Inhibitory Effect of Astaxanthin on Gene Expression Changes in Helicobacter pylori-Infected Human Gastric Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Line and Culture Conditions
2.2. Bacterial Strain
2.3. Treatment of AGS Cells with ASTX and H. pylori Infection
2.4. Preparation of Total RNA Extracts and Library Construction
2.5. RNA-Sequencing and Bioinformatics Analysis
2.6. Validation of DEGs Using Real-Time Polymerase Chain Reaction (PCR)
2.7. Statistical Analyses
3. Results
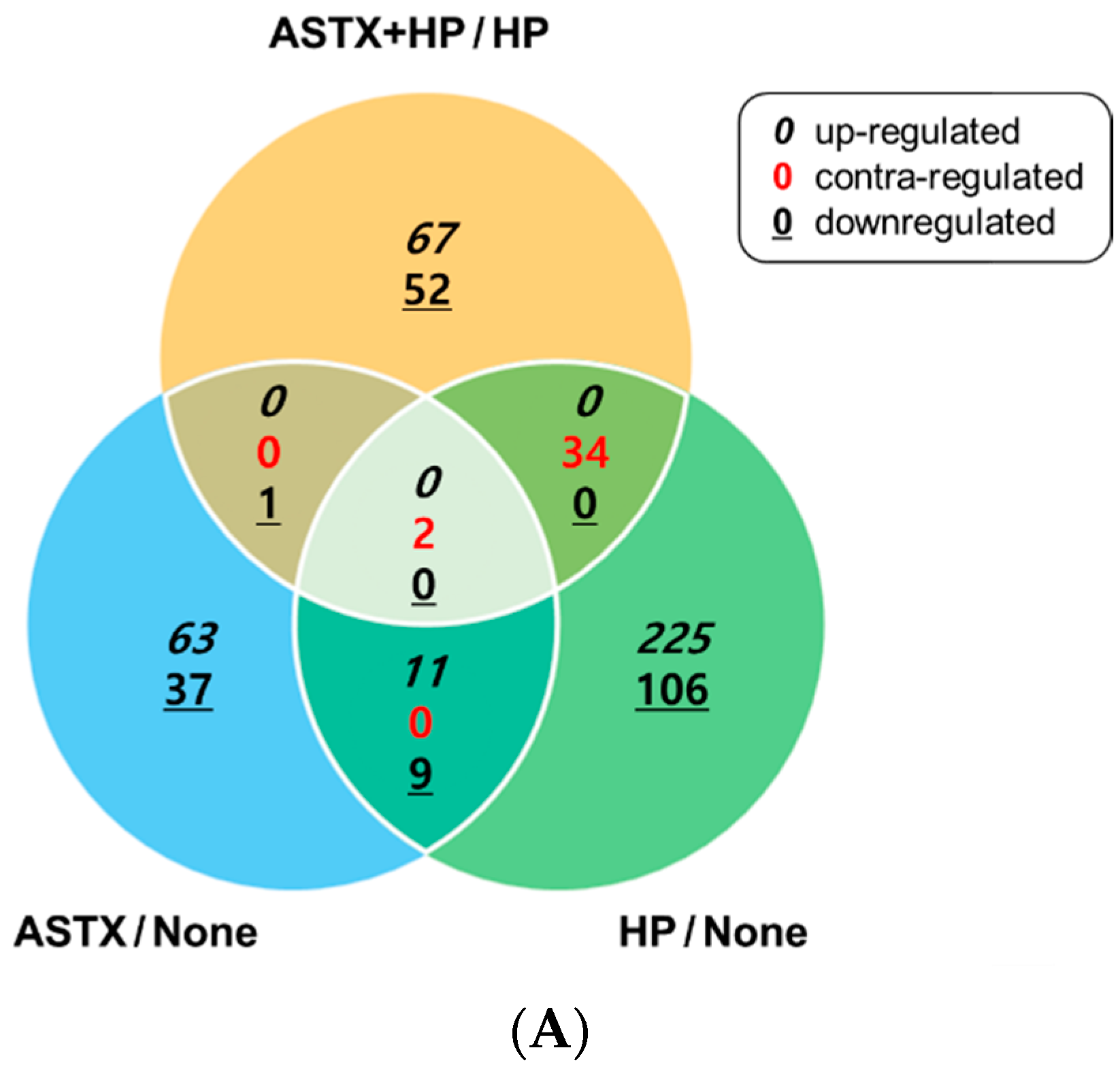
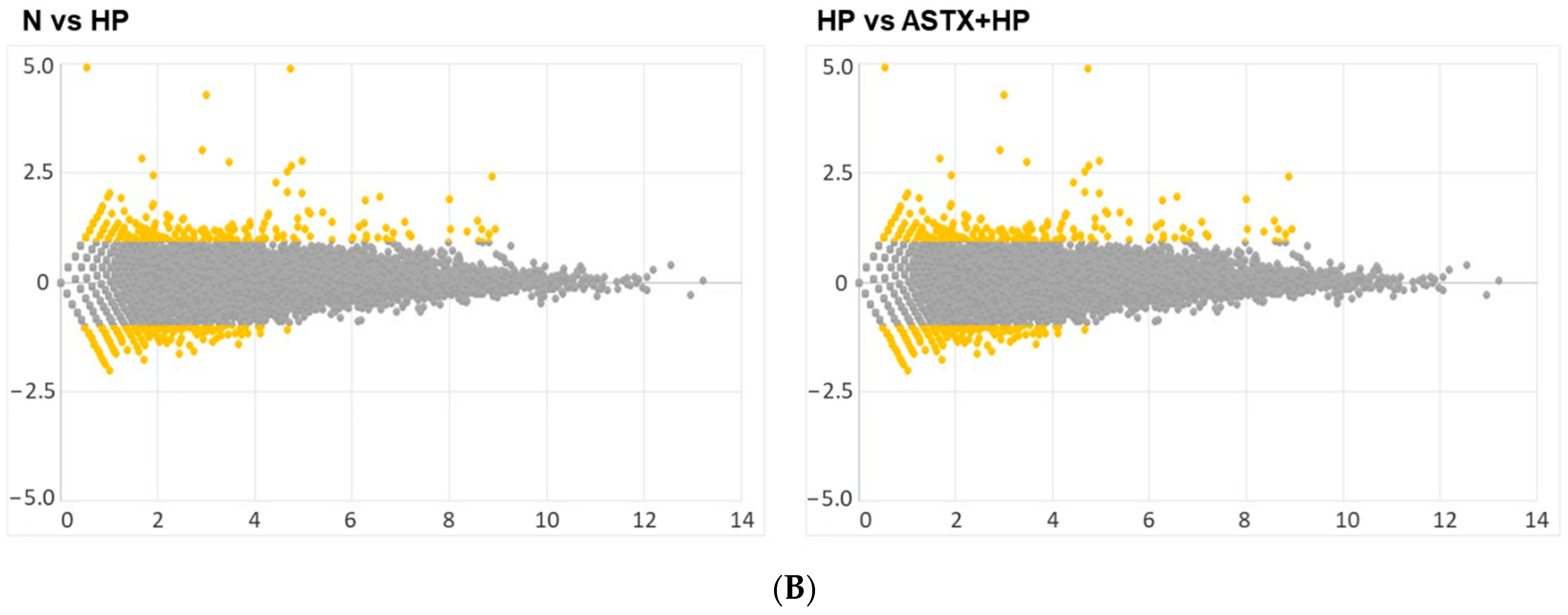
3.1. Gene Expression Profile of H. pylori-Infected and/or ASTX-Treated AGS Cells
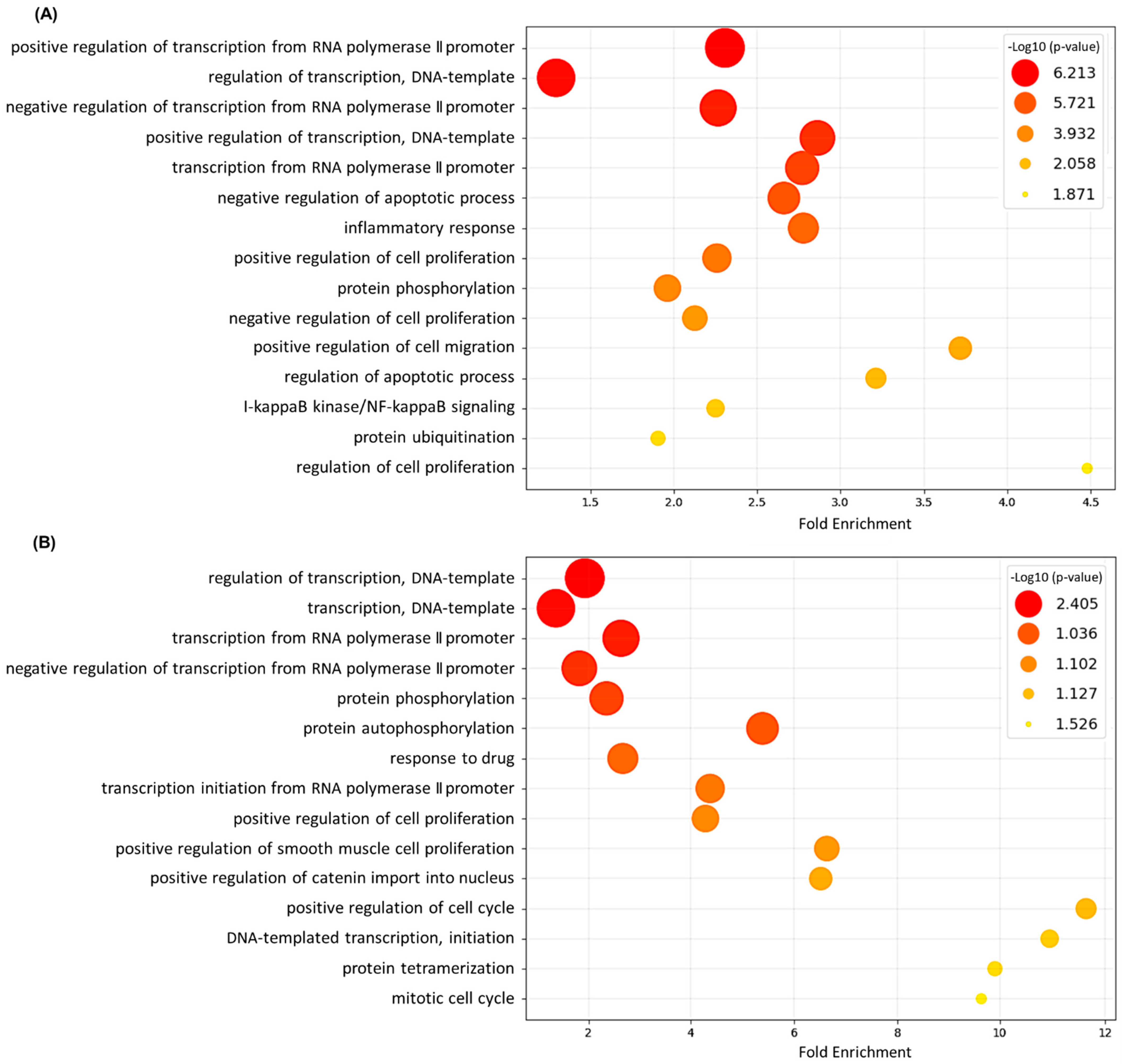
3.2. Gene Ontology (GO) Annotation and Functional Analysis of DEGs
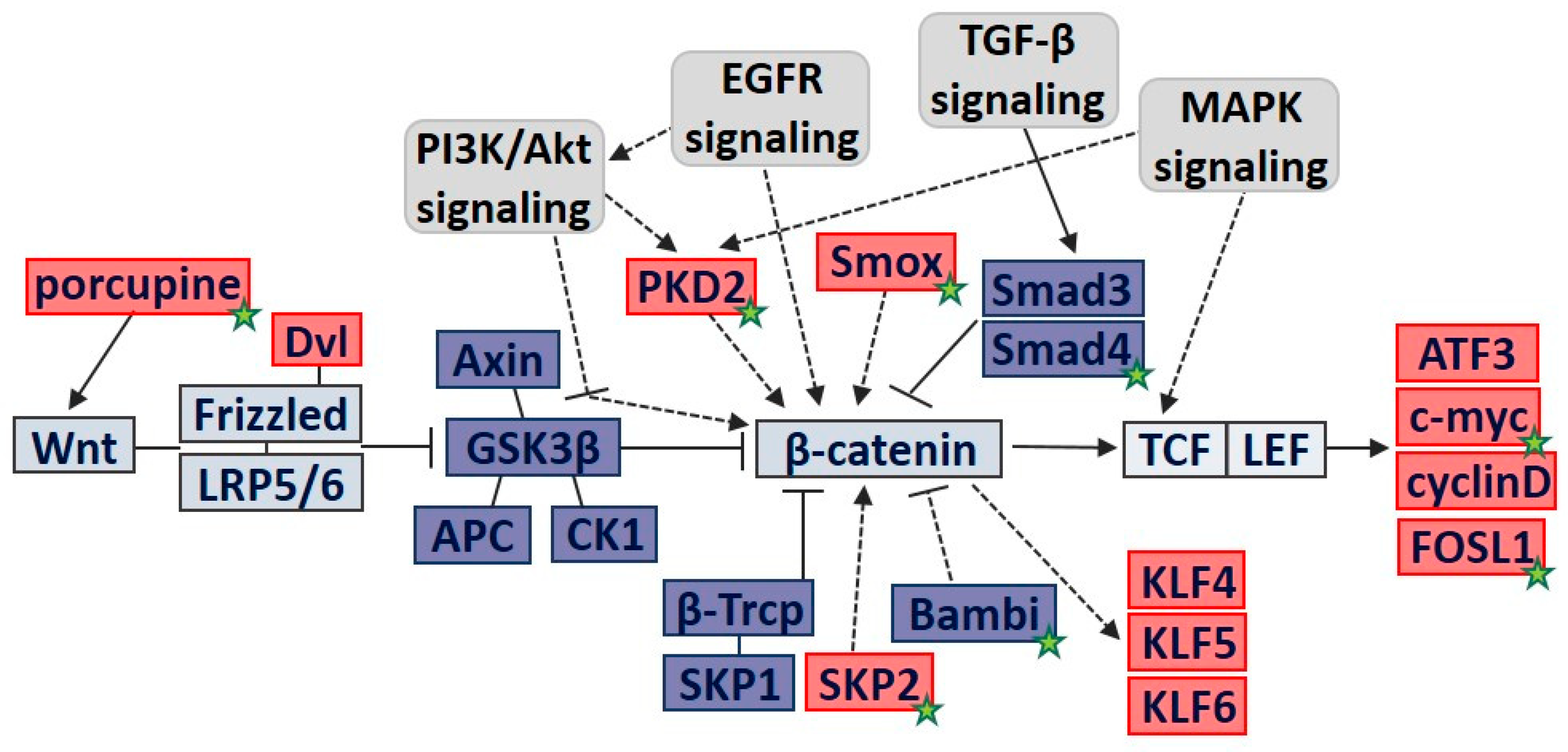
3.3. Molecular Pathway Analysis of DEGs Involved in Cell Proliferation
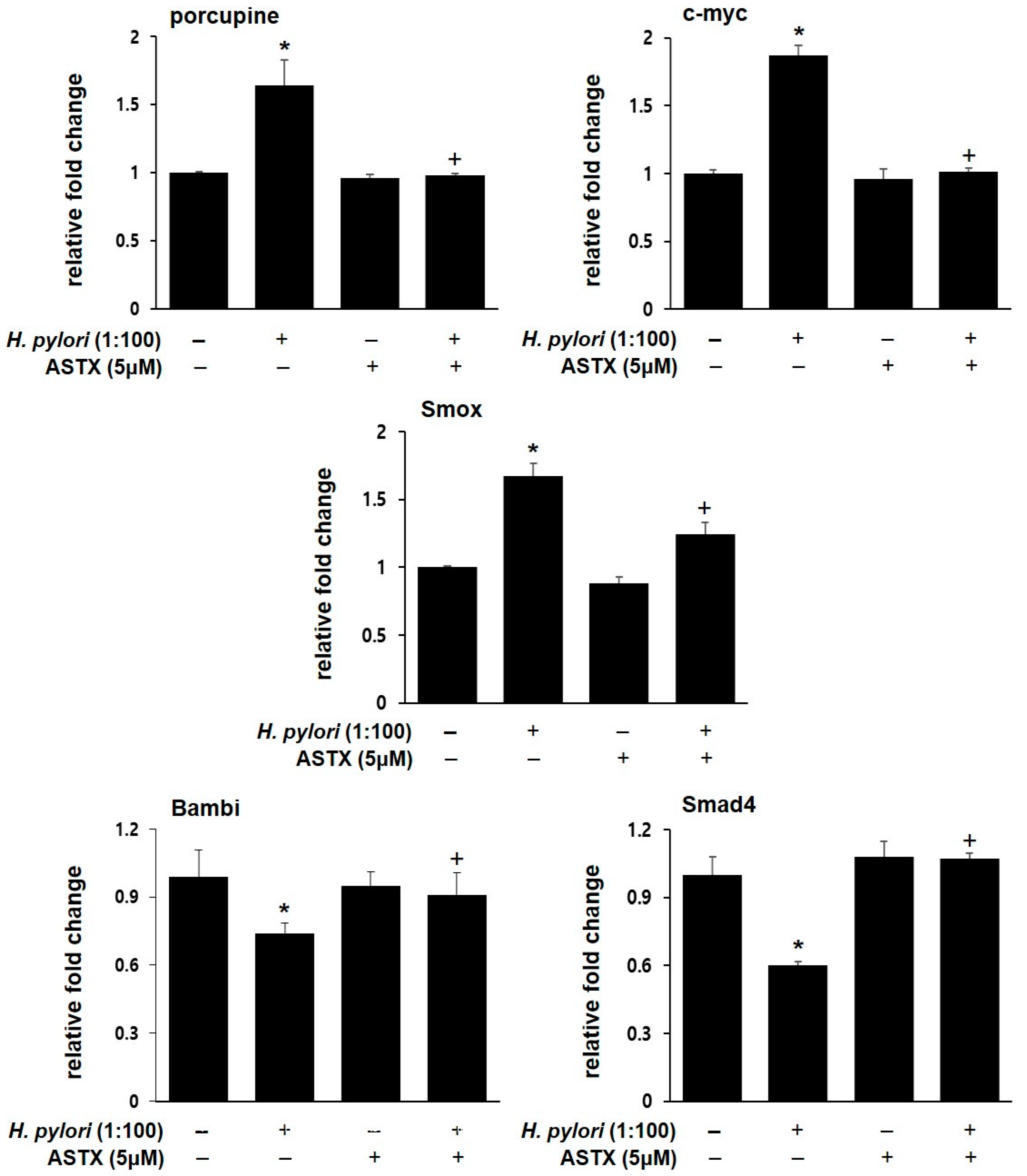
3.4. PCR Validation of Key Regulatory Genes Involved in H. pylori-Induced β-catenin Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thrift, A.P.; El-Serag, H.B. Burden of gastric cancer. Clin. Gastroenterol. Hepatol. 2020, 18, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Ghotaslou, R.; Leylabadlo, H.E.; Nasiri, M.J.; Dabiri, H.; Hashemi, A. Risk of gastric cancer in association with Helicobacter pylori different virulence factors: A systematic review and meta-analysis. Microb. Pathog. 2018, 118, 214–219. [Google Scholar]
