Therapeutic Effects of Dietary Soybean Genistein on Triple-Negative Breast Cancer via Regulation of Epigenetic Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
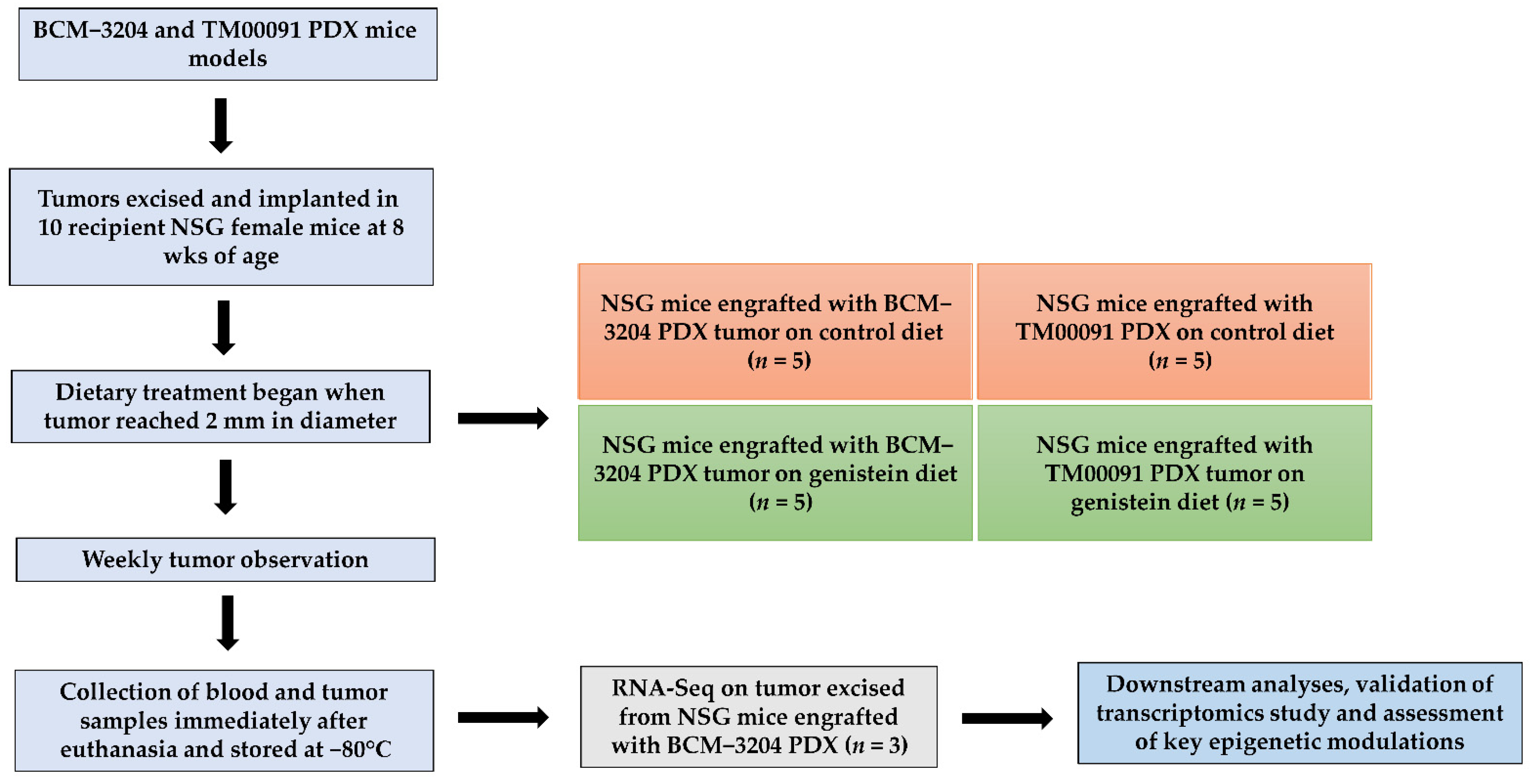
2.1. Animals
2.2. Animal Diets
2.3. Tumor Implantation, Observation, and Collection
2.4. RNA Sequencing (RNA-Seq) Analysis
2.5. Differentially Expressed Gene (DEG) Analysis
2.6. Gene Set Function Enrichment
2.7. Quantitative Real-Time PCR
2.8. Western Blot Analysis
2.9. DNA Methyltransferase (DNMT) and Histone Deacetylase (HDAC) Activity Assay
2.10. Global DNA Methylation, Hydroxymethylation and Histone Methylation Analysis
2.11. Statistical Analysis
3. Results
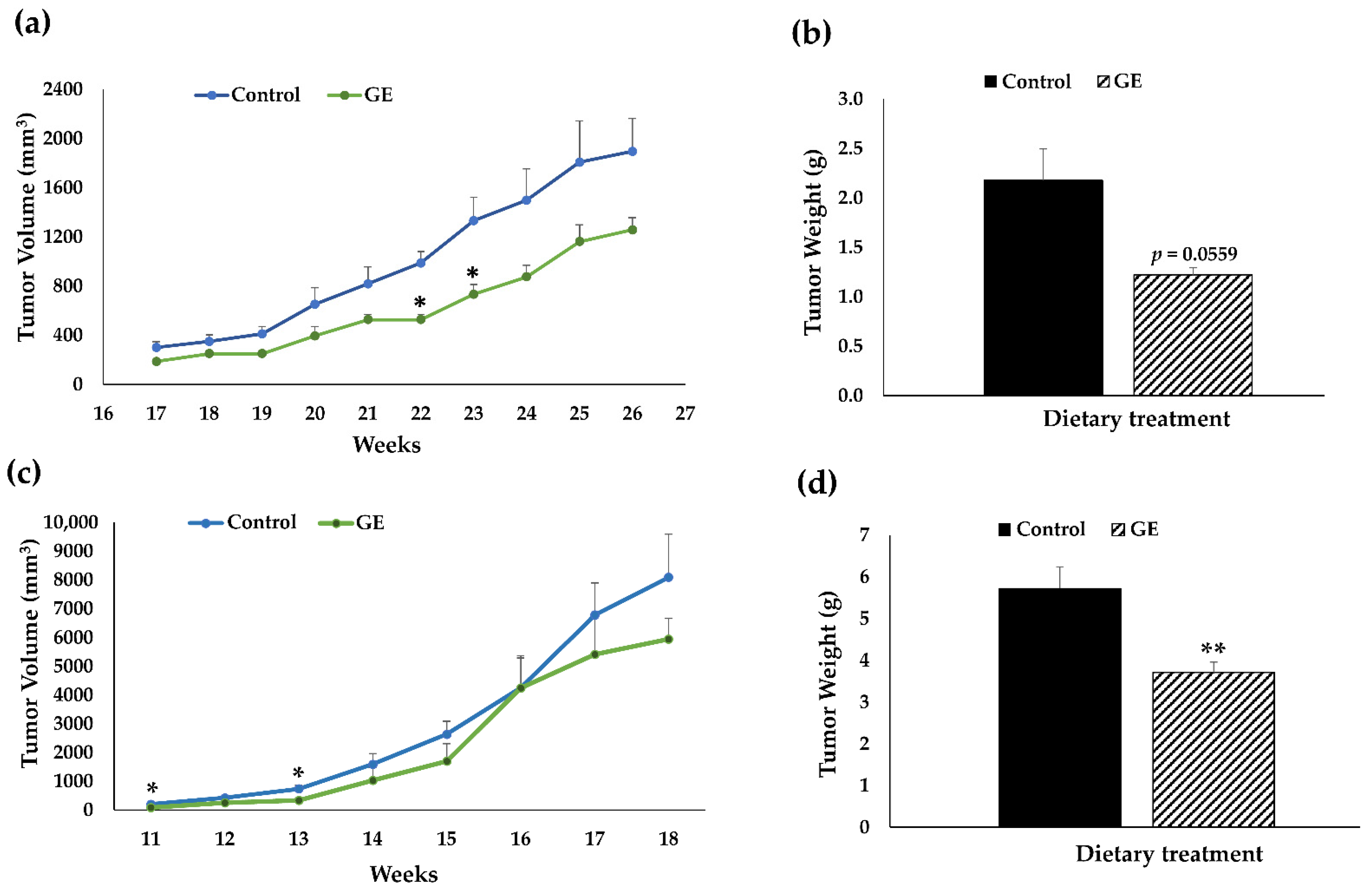
3.1. Dietary GE Inhibited TNBC Growth in PDX Models
3.2. Genome-Wide Transcriptomic Alterations Induced by GE Administration in BCM-3204 PDX Model
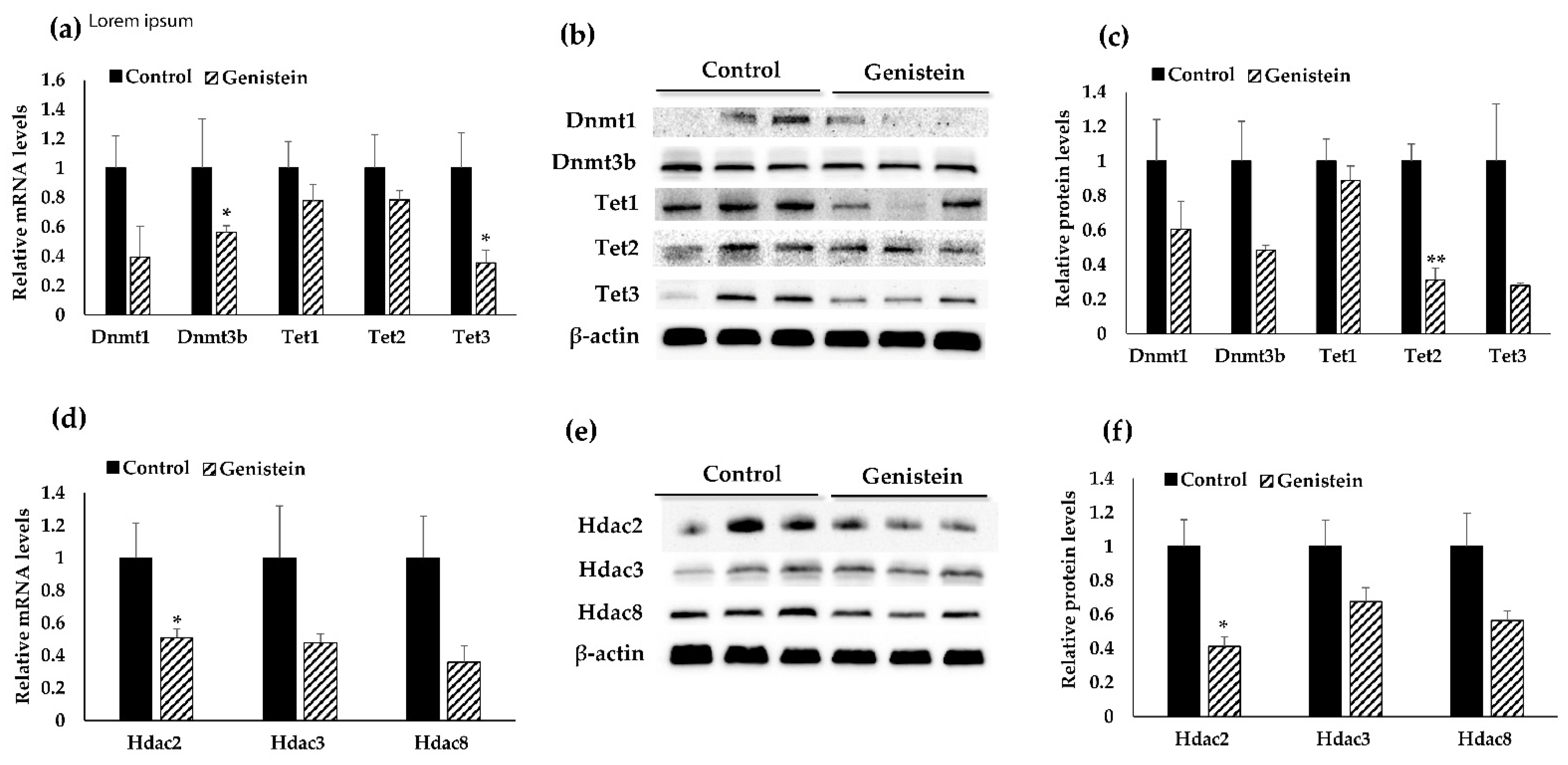
3.3. Validation Analyses of Target Gene Expression at Transcriptional and Translational Levels
3.4. Cd74-Regulated Signaling Pathway May Contribute to GE Diet-Induced Therapeutic Effects against TNBC
3.5. Dietary GE Treatment Resulted in Expression Changes in Multiple Epigenetic-Related Genes
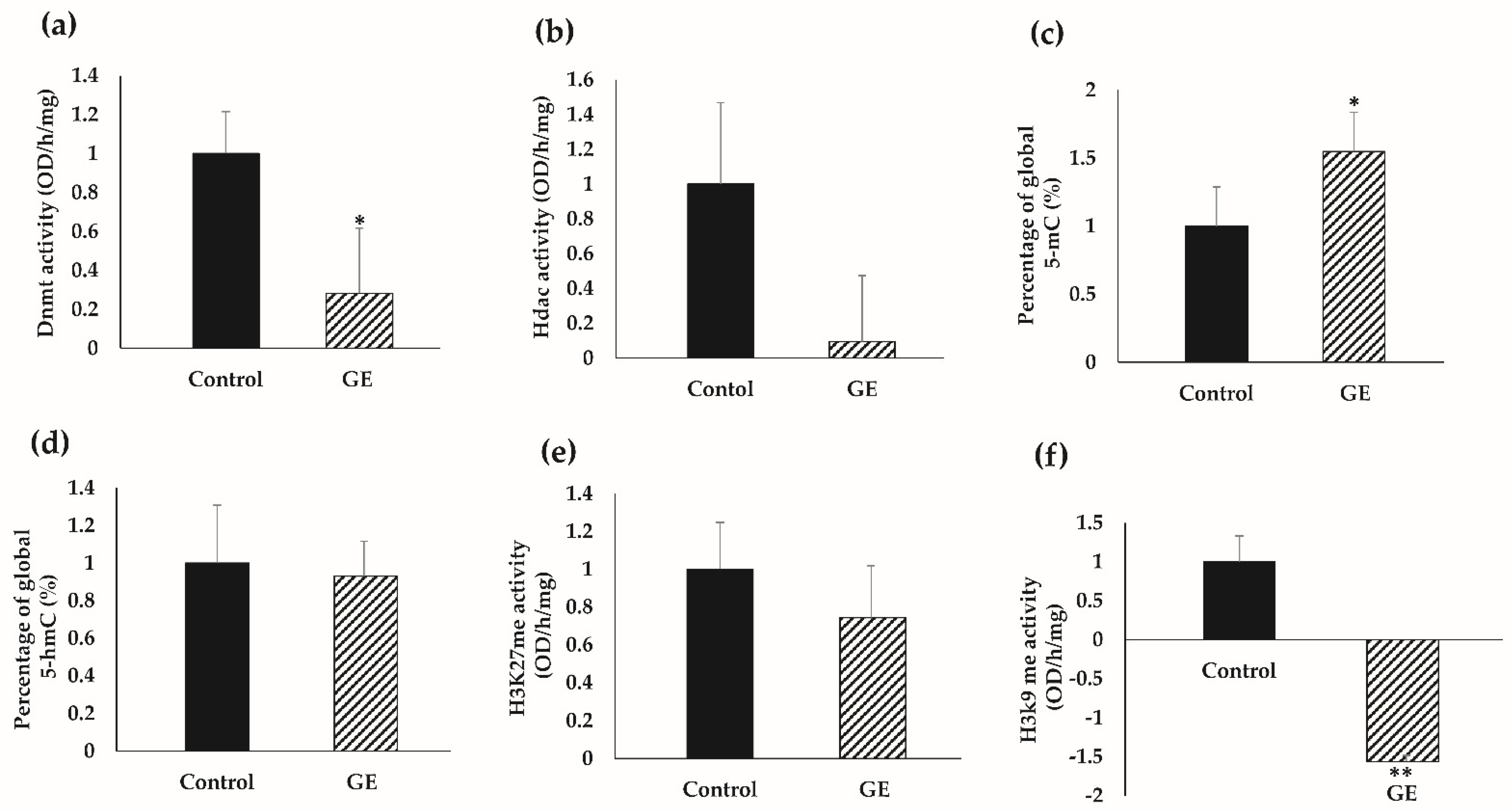
3.6. GE Influenced Global Epigenetic Profiles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Cd74 | Cluster of Differentiation 74 |
| cDNA | Complementary DNA |
| DEGs | Differentially expressed genes |
| Dnmts | DNA methytransferases |
| ER | Estrogen receptor |
| FastQC | FastQ quality control |
| FC | Fold change |
| FDR | False-discovery rate |
| Fzd9 | Frizzled 9 |
| GE | Genistein |
| GO | Gene Ontology |
| Hdac | Histone deacetylase |
| HER-2 | Human epidermal growth factor receptor 2 |
| hTERT | Human telomerase reverse transcriptase |
| Ifi44 | Interferon-Induced Protein 44 |
| Lpl | Lipoprotein lipase |
| NF-κB | Nuclear factor kappa B |
| NOD | Nonobese diabetic |
| NSG | NOD/SCID/IL-2γ-receptor null |
| PCoA | Principal Coordinates Analysis |
| PDX | Patient-derived xenograft |
| PR | Progesterone receptor |
| RNA-seq | RNA sequencing |
| Sat1 | Spermidine/spermine-N1-acetyltransferase 1 |
| TAp63 | Tumor protein p63 |
| Tet | Ten-eleven translocation |
| TNBC | Triple negative breast cancer |
| Wwc1 | WW and C2 domain containing 1 |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef]
- Morris, G.J.; Naidu, S.; Topham, A.K.; Guiles, F.; Xu, Y.; McCue, P.; Schwartz, G.F.; Park, P.K.; Rosenberg, A.L.; Brill, K.; et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and End Results database. Cancer 2007, 110, 876–884. [Google Scholar] [CrossRef]
