Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Samples
2.2. Saponification of Olive Oils
2.3. Analysis of Fatty Acids by Gas Chromatography (GC)
2.4. Coupling of Fatty Acids
2.5. RAW264.7 Macrophage Culture
2.6. Cell Viability
2.7. RNA Isolation and cDNA Synthesis
2.8. Quantitative Real-Time RT-PCR
2.9. Immunoblotting
2.10. Quantification of Nitric Oxide Formation Using Griess Assay
2.11. Analysis of PGE2 and Arachidonic Acid (AA) by UPLC-MS/MS
2.12. Statistics
3. Results
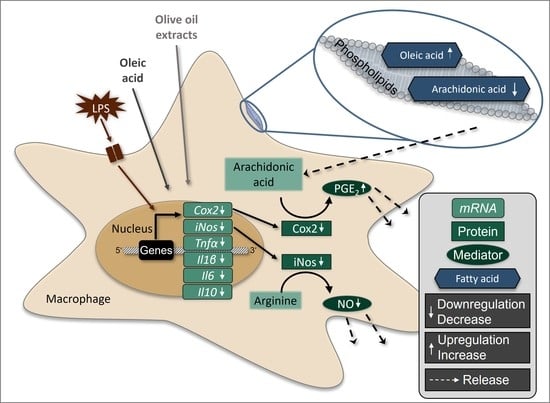
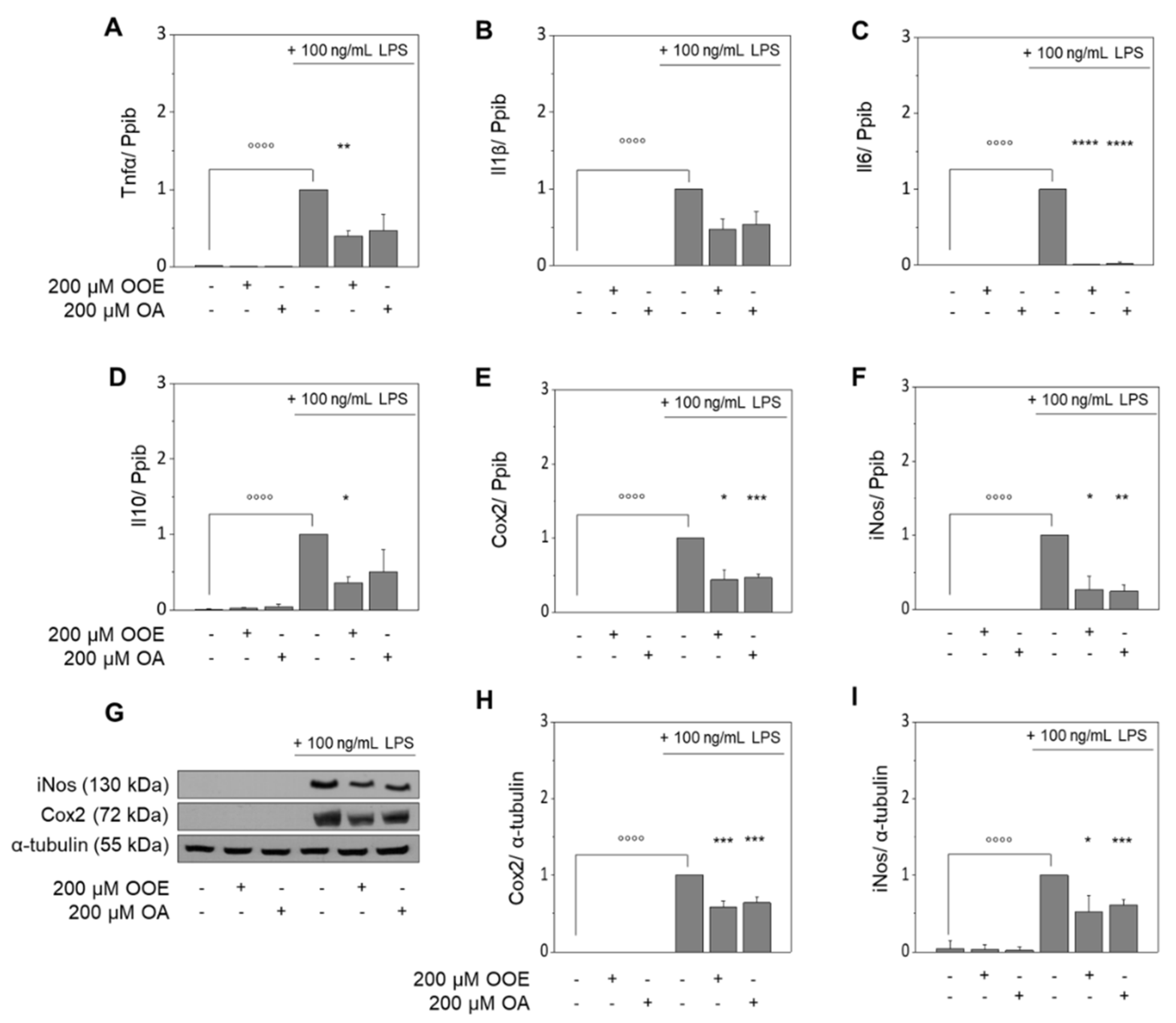
3.1. OOE and OA Inhibit the LPS-Induced Expression of Target Genes in RAW264.7 Macrophages
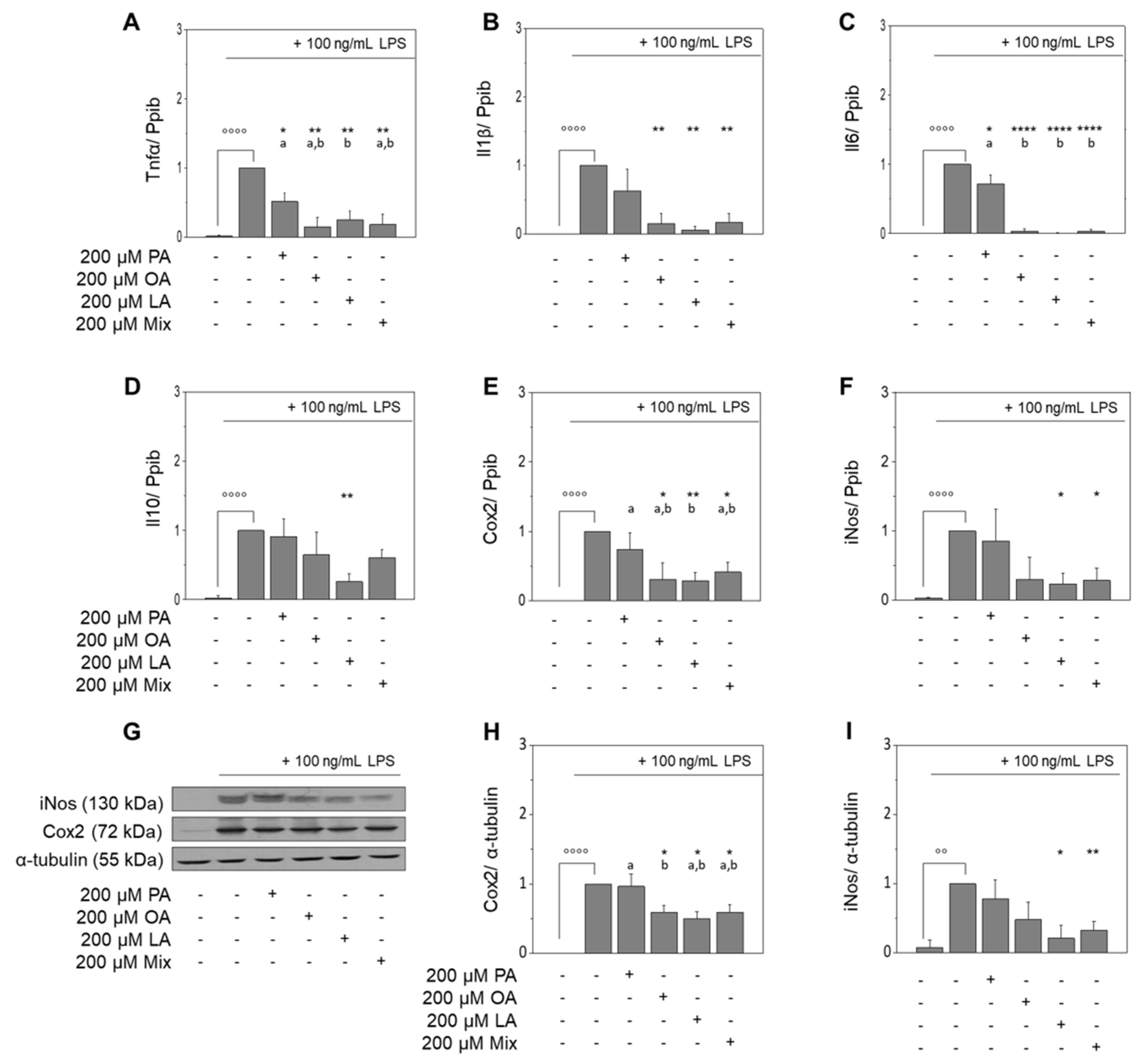
3.2. Fatty Acids of EVOO Inhibit the LPS-Induced Expression of Target Genes in RAW264.7 Macrophages
3.3. Regulation of Nitric Oxide Formation in Murine RAW264.7 Macrophages Depends on Fatty Acid Saturation and Is Dose-Dependent
3.4. Formation of PGE2 in LPS-Stimulated Murine Macrophages Is Augmented by OOE and FA
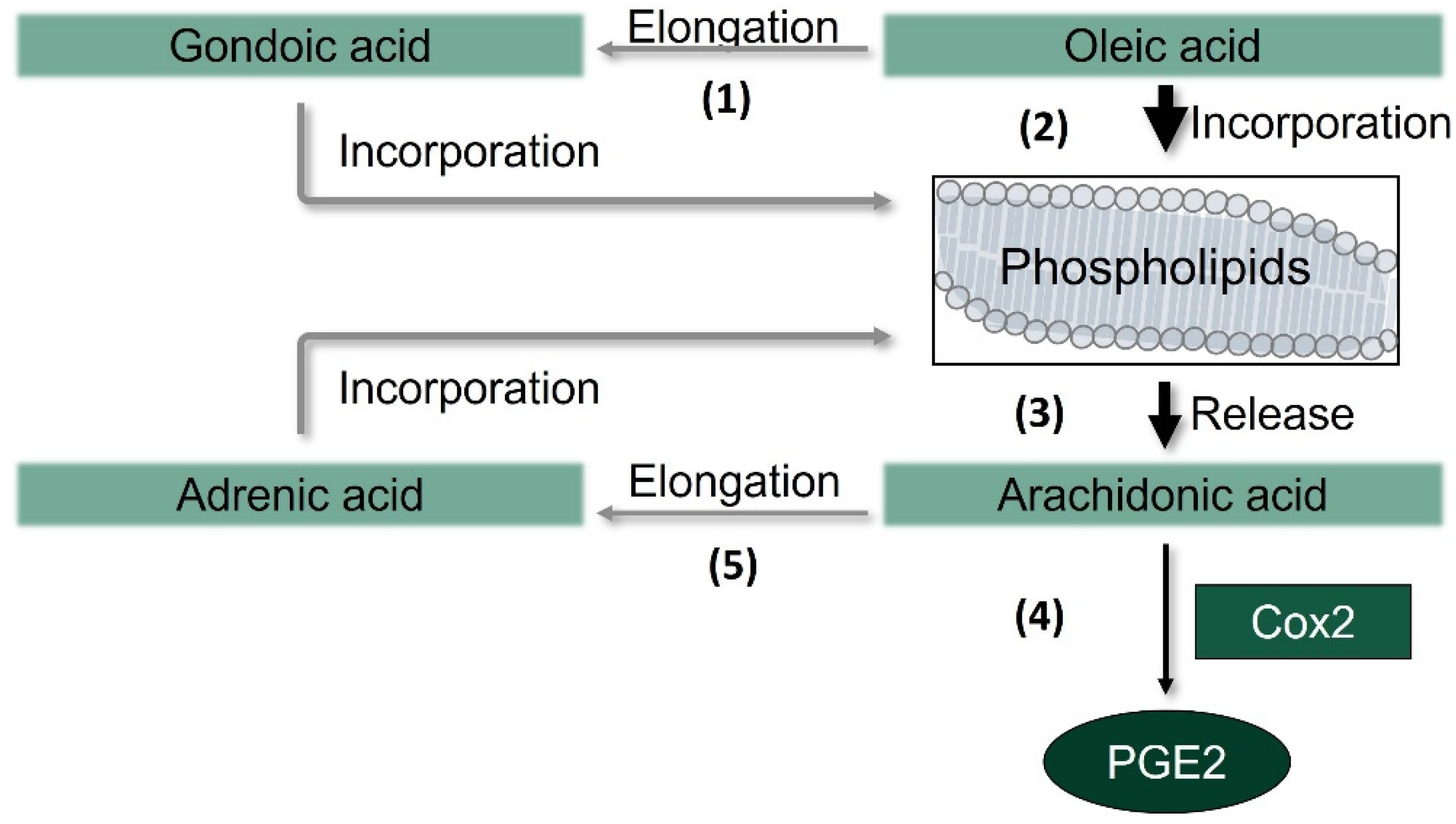
3.5. Oleic Acid Changes the Composition of Cellular Lipids and Phospholipids in RAW264.7 Macrophages
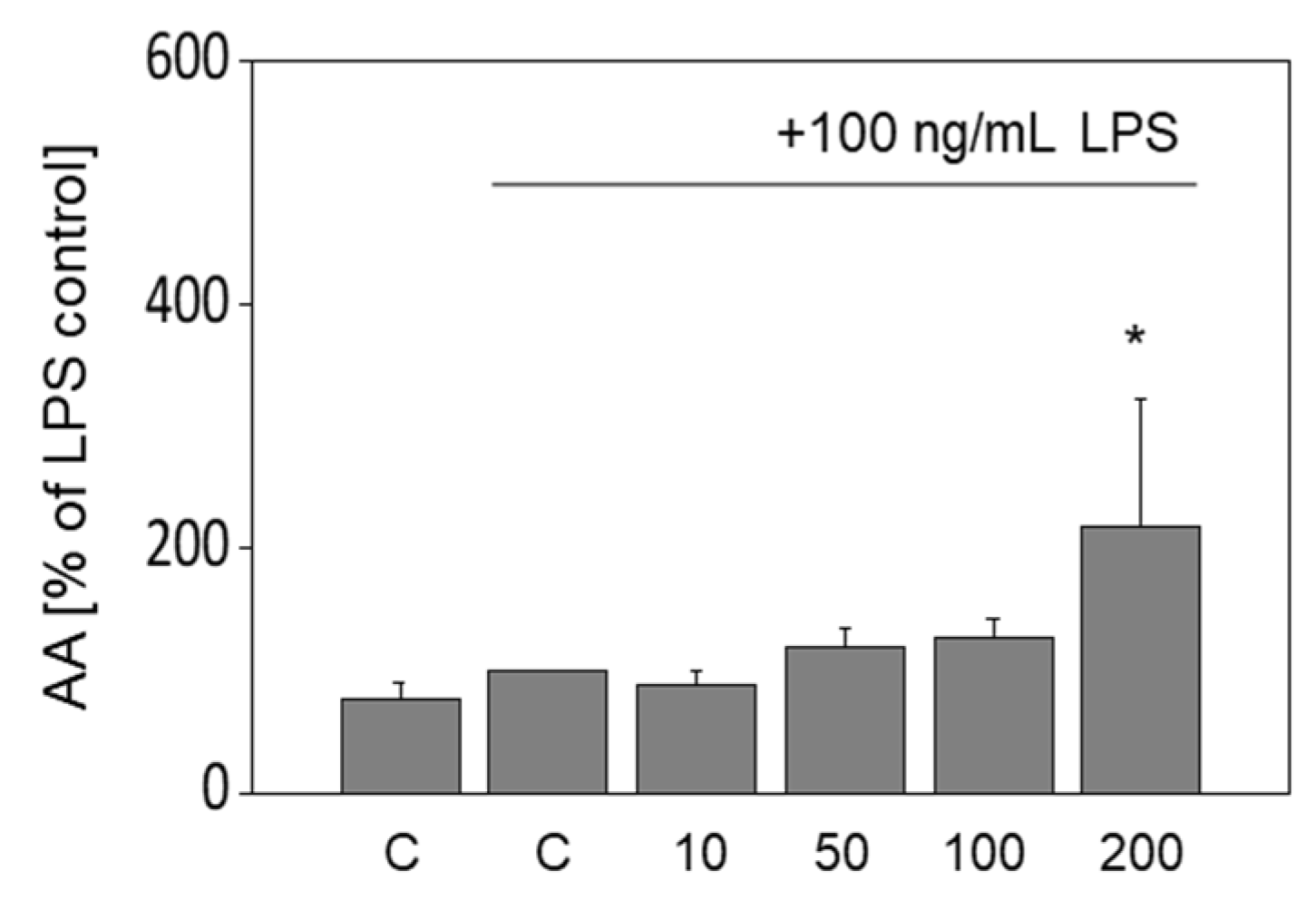
3.6. Release of Arachidonic Acid in LPS-Stimulated Murine Macrophages Is Increased by OA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet. N. Engl. J. Med. 2013, 368, 1279–1290. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.; Ioannidis, J.P.A. PREDIMED trial of Mediterranean diet: Retracted, republished, still trusted? BMJ 2019, 364, l341. [Google Scholar] [CrossRef]
- Finicelli, M.; Squillaro, T.; Di Cristo, F.; Di Salle, A.; Melone, M.A.B.; Galderisi, U.; Peluso, G. Metabolic syndrome, Mediterranean diet, and polyphenols: Evidence and perspectives. J. Cell. Physiol. 2019, 234, 5807–5826. [Google Scholar] [CrossRef]