- Díaz, P.; Valenzuela Valderrama, M.; Bravo, J.; Quest, A.F. Helicobacter pylori and gastric cancer: Adaptive cellular mechanisms involved in disease progression. Front. Microbiol. 2018, 9, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.Z.; Goldberg, J.B.; Hatakeyama, M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010, 6, 851–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polk, D.B.; Peek, R.M. Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 2010, 10, 403–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, V.S.; Ng, S.S.; Boersema, P.J.; Low, T.Y.; Karthaus, W.R.; Gerlach, J.P.; Mohammed, S.; Heck, A.J.; Maurice, M.M.; Mahmoudi, T.; et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012, 149, 1245–1256. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Xin, N.; Wang, W.; Zhao, C. Wnt/β-catenin, an oncogenic pathway targeted by H.pylori in gastric carcinogenesis. Oncotarget 2015, 6, 35579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Lim, J.W.; Kim, H. Astaxanthin inhibits Helicobacter pylori-induced inflammatory and oncogenic responses in gastric mucosal tissues of mice. J. Cancer Prev. 2020, 25, 244. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lim, J.W.; Kim, H. β-carotene inhibits expression of c-myc and cyclin E in Helicobacter pylori-infected gastric epithelial cells. J. Cancer Prev. 2019, 24, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambati, R.R.; Phang, S.M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, extraction, stability, biological activities and its commercial applications—A review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef]