- Alluri, P.; Newman, L.A. Basal-like and triple-negative breast cancers: Searching for positives among many negatives. Surg. Oncol. Clin. N. Am. 2014, 23, 567–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenwell, M.; Rahman, P.K.S.M. Medicinal Plants: Their Use in Anticancer Treatment. Int. J. Pharm. Sci. Res. 2015, 6, 4103–4112. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.Y.; Tseng, M.; Daly, M.B. Correlates of soy food consumption in women at increased risk for breast cancer. J. Am. Diet. Assoc. 2005, 105, 1552–1558. [Google Scholar] [CrossRef] [Green Version]
- Barnes, S. Effect of genistein on in vitro and in vivo models of cancer. J. Nutr. 1995, 125, 777S–783S. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, N.; Akhtar, J.; Singh, S.P.; Ahsan, F. An Overview on Genistein and its Various Formulations. Drug Res. 2019, 69, 305–313. [Google Scholar] [CrossRef]
- Varinska, L.; Gal, P.; Mojzisova, G.; Mirossay, L.; Mojzis, J. Soy and breast cancer: Focus on angiogenesis. Int. J. Mol. Sci. 2015, 16, 11728–11749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliacci, M.C.; Smacchia, M.; Migliorati, G.; Grignani, F.; Riccardi, C.; Nicoletti, I. Growth-inhibitory effects of the natural phyto-oestrogen genistein in MCF-7 human breast cancer cells. Eur. J. Cancer 1994, 30, 1675–1682. [Google Scholar] [CrossRef]
- Vidya, M. Genistein: Its Role in Breast Cancer Growth and Metastasis. Curr. Drug Metab. 2020, 21, 6–10. [Google Scholar] [CrossRef]
- Tollefsbol, T.O. Methods of epigenetic analysis. Methods Mol. Biol. 2004, 287, 1–8. [Google Scholar] [CrossRef]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, L.; Andrews, L.G.; Tollefsbol, T.O. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int. J. Cancer 2009, 125, 286–296. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Meeran, S.M.; Patel, S.N.; Chen, H.; Hardy, T.M.; Tollefsbol, T.O. Epigenetic reactivation of estrogen receptor-α (ERα) by genistein enhances hormonal therapy sensitivity in ERα-negative breast cancer. Mol. Cancer 2013, 12, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Chen, H.; Hardy, T.M.; Tollefsbol, T.O. Epigenetic regulation of multiple tumor-related genes leads to suppression of breast tumorigenesis by dietary genistein. PLoS ONE 2013, 8, e54369. [Google Scholar] [CrossRef] [Green Version]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-derived xenograft models: An emerging platform for translational cancer research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.; Lee, C.H.; Seol, H.S.; Choi, Y.S.; Kim, E.; Lee, E.J.; Rhee, J.K.; Singh, S.R.; Jun, E.S.; Han, B.; et al. Generation and molecular characterization of pancreatic cancer patient-derived xenografts reveals their heterologous nature. Oncotarget 2016, 7, 62533–62546. [Google Scholar] [CrossRef] [Green Version]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richmond, A.; Su, Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis. Model Mech. 2008, 1, 78–82. [Google Scholar] [CrossRef] [Green Version]