- Schwingshackl, L.; Christoph, M.; Hoffmann, G.F. Effects of Olive Oil on Markers of Inflammation and Endothelial Function—A Systematic Review and Meta-Analysis. Nutrients 2015, 7, 7651–7675. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Wilmot, K.A.; Ghasemzadeh, N.; Molloy, D.L.; Burkman, G.; Mekonnen, G.; Gongora, M.C.; Quyyumi, A.A.; Sperling, L.S. Mediterranean Dietary Patterns and Cardiovascular Health. Annu. Rev. Nutr. 2015, 35, 425–449. [Google Scholar] [CrossRef]
- Buckley, C.D.; Gilroy, D.; Serhan, C.N. Proresolving Lipid Mediators and Mechanisms in the Resolution of Acute Inflammation. Immunity 2014, 40, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Calder, P.C. Fatty acids and inflammation: The cutting edge between food and pharma. Eur. J. Pharmacol. 2011, 668, S50–S58. [Google Scholar] [CrossRef]
- Hamidzadeh, K.; Christensen, S.M.; Dalby, E.; Chandrasekaran, P.; Mosser, D.M. Macrophages and the Recovery from Acute and Chronic Inflammation. Annu. Rev. Physiol. 2017, 79, 567–592. [Google Scholar] [CrossRef] [Green Version]
- Ait-Oufella, H.; Taleb, S.; Mallat, Z.; Tedgui, A. Recent Advances on the Role of Cytokines in Atherosclerosis. Arter. Thromb. Vasc. Biol. 2011, 31, 969–979. [Google Scholar] [CrossRef] [Green Version]
- Aktan, F. iNOS-mediated nitric oxide production and its regulation. Life Sci. 2004, 75, 639–653. [Google Scholar] [CrossRef]
- Simmons, D.L.; Botting, R.M.; Hla, T. Cyclooxygenase Isozymes: The Biology of Prostaglandin Synthesis and Inhibition. Pharmacol. Rev. 2004, 56, 387–437. [Google Scholar] [CrossRef] [Green Version]
- Rastrelli, L.; Passi, S.; Ippolito, F.; Vacca, G.; De Simone, F. Rate of Degradation of α-Tocopherol, Squalene, Phenolics, and Polyunsaturated Fatty Acids in Olive Oil during Different Storage Conditions. J. Agric. Food Chem. 2002, 50, 5566–5570. [Google Scholar] [CrossRef]