- Hussein, G.; Sankawa, U.; Goto, H.; Matsumoto, K.; Watanabe, H. Astaxanthin, a carotenoid with potential in human health and nutrition. J. Nat. Prod. 2006, 69, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.C.; Yusoff, F.M.; Shariff, M.; Kamarudin, M.S. Astaxanthin as feed supplement in aquatic animals. Rev. Aquac. 2018, 10, 738–773. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, J.J.; Lee, B.J.; Joo, M.K.; Chun, H.J.; Lee, S.W.; Bak, Y.T. Astaxanthin inhibits proliferation of human gastric cancer cell lines by interrupting cell cycle progression. Gut Liver 2016, 10, 369. [Google Scholar] [CrossRef] [Green Version]
- Su, X.Z.; Chen, R.; Wang, C.B.; Ouyang, X.L.; Jiang, Y.; Zhu, M.Y. Astaxanthin combine with human serum albumin to abrogate cell proliferation, migration, and drug-resistant in human ovarian carcinoma SKOV3 cells. Anti-Cancer Agents Med. Chem. 2019, 19, 792–801. [Google Scholar] [CrossRef]
- McCall, B.; McPartland, C.K.; Moore, R.; Frank-Kamenetskii, A.; Booth, B.W. Effects of astaxanthin on the proliferation and migration of breast cancer cells in vitro. Antioxidants 2018, 7, 135. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Lim, J.W.; Kim, H. Astaxanthin Inhibits Mitochondrial dysfunction and interleukin-8 expression in Helicobacter pylori-infected gastric epithelial cells. Nutrients 2018, 10, 1320. [Google Scholar] [CrossRef] [Green Version]
- Park, B.; Lim, J.W.; Kim, H. Lycopene treatment inhibits activation of Jak1/Stat3 and Wnt/β-catenin signaling and attenuates hyperproliferation in gastric epithelial cells. Nutr. Res. 2019, 70, 70–81. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, H. Transcriptome Analysis of the Inhibitory Effect of Astaxanthin on Helicobacter pylori-Induced Gastric Carcinoma Cell Motility. Mar. Drugs 2020, 18, 365. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Multi-layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/β-catenin signaling activation. Int. J. Mol. Med. 2018, 42, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Sierra, J.C.; Piazuelo, M.B.; Luis, P.B.; Barry, D.P.; Allaman, M.M.; Asim, M.; Sebrell, T.A.; Finley, J.L.; Rose, K.L.; Hill, S.; et al. Spermine oxidase mediates Helicobacter pylori-induced gastric inflammation, DNA damage, and carcinogenic signaling. Oncogene 2020, 39, 4465–4474. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Song, X.; Ma, H.; Liu, L.; Wen, X.; Yu, J.; Wang, L.; Hu, S. Knockdown of BAMBI inhibits β-catenin and transforming growth factor β to suppress metastasis of gastric cancer cells. Mol. Med. Rep. 2014, 10, 874–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khin, S.S.; Kitazawa, R.; Win, N.; Aye, T.T.; Mori, K.; Kondo, T.; Kitazawa, S. BAMBI gene is epigenetically silenced in subset of high-grade bladder cancer. Int. J. Cancer 2009, 125, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Marwitz, S.; Depner, S.; Dvornikov, D.; Merkle, R.; Szczygieł, M.; Müller-Decker, K.; Lucarelli, P.; Wäsch, M.; Mairbäurl, H.; Rabe, K.F.; et al. Downregulation of the TGFβ pseudoreceptor BAMBI in non–small cell lung cancer enhances TGFβ signaling and invasion. Cancer Res. 2016, 76, 3785–3801. [Google Scholar] [CrossRef] [Green Version]
- Powell, S.M.; Harper, J.C.; Hamilton, S.R.; Robinson, C.R.; Cummings, O.W. Inactivation of Smad4 in gastric carcinomas. Cancer Res. 1997, 57, 4221–4224. [Google Scholar] [PubMed]