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J.; Cai, S.; Clarke, R.B.; et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navone, N.M.; van Weerden, W.M.; Vessella, R.L.; Williams, E.D.; Wang, Y.; Isaacs, J.T.; Nguyen, H.M.; Culig, Z.; van der Pluijm, G.; Rentsch, C.A.; et al. Movember GAP1 PDX project: An international collection of serially transplantable prostate cancer patient-derived xenograft (PDX) models. Prostate 2018, 78, 1262–1282. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Rhee, H.; Kim, J.; Lee, S. Validity of patient-derived xenograft mouse models for lung cancer based on exome sequencing data. Genom. Inform. 2020, 18, e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, S.; Behring, M.; Kim, H.G.; Bajpai, P.; Chakravarthi, B.; Gupta, N.; Elkholy, A.; Al Diffalha, S.; Varambally, S.; Manne, U. Targeting P4HA1 with a Small Molecule Inhibitor in a Colorectal Cancer PDX Model. Transl. Oncol. 2020, 13, 100754. [Google Scholar] [CrossRef]
- Nelson, S.R.; Zhang, C.; Roche, S.; O’Neill, F.; Swan, N.; Luo, Y.; Larkin, A.; Crown, J.; Walsh, N. Modelling of pancreatic cancer biology: Transcriptomic signature for 3D PDX-derived organoids and primary cell line organoid development. Sci. Rep. 2020, 10, 2778. [Google Scholar] [CrossRef]
- Zhang, X.; Claerhout, S.; Prat, A.; Dobrolecki, L.E.; Petrovic, I.; Lai, Q.; Landis, M.D.; Wiechmann, L.; Schiff, R.; Giuliano, M.; et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013, 73, 4885–4897. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.; Royston, K.J.; Li, Y.; Stoll, M.L.; Skibola, C.F.; Wilson, L.S.; Barnes, S.; Morrow, C.D.; Tollefsbol, T.O. Impact of genistein on the gut microbiome of humanized mice and its role in breast tumor inhibition. PLoS ONE 2017, 12, e0189756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.-R.; Gugger, E.T.; Tanaka, T.; Guo, Y.; Blackburn, G.L.; Clinton, S.K. Soybean phytochemicals inhibit the growth of transplantable human prostate carcinoma and tumor angiogenesis in mice. J. Nutr. 1999, 129, 1628–1635. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Chen, M.; Wu, H.; Li, Y.; Tollefsbol, T.O. Maternal Epigenetic Regulation Contributes to Prevention of Estrogen Receptor-negative Mammary Cancer with Broccoli Sprout Consumption. Cancer Prev. Res. 2020, 13, 449–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Soneson, C.; Love, M.; Robinson, M. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Kabos, P.; Finlay-Schultz, J.; Li, C.; Kline, E.; Finlayson, C.; Wisell, J.; Manuel, C.A.; Edgerton, S.M.; Harrell, J.C.; Elias, A.; et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res. Treat. 2012, 135, 415–432. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Buckhaults, P.; Li, S.; Tollefsbol, T. Temporal Efficacy of a Sulforaphane-Based Broccoli Sprout Diet in Prevention of Breast Cancer through Modulation of Epigenetic Mechanisms. Cancer Prev. Res. 2018, 11, 451–464. [Google Scholar] [CrossRef] [Green Version]
- Arora, I.; Li, Y.; Sharma, M.; Crowley, M.R.; Crossman, D.K.; Li, S.; Tollefsbol, T.O. Systematic integrated analyses of methylomic and transcriptomic impacts of early combined botanicals on estrogen receptor-negative mammary cancer. Sci. Rep. 2021, 11, 9481. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Zhang, Y.; Li, N.; Liu, X.; Dong, J. CD74: A potential novel target for triple-negative breast cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2012, 33, 2273–2277. [Google Scholar] [CrossRef]
- Zeiner, P.S.; Zinke, J.; Kowalewski, D.J.; Bernatz, S.; Tichy, J.; Ronellenfitsch, M.W.; Thorsen, F.; Berger, A.; Forster, M.T.; Muller, A.; et al. CD74 regulates complexity of tumor cell HLA class II peptidome in brain metastasis and is a positive prognostic marker for patient survival. Acta Neuropathol. Commun. 2018, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Kuemmerle, N.B.; Rysman, E.; Lombardo, P.S.; Flanagan, A.J.; Lipe, B.C.; Wells, W.A.; Pettus, J.R.; Froehlich, H.M.; Memoli, V.A.; Morganelli, P.M.; et al. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol. Cancer Ther. 2011, 10, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.W.; Cheng, Y.; Liu, W.; Li, T.; Yegnasubramanian, S.; Zheng, S.L.; Xu, J.; Isaacs, W.B.; Chang, B.L. Genetic and epigenetic inactivation of LPL gene in human prostate cancer. Int. J. Cancer 2009, 124, 734–738. [Google Scholar] [CrossRef] [Green Version]
- Kirikoshi, H.; Sekihara, H.; Katoh, M. Expression profiles of 10 members of Frizzled gene family in human gastric cancer. Int. J. Oncol. 2001, 19, 767–771. [Google Scholar] [CrossRef]