- Carluccio, M.A.; Massaro, M.; Bonfrate, C.; Siculella, L.; Maffia, M.; Nicolardi, G.; Distante, A.; Storelli, C.; Caterina, R.D. Oleic Acid Inhibits Endothelial Activation: A Direct Vascular Antiatherogenic Mechanism of a Nutritional Component in the Mediterranean Diet. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.T.; Lee, J.Y.; Lee, J.; Kim, H.; Yoon, K.-S.; Choe, W.; Kang, I. Oleic acid reduces lipopolysaccharide-induced expression of iNOS and COX-2 in BV2 murine microglial cells: Possible involvement of reactive oxygen species, p38 MAPK, and IKK/NF-κB signaling pathways. Neurosci. Lett. 2009, 464, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.F.; Chau, Y.P.; Ni Kung, H.; Lu, K.S. The lipopolysaccharide-induced pro-inflammatory response in RAW264.7 cells is attenuated by an unsaturated fatty acid–bovine serum albumin complex and enhanced by a saturated fatty acid–bovine serum albumin complex. Inflamm. Res. 2012, 61, 151–160. [Google Scholar] [CrossRef] [PubMed]
- De Santis, S.; Liso, M.; Verna, G.; Curci, F.; Milani, G.; Faienza, M.; Franchini, C.; Moschetta, A.; Chieppa, M.; Clodoveo, M.; et al. Extra Virgin Olive Oil Extracts Modulate the Inflammatory Ability of Murine Dendritic Cells Based on Their Polyphenols Pattern: Correlation between Chemical Composition and Biological Function. Antioxidants 2021, 10, 1016. [Google Scholar] [CrossRef] [PubMed]
- Teres, S.; Barceló-Coblijn, G.; Benet, M.; Alvarez, R.; Bressani, R.; Halver, J.E.; Escriba, P.V. Oleic acid content is responsible for the reduction in blood pressure induced by olive oil. Proc. Natl. Acad. Sci. USA 2008, 105, 13811–13816. [Google Scholar] [CrossRef] [Green Version]
- Müller, A.K.; Schmölz, L.; Wallert, M.; Schubert, M.; Schlörmann, W.; Glei, M.; Lorkowski, S. In Vitro Digested Nut Oils Attenuate the Lipopolysaccharide-Induced Inflammatory Response in Macrophages. Nutrients 2019, 11, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krist, S. Lexikon der pflanzlichen Fette und Öle; Springer: Berlin/Heidelberg, Germany, 2013; Volume 2. [Google Scholar]
- Bligh, E.G.; Dyer, W.J. A RAPID METHOD OF TOTAL LIPID EXTRACTION AND PURIFICATION. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Hošek, J.; Závalová, V.; Kollár, P. Effect of solvent on cytotoxicity and bioavailability of fatty acids. Immunopharmacol. Immunotoxicol. 2010, 32, 462–465. [Google Scholar] [CrossRef]