- Xiao, B.; Zhu, E.D.; Li, N.; Lu, D.S.; Li, W.; Li, B.S.; Zhao, Y.L.; Mao, X.H.; Guo, G.; Yu, P.W.; et al. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol. Rep. 2012, 27, 559–566. [Google Scholar]
- Yousefi, B.; Mohammadlou, M.; Abdollahi, M.; Salek Farrokhi, A.; Karbalaei, M.; Keikha, M.; Kokhaei, P.; Valizadeh, S.; Rezaiemanesh, A.; Arabkari, V.; et al. Epigenetic changes in gastric cancer induction by Helicobacter pylori. J. Cell. Physiol. 2019, 234, 21770–21784. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, R.; Zhang, X.; Qiu, B.; Chen, T.; Li, Z.; Xu, Y.; Zhang, Z. Transcription factor 7 promotes the progression of perihilar cholangiocarcinoma by inducing the transcription of c-Myc and FOS-like antigen 1. EBioMedicine 2019, 45, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Kavitha, K.; Kowshik, J.; Kishore, T.K.K.; Baba, A.B.; Nagini, S. Astaxanthin inhibits NF-κB and Wnt/β-catenin signaling pathways via inactivation of Erk/MAPK and PI3K/Akt to induce intrinsic apoptosis in a hamster model of oral cancer. Biochim. Biophys. Acta 2013, 1830, 4433–4444. [Google Scholar] [CrossRef]
- Li, J.; Dai, W.; Xia, Y.; Chen, K.; Li, S.; Liu, T.; Zhang, R.; Wang, J.; Lu, W.; Zhou, Y.; et al. Astaxanthin inhibits proliferation and induces apoptosis of human hepatocellular carcinoma cells via Inhibition of NF-κB P65 and Wnt/β-catenin in vitro. Mar. Drugs 2015, 13, 6064–6081. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.L.; Li, M.R.; Chen, Z.; Liu, X.W.; Sheng, Q.; Zhou, H.M. Inhibition of the Wnt palmitoyltransferase porcupine suppresses cell growth and downregulates the Wnt/β-catenin pathway in gastric cancer. Oncol. Lett. 2013, 5, 1719–1723. [Google Scholar] [CrossRef] [Green Version]