- Zhang, Z.; Schittenhelm, J.; Guo, K.; Bühring, H.J.; Trautmann, K.; Meyermann, R.; Schluesener, H.J. Upregulation of frizzled 9 in astrocytomas. Neuropathol. Appl. Neurobiol. 2006, 32, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.; Edmiston, S.N.; May, R.; Kuan, P.F.; Chu, H.; Bryant, C.; Tse, C.K.; Swift-Scanlan, T.; Geradts, J.; Troester, M.A.; et al. DNA methylation profiling in the Carolina Breast Cancer Study defines cancer subclasses differing in clinicopathologic characteristics and survival. Breast Cancer Res. BCR 2014, 16, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brett-Morris, A.; Wright, B.M.; Seo, Y.; Pasupuleti, V.; Zhang, J.; Lu, J.; Spina, R.; Bar, E.E.; Gujrati, M.; Schur, R.; et al. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer Res. 2014, 74, 6925–6934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, H.; Wang, X.; Huang, W.; Dai, Y.; Yang, M.; Liang, H.; Wu, X.; Zhang, L.; Huang, W.; Yuan, L.; et al. Interferon-Induced Protein 44 Correlated With Immune Infiltration Serves as a Potential Prognostic Indicator in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 557157. [Google Scholar] [CrossRef]
- Zhao, M.; Zhou, Y.; Zhu, B.; Wan, M.; Jiang, T.; Tan, Q.; Liu, Y.; Jiang, J.; Luo, S.; Tan, Y.; et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 2016, 75, 1998–2006. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Fu, Y.; Tiirikainen, M.; Yu, H. Low expression of WWC1, a tumor suppressor gene, is associated with aggressive breast cancer and poor survival outcome. FEBS Open Bio. 2019, 9, 1270–1280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-H.; Lin, J.-Y. Recent advances of cluster of differentiation 74 in cancer. World J. Immunol. 2014, 4, 174–184. [Google Scholar] [CrossRef]
- Binsky, I.; Haran, M.; Starlets, D.; Gore, Y.; Lantner, F.; Harpaz, N.; Leng, L.; Goldenberg, D.M.; Shvidel, L.; Berrebi, A.; et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc. Natl. Acad. Sci. USA 2007, 104, 13408–13413. [Google Scholar] [CrossRef] [Green Version]
- Binsky, I.; Lantner, F.; Grabovsky, V.; Harpaz, N.; Shvidel, L.; Berrebi, A.; Goldenberg, D.M.; Leng, L.; Bucala, R.; Alon, R.; et al. TAp63 regulates VLA-4 expression and chronic lymphocytic leukemia cell migration to the bone marrow in a CD74-dependent manner. J. Immunol. 2010, 184, 4761–4769. [Google Scholar] [CrossRef] [Green Version]
- Khongthong, P.; Roseweir, A.K.; Edwards, J. The NF-KB pathway and endocrine therapy resistance in breast cancer. Endocr. Relat. Cancer 2019, 26, R369–R380. [Google Scholar] [CrossRef] [PubMed]
- Starlets, D.; Gore, Y.; Binsky, I.; Haran, M.; Harpaz, N.; Shvidel, L.; Becker-Herman, S.; Berrebi, A.; Shachar, I. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood 2006, 107, 4807–4816. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Kushihata, F.; Honda, K.; Mominoki, K.; Matsuda, S.; Kobayashi, N. Bcl-xL overexpression in human hepatocellular carcinoma. Int. J. Oncol. 2002, 21, 515–519. [Google Scholar] [CrossRef]