- Banning, A.; Florian, S.; Deubel, S.; Thalmann, S.; Müller-Schmehl, K.; Jacobasch, G.; Brigelius-Flohé, R. GPx2 Counteracts PGE2Production by Dampening COX-2 and mPGES-1 Expression in Human Colon Cancer Cells. Antioxidants Redox Signal. 2008, 10, 1491–1500. [Google Scholar] [CrossRef]
- Wallert, M.; Schmölz, L.; Koeberle, A.; Krauth, V.; Glei, M.; Galli, F.; Werz, O.; Birringer, M.; Lorkowski, S. α-Tocopherol long-chain metabolite α-13′-COOH affects the inflammatory response of lipopolysaccharide-activated murine RAW264.7 macrophages. Mol. Nutr. Food Res. 2015, 59, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Stolle, K.; Schnoor, M.; Fuellen, G.; Spitzer, M.; Cullen, P.; Lorkowski, S. Cloning, genomic organization, and tissue-specific expression of the RASL11B gene. Biochim. Biophys. Acta 2007, 1769, 514–524. [Google Scholar] [CrossRef]
- Stolle, K.; Schnoor, M.; Fuellen, G.; Spitzer, M.; Engel, T.; Spener, F.; Cullen, P.; Lorkowski, S. Cloning, cellular localization, genomic organization, and tissue-specific expression of the TGFβ1-inducible SMAP-5 gene. Gene 2005, 351, 119–130. [Google Scholar] [CrossRef]
- Schmölz, L.; Wallert, M.; Lorkowski, S. Optimized incubation regime for nitric oxide measurements in murine macrophages using the Griess assay. J. Immunol. Methods 2017, 449, 68–70. [Google Scholar] [CrossRef]
- Schaible, A.; Koeberle, A.; Northoff, H.; Lawrenz, B.; Weinigel, C.; Barz, D.; Werz, O.; Pergola, C. High capacity for leukotriene biosynthesis in peripheral blood during pregnancy. Prostaglandins Leukot. Essent. Fat. Acids 2013, 89, 245–255. [Google Scholar] [CrossRef]