- Casero, R.A.; Stewart, T.M.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Jadallah, S.; Toubaji, A.; Lecksell, K.; Hicks, J.L.; Kowalski, J.; Bova, G.S.; Marzo, A.M.; Netto, G.J.; Casero, R.A. Increased spermine oxidase expression in human prostate cancer and prostatic intraepithelial neoplasia tissues. Prostate 2008, 68, 766–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Chaturvedi, R.; Cheng, Y.; Bussiere, F.I.; Asim, M.; Yao, M.D.; Potosky, D.; Meltzer, S.J.; Rhee, J.G.; Kim, S.S.; et al. Spermine oxidation induced by Helicobacter pylori results in apoptosis and DNA damage: Implications for gastric carcinogenesis. Cancer Res. 2004, 64, 8521–8525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, R.; Asim, M.; Romero–Gallo, J.; Barry, D.P.; Hoge, S.; De Sablet, T.; Delgado, A.G.; Wroblewski, L.E.; Piazuelo, M.B.; Yan, F.; et al. Spermine oxidase mediates the gastric cancer risk associated with Helicobacter pylori CagA. Gastroenterology 2011, 141, 1696–1708. [Google Scholar] [CrossRef] [Green Version]
- Sekiya, T.; Adachi, S.; Kohu, K.; Yamada, T.; Higuchi, O.; Furukawa, Y.; Nakamura, Y.; Nakamura, T.; Tashiro, K.; Kuhara, S.; et al. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-β signaling, as a target of the β-catenin pathway in colorectal tumor cells. J. Biol. Chem. 2004, 279, 6840–6846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Lee, M.J.; Zhang, J.; Yu, G.R.; Kim, D.G. C-terminal-truncated HBV X promotes hepato-oncogenesis through inhibition of tumor-suppressive β-catenin/BAMBI signaling. Exp. Mol. Med. 2016, 48, e275. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.H.; Kim, S.H.; Lee, J.H.; Choi, Y.L.; Kim, Y.C.; Park, T.S.; Hong, Y.C.; Wu, C.F.; Shin, Y.K. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin. Cancer Res. 2007, 13, 102–110. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.L.; Li, Z.; Wang, W.Z.; Chen, Z.; Zhang, L.; Li, Q.; Wei, S.; Li, B.W.; Xu, J.H.; Chen, L.; et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J. Gastroenterol. 2018, 53, 725–739. [Google Scholar] [CrossRef] [Green Version]
- Faraone, I.; Sinisgalli, C.; Ostuni, A.; Armentano, M.F.; Carmosino, M.; Milella, L.; Russo, D.; Labanca, F.; Khan, H. Astaxanthin anticancer effects are mediated through multiple molecular mechanisms: A systematic review. Pharmacol. Res. 2020, 155, 104689. [Google Scholar] [CrossRef] [PubMed]
- El Omari, N.; Bakha, M.; Imtara, H.; Guaouguaoua, F.E.; Balahbib, A.; Zengin, G.; Bouyahya, A. Anticancer mechanisms of phytochemical compounds: Focusing on epigenetic targets. Environ. Sci. Pollut. Res. 2021, 28, 1–35. [Google Scholar] [CrossRef]

| Gene Symbol | Fold Change | Gene Expression Value | Encoded Protein | ||||
|---|---|---|---|---|---|---|---|
| HP/ None | ASTX + HP/HP | Normalized Data (Log2) | |||||
| None | HP | ASTX | ASTX + HP | ||||
| BAMBI | 0.543 | 1.537 | 6.563 | 5.682 | 6.488 | 6.302 | BMP and activin membrane-bound inhibitor |
| FOSL1 | 2.107 | 0.669 | 4.050 | 5.125 | 4.603 | 4.513 | Fos-like 1 |
| TSPAN31 | 1.517 | 0.513 | 3.900 | 4.501 | 4.352 | 3.537 | tetraspanin 31 |
| E2F7 | 0.640 | 1.635 | 5.146 | 4.501 | 5.185 | 5.210 | E2F transcription factor 7 |
| ATXN1L | 0.630 | 1.514 | 4.629 | 3.964 | 4.423 | 4.562 | ataxin 1 like |
