. Open Life Sci. 2021, 16, 277–286. [Google Scholar] [CrossRef]
- Berta, G.; Chebeñová, V.; Brežná, B.; Pangallq, D.; Valík, L.; Kuchta, T. Identification of lactic acid bacteria in Slovakian bryndza cheese. J. Food Nutr. Res. 2009, 48, 65–71. [Google Scholar]
- Bull, F.C.; Al-Ansari, S.S.; Biddle, S.; Borodulin, K.; Buman, M.P.; Cardon, G.; Carty, C.; Chaput, J.P.; Chastin, S.; Chou, R.; et al. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. Br. J. Sports Med. 2020, 54, 1451–1462. [Google Scholar] [CrossRef]
- Weir, J.D.V. New methods for calculating metabolic rate with special reference to protein metabolism. J. Physiol. 1949, 109, 1–9. [Google Scholar] [CrossRef]
- Zheng, W.; Tsompana, M.; Ruscitto, A.; Sharma, A.; Genco, R.; Sun, Y.; Buck, M.J. An accurate and efficient experimental approach for characterization of the complex oral microbiota. Microbiome 2015, 3, 48. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.; Robeson, M.; Kaehler, B.; Dillon, M. Bokulich-Lab/RESCRIPt: 2020.6.1. 2020. Available online: https://zenodo.org/record/3945228#.YKYWs91nrIU (accessed on 20 May 2021).
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- R Core Team. European Environment Agency. 2020. Available online: https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (accessed on 14 April 2021).
- CRAN. Package Emmeans. Available online: https://cran.r-project.org/web/packages/emmeans/index.html (accessed on 25 April 2021).
- CRAN. Package RandomForestSRC. Available online: https://cran.r-project.org/web/packages/randomForestSRC/index.html (accessed on 14 April 2021).
- Turck, N.; Vutskits, L.; Sanchez-Pena, P.; Robin, X.; Hainard, A.; Gex-Fabry, M.; Fouda, C.; Bassem, H.; Mueller, M.; Lisacek, F.; et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar]
- Tsiamis, G.; Badger, J.; Yildirim, S.; Judith Marcos-Zambrano, L.; Truu, J.; Turukalo, L.T.; de Santa Pau, C.E.; Karaduzovic-Hadziabdic, K.; Loncar Turukalo, T.; Przymus, P.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 313. [Google Scholar]
- Huang, W.C.; Pan, C.H.; Wei, C.C.; Huang, H.Y. Lactobacillus plantarum ps128 improves physiological adaptation and performance in triathletes through gut microbiota modulation. Nutrients 2020, 12, 2315. [Google Scholar] [CrossRef] [PubMed]
- ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data (BETA). Available online: https://biit.cs.ut.ee/clustvis/ (accessed on 17 May 2021).
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Headland, M.L.; Clifton, P.M.; Keogh, J.B. Effect of intermittent compared to continuous energy restriction on weight loss and weight maintenance after 12 months in healthy overweight or obese adults. Int. J. Obes. 2019, 43, 2028–2036. [Google Scholar] [CrossRef] [PubMed]
- Geiker, N.R.W.; Ritz, C.; Pedersen, S.D.; Larsen, T.M.; Hill, J.O.; Astrup, A. A weight-loss program adapted to the menstrual cycle increases weight loss in healthy, overweight, premenopausal women: A 6-mo randomized controlled trial. Am. J. Clin. Nutr. 2016, 104, 15–20. [Google Scholar] [CrossRef]
- Fathi, Y.; Faghih, S.; Zibaeenezhad, M.J.; Tabatabaei, S.H.R. Kefir drink leads to a similar weight loss, compared with milk, in a dairy-rich non-energy-restricted diet in overweight or obese premenopausal women: A randomized controlled trial. Eur. J. Nutr. 2016, 55, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Josse, A.R.; Atkinson, S.A.; Tarnopolsky, M.A.; Phillips, S.M. Increased Consumption of Dairy Foods and Protein during Diet- and Exercise-Induced Weight Loss Promotes Fat Mass Loss and Lean Mass Gain in Overweight and Obese Premenopausal Women. J. Nutr. 2011, 141, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Freeland, K.R.; Wilson, C.; Wolever, T.M.S. Adaptation of colonic fermentation and glucagon-like peptide-1 secretion with increased wheat fibre intake for 1 year in hyperinsulinaemic human subjects. Br. J. Nutr. 2010, 103, 82–90. [Google Scholar] [CrossRef]