- Werz, O.; Gerstmeier, J.; Libreros, S.; De La Rosa, X.; Werner, M.; Norris, P.; Chiang, N.; Serhan, C.N. Human macrophages differentially produce specific resolvin or leukotriene signals that depend on bacterial pathogenicity. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pein, H.; Ville, A.; Pace, S.; Temml, V.; Garscha, U.; Raasch, M.; Alsabil, K.; Viault, G.; Dinh, C.-P.; Guilet, D.; et al. Endogenous metabolites of vitamin E limit inflammation by targeting 5-lipoxygenase. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Piroddi, M.; Albini, A.; Fabiani, R.; Giovannelli, L.; Luceri, C.; Natella, F.; Rosignoli, P.; Rossi, T.; Taticchi, A.; Servili, M.; et al. Nutrigenomics of extra-virgin olive oil: A review. BioFactors 2016, 43, 17–41. [Google Scholar] [CrossRef] [PubMed]
- Estruch, R.; Ros, E.; Salas-Salvadó, J.; Covas, M.-I.; Corella, D.; Arós, F.; Gómez-Gracia, E.; Ruiz-Gutiérrez, V.; Fiol, M.; Lapetra, J.; et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N. Engl. J. Med. 2018, 378, e34. [Google Scholar] [CrossRef]
- Cárdeno, A.; Sánchez-Hidalgo, M.; Aparicio-Soto, M.; Sánchez-Fidalgo, S.; Alarcón-De-La-Lastra, C. Extra virgin olive oil polyphenolic extracts downregulate inflammatory responses in LPS-activated murine peritoneal macrophages suppressing NFkB and MAPK signalling pathways. Food Function 2014, 5, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Huang, G.; Wang, Z.; Zhuang, S.; Xu, L.; Song, B.; Xiong, Y.; Guan, S. Tyrosol exhibits negative regulatory effects on LPS response and endotoxemia. Food Chem. Toxicol. 2013, 62, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Casas, R.; Estruch, R.; Sacanella, E. The Protective Effects of Extra Virgin Olive Oil on Immune-mediated Inflammatory Responses. Endocr. Metab. Immune Disord. Drug Targets 2018, 18, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Morinelli, T.A.; Meier, K.E.; Rosenzweig, S.A.; Egan, B.M. Oleic Acid–Induced Mitogenic Signaling in Vascular Smooth Muscle Cells. Circ. Res. 1996, 79, 611–619. [Google Scholar] [CrossRef]
- Jump, D.B.; Clarke, S.D. Regulation of gene expression by dietary fat. Annu. Rev. Nutr. 1999, 19, 63–90. [Google Scholar] [CrossRef]