| SMAD4 | 0.661 | 1.437 | 4.853 | 4.215 | 4.663 | 4.838 | SMAD family member 4 |
| ZNF675 | 0.642 | 1.538 | 4.340 | 3.700 | 4.848 | 4.321 | zinc finger protein 675 |
| SERTAD3 | 1.637 | 0.623 | 3.323 | 4.034 | 3.166 | 3.353 | SERTA domain-containing protein 3 |
| TGFB1 | 1.580 | 0.717 | 3.464 | 4.123 | 3.795 | 3.482 | transforming growth factor-β1 |
| SMOX | 1.537 | 0.706 | 4.665 | 5.208 | 4.985 | 4.640 | spermine oxidase |
| PKD2 | 0.585 | 1.680 | 4.809 | 4.034 | 4.764 | 4.783 | polycystin 2 |
| SKP2 | 0.548 | 1.524 | 6.606 | 5.739 | 6.205 | 6.347 | S-phase kinase-associated protein 2 |
| MYC | 1.606 | 0.665 | 4.819 | 5.502 | 5.058 | 5.096 | c-myc |
| PORCN | 1.648 | 0.626 | 3.797 | 4.518 | 3.816 | 3.956 | porcupine |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.H.; Kim, H. Inhibitory Effect of Astaxanthin on Gene Expression Changes in Helicobacter pylori-Infected Human Gastric Epithelial Cells. Nutrients 2021, 13, 4281. https://doi.org/10.3390/nu13124281
Kim SH, Kim H. Inhibitory Effect of Astaxanthin on Gene Expression Changes in Helicobacter pylori-Infected Human Gastric Epithelial Cells. Nutrients. 2021; 13(12):4281. https://doi.org/10.3390/nu13124281
Chicago/Turabian StyleKim, Suhn Hyung, and Hyeyoung Kim. 2021. "Inhibitory Effect of Astaxanthin on Gene Expression Changes in Helicobacter pylori-Infected Human Gastric Epithelial Cells" Nutrients 13, no. 12: 4281. https://doi.org/10.3390/nu13124281
APA StyleKim, S. H., & Kim, H. (2021). Inhibitory Effect of Astaxanthin on Gene Expression Changes in Helicobacter pylori-Infected Human Gastric Epithelial Cells. Nutrients, 13(12), 4281. https://doi.org/10.3390/nu13124281