- Chambers, E.S.; Byrne, C.S.; Morrison, D.J.; Murphy, K.G.; Preston, T.; Tedford, C.; Garcia-Perez, I.; Fountana, S.; Serrano-Contreras, J.I.; Holmes, E.; et al. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: A randomised cross-over t. Gut 2019, 68, 1430–1438. [Google Scholar] [CrossRef]
- Pérez-Santiago, J.; Gianella, S.; Massanella, M.; Spina, C.A.; Karris, M.Y.; Var, S.R.; Patel, D.; Jordan, P.S.; Young, J.A.; Little, S.J.; et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. Aids 2013, 27, 1921–1931. [Google Scholar] [CrossRef]
- Nissen, L.; Chingwaru, W.; Sgorbati, B.; Biavati, B.; Cencic, A. Gut health promoting activity of new putative probiotic/protective Lactobacillus spp. strains: A functional study in the small intestinal cell model. Int. J. Food Microbiol. 2009, 135, 288–294. [Google Scholar] [CrossRef]
- Bellikci-Koyu, E.; Sarer-Yurekli, B.P.; Akyon, Y.; Aydin-Kose, F.; Karagozlu, C.; Ozgen, A.G.; Brinkmann, A.; Nitsche, A.; Ergunay, K.; Yilmaz, E.; et al. Effects of regular kefir consumption on gut microbiota in patients with metabolic syndrome: A parallel-group, randomized, controlled study. Nutrients 2019, 11, 2089. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Safety Demonstration of Microbial Food Cultures (MFC) in Fermented Food Products. Bull. Int. Dairy Fed. 2012, 455, 2. [Google Scholar]
- Swarte, J.C.; Eelderink, C.; Douwes, R.M.; Said, M.Y.; Hu, S.; Post, A.; Westerhuis, R.; Bakker, S.J.L.; Harmsen, H.J.M. Effect of high versus low dairy consumption on the gut microbiome: Results of a randomized, cross-over study. Nutrients 2020, 12, 2129. [Google Scholar] [CrossRef]
- Hols, P.; Kleerebezem, M.; Schanck, A.N.; Ferain, T.; Hugenholtz, J.; Delcour, J.; De Vos, W.M. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 1999, 17, 588–592. [Google Scholar] [CrossRef]
- Wieërs, G.; Belkhir, L.; Enaud, R.; Leclercq, S.; Philippart de Foy, J.M.; Dequenne, I.; de Timary, P.; Cani, P.D. How Probiotics Affect the Microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 454. [Google Scholar] [CrossRef]
- Nishiyama, K.; Kobayashi, T.; Sato, Y.; Watanabe, Y.; Kikuchi, R.; Kanno, R.; Koshizuka, T.; Miyazaki, N.; Ishioka, K.; Suzutani, T. A double-blind controlled study to evaluate the effects of yogurt enriched with lactococcus lactis 11/19-b1 and bifidobacterium lactis on serum low-density lipoprotein level and antigen-specific interferon-γ releasing ability. Nutrients 2018, 10, 1778. [Google Scholar] [CrossRef]
- Beltrán-Barrientos, L.M.; González-Córdova, A.F.; Hernández-Mendoza, A.; Torres-Inguanzo, E.H.; Astiazarán-García, H.; Esparza-Romero, J.; Vallejo-Cordoba, B. Randomized double-blind controlled clinical trial of the blood pressure–lowering effect of fermented milk with Lactococcus lactis: A pilot study2. J. Dairy Sci. 2018, 101, 2819–2825. [Google Scholar] [CrossRef]
- Burton, K.J.; Rosikiewicz, M.; Pimentel, G.; Bütikofer, U.; Von Ah, U.; Voirol, M.J.; Croxatto, A.; Aeby, S.; Drai, J.; McTernan, P.G.; et al. Probiotic yogurt and acidified milk similarly reduce postprandial inflammation and both alter the gut microbiota of healthy, young men. Br. J. Nutr. 2017, 117, 1312–1322. [Google Scholar] [CrossRef]
- Ito, M.; Kusuhara, S.; Yokoi, W.; Sato, T.; Ishiki, H.; Miida, S.; Matsui, A.; Nakamori, K.; Nonaka, C.; Miyazaki, K. Streptococcus thermophilus fermented milk reduces serum MDA-LDL and blood pressure in healthy and mildly hypercholesterolaemic adults. Benef. Microbes 2017, 8, 171–178. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrión, J.M.; Cuñé, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef]
- Gomes, A.C.; de Sousa, R.G.M.; Botelho, P.B.; Gomes, T.L.N.; Prada, P.O.; Mota, J.F. The additional effects of a probiotic mix on abdominal adiposity and antioxidant Status: A double-blind, randomized trial. Obesity 2017, 25, 30–38. [Google Scholar] [CrossRef]