- Clore, J.N.; Allred, J.; White, D.; Li, J.; Stillman, J. The role of plasma fatty acid composition in endogenous glucose production in patients with type 2 diabetes mellitus. Metabolism 2002, 51, 1471–1477. [Google Scholar] [CrossRef]
- De Lima, T.M.; Lima, L.D.S.; Scavone, C.; Curi, R. Fatty acid control of nitric oxide production by macrophages. FEBS Lett. 2006, 580, 3287–3295. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, W. Docosahexaenoic acid suppresses nitric oxide production and inducible nitric oxide synthase expression in interferon-? plus lipopolysaccharide-stimulated murine macrophages by inhibiting the oxidative stress. Free. Radic. Biol. Med. 2003, 34, 1006–1016. [Google Scholar] [CrossRef]
- Lee, J.Y.; Sohn, K.H.; Rhee, S.H.; Hwang, D. Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J. Biol. Chem. 2001, 276, 16683–16689. [Google Scholar] [CrossRef] [Green Version]
- Wanidworanun, C.; Strober, W. Predominant role of tumor necrosis factor-alpha in human monocyte IL-10 synthesis. J. Immunol. 1993, 151, 6853–6861. [Google Scholar]
- de Lima-Salgado, T.M.; Alba-Loureiro, T.C.; do Nascimento, C.S.; Nunes, M.T.; Curi, R. Molecular mechanisms by which saturated fatty acids modulate TNF-alpha expression in mouse macrophage lineage. Cell Biochem. Biophys. 2011, 59, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.L.; Lamon-Fava, S.; Matthan, N.R.; Wu, D.; Lichtenstein, A.H. EPA and DHA Exposure Alters the Inflammatory Response but not the Surface Expression of Toll-like Receptor 4 in Macrophages. Lipids 2015, 50, 121–129. [Google Scholar] [CrossRef] [Green Version]
- Weldon, S.M.; Mullen, A.C.; Loscher, C.E.; Hurley, L.A.; Roche, H.M. Docosahexaenoic acid induces an anti-inflammatory profile in lipopolysaccharide-stimulated human THP-1 macrophages more effectively than eicosapentaenoic acid. J. Nutr. Biochem. 2007, 18, 250–258. [Google Scholar] [CrossRef]