- Karczmarek-Borowska, B.; Filip, A.; Wojcierowski, J.; Smoleń, A.; Korobowicz, E.; Korszen-Pilecka, I.; Zdunek, M. Estimation of prognostic value of Bcl-xL gene expression in non-small cell lung cancer. Lung Cancer 2006, 51, 61–69. [Google Scholar] [CrossRef]
- Olopade, O.I.; Adeyanju, M.O.; Safa, A.R.; Hagos, F.; Mick, R.; Thompson, C.B.; Recant, W.M. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 1997, 3, 230–237. [Google Scholar]
- Li, N.; Singh, S.; Cherukuri, P.; Li, H.; Yuan, Z.; Ellisen, L.W.; Wang, B.; Robbins, D.; DiRenzo, J. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells 2008, 26, 1253–1264. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Bai, Q.; Zou, L.Y.; Zhang, Q.Y.; Zhou, Y.; Chang, H.; Yi, L.; Zhu, J.D.; Mi, M.T. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosomes Cancer 2014, 53, 422–431. [Google Scholar] [CrossRef]
- Satoh, H.; Nishikawa, K.; Suzuki, K.; Asano, R.; Virgona, N.; Ichikawa, T.; Hagiwara, K.; Yano, T. Genistein, a soy isoflavone, enhances necrotic-like cell death in a breast cancer cell treated with a chemotherapeutic agent. Res. Commun. Mol. Pathol. Pharmacol. 2003, 113, 149–158. [Google Scholar]
- Gong, L.; Li, Y.; Nedeljkovic-Kurepa, A.; Sarkar, F.H. Inactivation of NF-kappaB by genistein is mediated via Akt signaling pathway in breast cancer cells. Oncogene 2003, 22, 4702–4709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pintova, S.; Dharmupari, S.; Moshier, E.; Zubizarreta, N.; Ang, C.; Holcombe, R.F. Genistein combined with FOLFOX or FOLFOX–Bevacizumab for the treatment of metastatic colorectal cancer: Phase I/II pilot study. Cancer Chemother. Pharmacol. 2019, 84, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Kimura, M.; Kawasaki, T.; Sato, S.; Tomita, Y. MHC class II antigen-associated invariant chain on renal cell cancer may contribute to the anti-tumor immune response of the host. Cancer Lett. 1997, 115, 121–127. [Google Scholar] [CrossRef]
- Datta, M.W.; Shahsafaei, A.; Nadler, L.M.; Freeman, G.J.; Dorfman, D.M. Expression of MHC class II-associated invariant chain (Ii;CD74) in thymic epithelial neoplasms. Appl. Immunohistochem. Mol. Morphol. AIMM 2000, 8, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, R.J.; Wilson, J.M.; Scott, N.; Coletta, P.L.; Hull, M.A. Differential CD74 (major histocompatibility complex Class II invariant chain) expression in mouse and human intestinal adenomas. Eur. J. Cancer 2009, 45, 1654–1663. [Google Scholar] [CrossRef]