- Kullisaar, T.; Zilmer, K.; Salum, T.; Rehema, A.; Zilmer, M. The use of probiotic L. fermentum ME-3 containing Reg’Activ Cholesterol supplement for 4 weeks has a positive influence on blood lipoprotein profiles and inflammatory cytokines: An open-label preliminary study. Nutr. J. 2016, 15, 93. [Google Scholar] [CrossRef]
- Tochikubo, O.; Nakamura, H.; Jinzu, H.; Nagao, K.; Yoshida, H.; Kageyama, N.; Miyano, H. Weight loss is associated with plasma free amino acid alterations in subjects with metabolic syndrome. Nutr. Diabetes 2016, 6, e197. [Google Scholar] [CrossRef]
- Rafiq, S.; Huma, N.; Pasha, I.; Sameen, A.; Mukhtar, O.; Khan, M.I. Chemical composition, nitrogen fractions and amino acids profile of milk from different animal species. Asian Australas. J. Anim. Sci. 2016, 29, 1022–1028. [Google Scholar] [CrossRef]
- Rigamonti, A.E.; Leoncini, R.; De Col, A.; Tamini, S. The Appetite—Suppressant and GLP-1-Stimulating Effects of Whey Proteins in Obese Subjects are Associated with Increased Circulating Levels of Specific Amino Acids. Nutrients 2020, 12, 775. [Google Scholar] [CrossRef]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Laiola, M.; Paparo, L.; Calignano, A.; De Caro, C.; Coretti, L.; Chiariotti, L.; Gilbert, J.A.; et al. Specific Signatures of the Gut Microbiota and Increased Levels of Butyrate in Children Treated with Fermented Cow’s Milk Containing Heat-Killed Lactobacillus paracasei CBA L74. Appl. Environ. Microbiol. 2017, 83, 19. [Google Scholar] [CrossRef]
- Kim, H.Y.; Park, K.Y. Clinical trials of kimchi intakes on the regulation of metabolic parameters and colon health in healthy Korean young adults. J. Funct. Foods 2018, 47, 325–333. [Google Scholar] [CrossRef]
- Naderpoor, N.; Mousa, A.; Gomez-arango, L.F.; Barrett, H.L.; Nitert, M.D.; Courten, B. De Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. J. Clin. Med. 2019, 8, 452. [Google Scholar] [CrossRef]
- Moreno-indias, I.; Sánchez-alcoholado, L.; García-fuentes, E.; Cardona, F. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am. J. Transl. Res. 2016, 8, 5672–5684. [Google Scholar]
- Frost, F.; Storck, L.J.; Kacprowski, T.; Gärtner, S.; Rühlemann, M.; Bang, C.; Franke, A.; Völker, U.; Aghdassi, A.A.; Steveling, A.; et al. A structured weight loss program increases gut microbiota phylogenetic diversity and reduces levels of Collinsella in obese type 2 diabetics: A pilot study. PLoS ONE 2019, 14, e0219489. [Google Scholar] [CrossRef] [PubMed]
- Fragiadakis, G.K.; Wastyk, H.C.; Robinson, J.L.; Sonnenburg, E.D.; Sonnenburg, J.L.; Gardner, C.D. Long-term dietary intervention reveals resilience of the gut microbiota despite changes in diet and weight. Am. J. Clin. Nutr. 2020, 111, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Cruz, M.; Flores-López, A.G.; Aguilar-López, M.; Sánchez-Tapia, M.; Medina-Vera, I.; Díaz, D.; Tovar, A.R.; Torres, N. Improvement of Lipoprotein Profile and Metabolic Endotoxemia by a Lifestyle Intervention That Modifies the Gut Microbiota in Subjects With Metabolic Syndrome. J. Am. Heart Assoc. 2019, 8, e012401. [Google Scholar] [CrossRef] [PubMed]
- Estaki, M.; Pither, J.; Baumeister, P.; Little, J.P.; Gill, S.K.; Ghosh, S.; Ahmadi-Vand, Z.; Marsden, K.R.; Gibson, D.L. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome 2016, 4, 42. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; De los Reyes-Gavilán, C.G.; Salazar, N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Lippert, K.; Kedenko, L.; Antonielli, L.; Kedenko, I.; Gemeier, C.; Leitner, M.; Kautzky-Willer, A.; Paulweber, B.; Hackl, E. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes 2017, 8, 545–556. [Google Scholar] [CrossRef]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-Del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E.; et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014, 8, 2218–2230. [Google Scholar] [CrossRef]
- Zeng, H.; Ishaq, S.L.; Zhao, F.Q.; Wright, A.D.G. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016, 35, 30–36. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).