- Zhao, G.; Etherton, T.D.; Martin, K.R.; Heuvel, J.P.V.; Gillies, P.J.; West, S.G.; Kris-Etherton, P.M. Anti-inflammatory effects of polyunsaturated fatty acids in THP-1 cells. Biochem. Biophys. Res. Commun. 2005, 336, 909–917. [Google Scholar] [CrossRef]
- Jin, J.; Lu, Z.; Li, Y.; Cowart, L.A.; Lopes-Virella, M.F.; Huang, Y. Docosahexaenoic acid antagonizes the boosting effect of palmitic acid on LPS inflammatory signaling by inhibiting gene transcription and ceramide synthesis. PLoS ONE 2018, 13, e0193343. [Google Scholar] [CrossRef] [Green Version]
- Tsatsanis, C.; Androulidaki, A.; Venihaki, M.; Margioris, A.N. Signalling networks regulating cyclooxygenase-2. Int. J. Biochem. Cell Biol. 2006, 38, 1654–1661. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. The relationship between the fatty acid composition of immune cells and their function. Prostaglandins, Leukot. Essent. Fat. Acids 2008, 79, 101–108. [Google Scholar] [CrossRef]
- Kita, Y.; Shindou, H.; Shimizu, T. Cytosolic phospholipase A(2) and lysophospholipid acyltransferases. Biochim. Biophys Acta Mol Cell Biol Lipids 2019, 1864, 838–845. [Google Scholar] [CrossRef]
- Tsai, P.J.; Huang, W.C.; Lin, S.W.; Chen, S.N.; Shen, H.J.; Chang, H.; Chuang, L.T. Juniperonic Acid Incorporation into the Phospholipids of Murine Macrophage Cells Modulates Pro-Inflammatory Mediator Production. Inflammation 2018, 41, 1200–1214. [Google Scholar] [CrossRef]


| FAME | OA (µM) | |||||
|---|---|---|---|---|---|---|
| 0 | 0 | 10 | 50 | 100 | 200 | |
| LPS (0.1 µg/mL) | ||||||
| C16:0 (PA) 2 | 28.1 ± 4.8 | 32.0 ± 1.0 | 28.9 ± 1.9 | 25.0 ± 3.0 | 21.4 ± 7.6 | 16.3 ± 5.7 ** |
| C16:1 (POA) 3 | 2.2 ± 0.5 | 2.2 ± 0.2 | 1.9 ± 0.3 | 0.8 ± 0.0 *** | 0.6 ± 0.1 *** | 0.5 ± 0.1 *** |
| C18:0 (SA) 4 | 28.0 ± 9.8 | 32.5 ± 2.7 | 28.1 ± 1.7 | 26.1 ± 2.3 | 22.9 ± 2.8 | 20.9 ± 4.8 * |
| C18:1 (OA) 5 | 18.5 ± 3.1 | 15.6 ± 1.7 | 20.9 ± 2.4 | 30.3 ± 2.8 | 40.2 ± 9.8 ** | 47.2 ± 12.8 *** |
| C18:2 (LA) 6 | 1.6 ± 0.5 | 1.3 ± 0.4 | 1.4 ± 0.2 | 1.3 ± 0.3 | 1.4 ± 0.7 | 1.9 ± 1.8 |
| C20:1 (GA) 7 | 0.7 ± 0.1 | 0.5 ± 0.1 | 0.8 ± 0.2 | 1.2 ± 0.2 | 1.5 ± 0.4 * | 1.6 ± 0.6 ** |