- Casciano, J.C.; Perry, C.; Cohen-Nowak, A.J.; Miller, K.D.; Vande Voorde, J.; Zhang, Q.; Chalmers, S.; Sandison, M.E.; Liu, Q.; Hedley, A.; et al. MYC regulates fatty acid metabolism through a multigenic program in claudin-low triple negative breast cancer. Br. J. Cancer 2020, 122, 868–884. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.F.; Sung, V.Y.C.; Kuzmin, E.; Couzens, A.L.; de Verteuil, D.A.; Ratcliffe, C.D.H.; Coelho, P.P.; Johnson, R.M.; Samavarchi-Tehrani, P.; Gruosso, T.; et al. KIBRA (WWC1) Is a Metastasis Suppressor Gene Affected by Chromosome 5q Loss in Triple-Negative Breast Cancer. Cell Rep. 2018, 22, 3191–3205. [Google Scholar] [CrossRef] [Green Version]
- Lantner, F.; Starlets, D.; Gore, Y.; Flaishon, L.; Yamit-Hezi, A.; Dikstein, R.; Leng, L.; Bucala, R.; Machluf, Y.; Oren, M.; et al. CD74 induces TAp63 expression leading to B-cell survival. Blood 2007, 110, 4303–4311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, K.; Flanagan, J.M. Is there a link between genome-wide hypomethylation in blood and cancer risk? Cancer Prev. Res. 2012, 5, 1345–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curry, E.; Green, I.; Chapman-Rothe, N.; Shamsaei, E.; Kandil, S.; Cherblanc, F.L.; Payne, L.; Bell, E.; Ganesh, T.; Srimongkolpithak, N.; et al. Dual EZH2 and EHMT2 histone methyltransferase inhibition increases biological efficacy in breast cancer cells. Clin. Epigenetics 2015, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Zhao, T.T.; Jin, F.; Li, J.G.; Xu, Y.Y.; Dong, H.T.; Liu, Q.; Xing, P.; Zhu, G.L.; Xu, H.; Miao, Z.F. Dietary isoflavones or isoflavone-rich food intake and breast cancer risk: A meta-analysis of prospective cohort studies. Clin. Nutr. 2019, 38, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Spicer, D.; Garcia, A.; Tseng, C.-C.; Hovanessian-Larsen, L.; Sheth, P.; Martin, S.E.; Hawes, D.; Russell, C.; MacDonald, H.; et al. Double-Blind Randomized 12-Month Soy Intervention Had No Effects on Breast MRI Fibroglandular Tissue Density or Mammographic Density. Cancer Prev. Res. 2015, 8, 942–951. [Google Scholar] [CrossRef] [Green Version]
- Delmanto, A.; Nahas-Neto, J.; Traiman, P.; Uemura, G.; Pessoa, E.C.; Nahas, E.A. Effects of soy isoflavones on mammographic density and breast parenchyma in postmenopausal women: A randomized, double-blind, placebo-controlled clinical trial. Menopause 2013, 20, 1049–1054. [Google Scholar] [CrossRef]
- Atteritano, M.; Pernice, F.; Mazzaferro, S.; Mantuano, S.; Frisina, A.; D’Anna, R.; Cannata, M.L.; Bitto, A.; Squadrito, F.; Frisina, N.; et al. Effects of phytoestrogen genistein on cytogenetic biomarkers in postmenopausal women: 1 year randomized, placebo-controlled study. Eur. J. Pharmacol. 2008, 589, 22–26. [Google Scholar] [CrossRef]