| C20:4 (AA) 8 | 1.4 ± 0.4 | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.6 ± 0.1 | 0.3 ± 0.1 ** | 0.3 ± 0.1 ** |
| C22:4 (AdA) 9 | 0.0 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.1 | 0.2 ± 0.0 | 0.2 ± 0.1 | 0.2 ± 0.1 |
| FAME | 0 µM OA | 200 µM OA | 0 µM OA + LPS (0.1 µg/mL) | 200 µM OA + LPS (0.1 µg/mL) |
|---|---|---|---|---|
| C16:0 (PA) 2 | 19.4 ± 1.7 | 14.0 ± 3.2 * | 20.7 ± 6.5 | 12.8 ± 3.8 * |
| C16:1 (POA) 3 | 1.6 ± 0.7 | 0.9 ± 0.6 | 1.5 ± 0.4 | 0.7 ± 0.3 * |
| C18:0 (SA) 4 | 13.4 ± 2.6 | 9.4 ± 1.5 | 13.7 ± 4.0 | 9.9 ± 4.7 |
| C18:1 (OA) 5 | 13.9 ± 5.2 | 22.4 ± 5.3 * | 12.6 ± 5.1 | 33.3 ± 12.3 * |
| C18:2 (LA) 6 | 2.0 ± 1.5 | 2.4 ± 0.6 * | 1.3 ± 0.5 | 1.6 ± 0.6 |
| C20:1 (GA) 7 | 0.8 ± 0.3 | 1.7 ± 1.0 | 2.1 ± 4.0 | 1.2 ± 0.6 |
| C20:4 (AA) 8 | 0.7 ± 0.5 | 0.6 ± 0.6 | 0.3 ± 0.4 | 0.2 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, A.K.; Albrecht, F.; Rohrer, C.; Koeberle, A.; Werz, O.; Schlörmann, W.; Glei, M.; Lorkowski, S.; Wallert, M. Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2. Nutrients 2021, 13, 4437. https://doi.org/10.3390/nu13124437
Müller AK, Albrecht F, Rohrer C, Koeberle A, Werz O, Schlörmann W, Glei M, Lorkowski S, Wallert M. Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2. Nutrients. 2021; 13(12):4437. https://doi.org/10.3390/nu13124437
Chicago/Turabian StyleMüller, Anke Katharina, Franziska Albrecht, Carsten Rohrer, Andreas Koeberle, Oliver Werz, Wiebke Schlörmann, Michael Glei, Stefan Lorkowski, and Maria Wallert. 2021. "Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2" Nutrients 13, no. 12: 4437. https://doi.org/10.3390/nu13124437
APA StyleMüller, A. K., Albrecht, F., Rohrer, C., Koeberle, A., Werz, O., Schlörmann, W., Glei, M., Lorkowski, S., & Wallert, M. (2021). Olive Oil Extracts and Oleic Acid Attenuate the LPS-Induced Inflammatory Response in Murine RAW264.7 Macrophages but Induce the Release of Prostaglandin E2. Nutrients, 13(12), 4437. https://doi.org/10.3390/nu13124437