- Liu, R.; Yu, X.; Chen, X.; Zhong, H.; Liang, C.; Xu, X.; Xu, W.; Cheng, Y.; Wang, W.; Yu, L.; et al. Individual factors define the overall effects of dietary genistein exposure on breast cancer patients. Nutr. Res. 2019, 67, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Nihal, M.; Ahmad, N. Dose translation from animal to human studies revisited. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 659–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
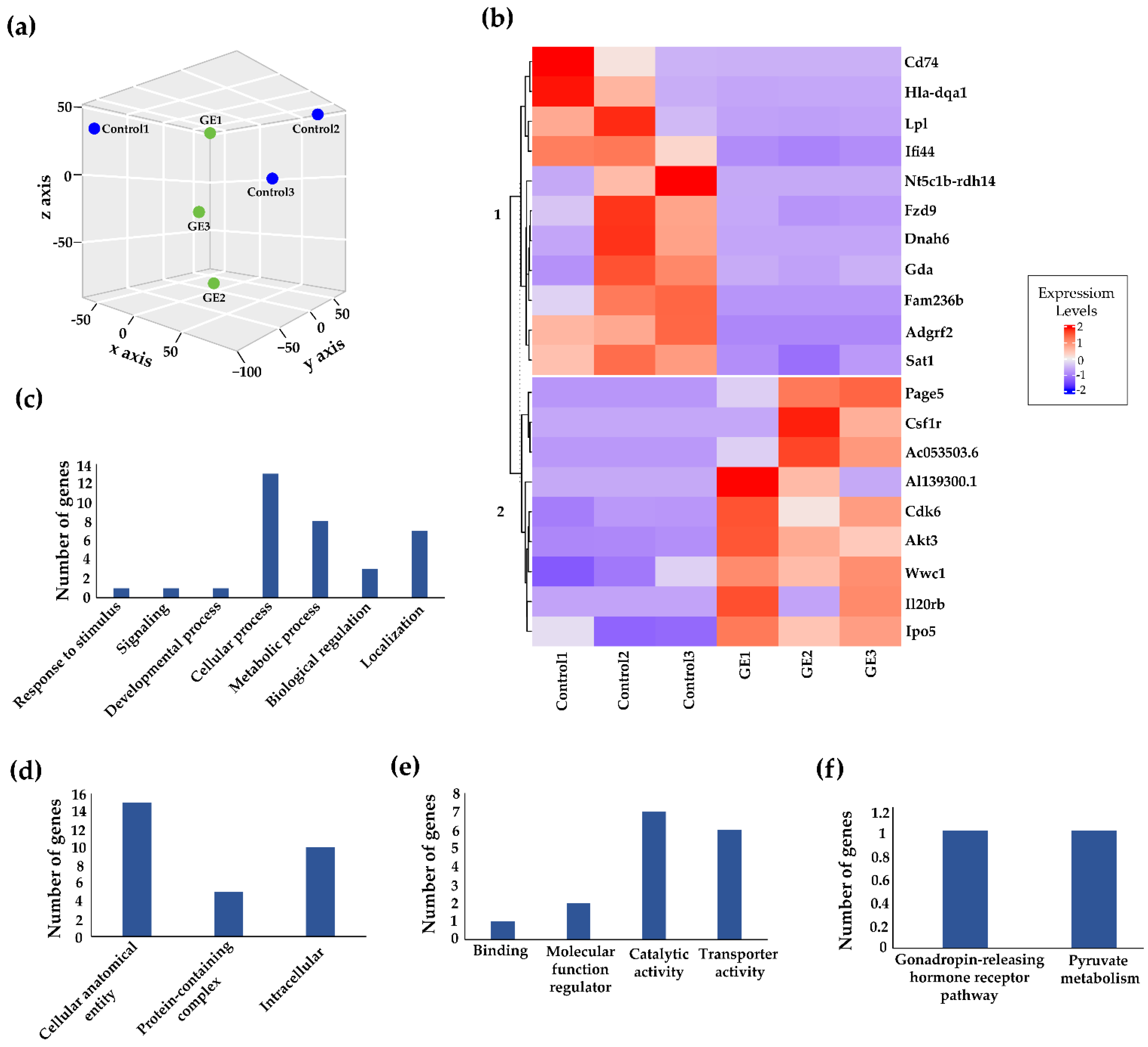

| Gene Symbol | Gene Expression Fold Change (log2FC) | Average Differential Expression | p-Value for Differential Expression | False Discovery Rate (FDR) |
|---|---|---|---|---|
| Cd74 | −4.760 | 2.879 | 2.20 × 10−7 | 0.003 |
| Lpl | −2.806 | 2.294 | 2.26 × 10−6 | 0.017 |
| Ifi44 | −2.191 | 2.100 | 8.23 × 10−6 | 0.027 |
| Dnah6 | −6.634 | −3.412 | 5.63 × 10−6 | 0.027 |
| Il20rb | 6.115 | −3.578 | 9.12 × 10−6 | 0.027 |
| Wwc1 | 0.903 | 5.994 | 1.27 × 10−5 | 0.028 |
| Sat1 | −0.823 | 6.315 | 1.49 × 10−5 | 0.028 |
| Al139300.1 | 5.984 | −3.559 | 1.32 × 10−5 | 0.028 |
| Akt3 | 1.335 | 5.583 | 2.34 × 10−5 | 0.037 |
| Ipo5 | 0.935 | 7.051 | 2.46 × 10−5 | 0.037 |
| Gda | −1.759 | 2.098 | 3.27 × 10−5 | 0.039 |
| Fzd9 | −1.186 | 4.141 | 4.89 × 10−5 | 0.039 |
| Cdk6 | 0.903 | 9.485 | 4.07 × 10−5 | 0.039 |
| Hla-dqa1 | −5.147 | −1.468 | 4.60 × 10−5 | 0.039 |
| Adgrf2 | −4.405 | −3.498 | 3.85 × 10−5 | 0.039 |
| Page5 | 4.595 | −3.685 | 3.63 × 10−5 | 0.039 |
| Fam236b | −4.509 | −3.722 | 4.96 × 10−5 | 0.039 |
| Ac053503.6 | 4.624 | −3.537 | 5.12 × 10−5 | 0.039 |
| Csf1r | 5.257 | −3.778 | 3.26 × 10−5 | 0.039 |
| Nt5c1b-rdh14 | −4.914 | −3.895 | 5.25 × 10−5 | 0.039 |
| Gene | Function in Cancer | Epigenetic Regulation | RNA-Seq |
|---|---|---|---|
| Cluster of Differentiation 74 (Cd74) | Cd74 is overexpressed in breast cancer patients. It is also found to be significantly correlated with lymph node metastasis in TNBC [38] | Epigenetic mechanisms play a role in Cd74 expression via Cd74 promoter methylation [39] | DEG, significant decrease (4.76 fold) |
| Lipoprotein lipase (Lpl) | Breast cancer and sarcoma cells express and secrete active Lpl enzyme to acquire fatty acids from the blood circulation, which facilitate growth of these cells [40] | Epigenetic changes at the promoter regions may alter expression of the Lpl gene and may play an important role in prostate cancer development [41] | DEG, significant decrease (2.8 fold) |
| Frizzled 9 (Fzd9) | Oncogene. Overexpressed Fzd9 is found in various types of cancer [42,43] | Hypermethylated Fzd9 is associated with hormone receptor positive, luminal A, or p53 wild-type breast cancers [44] | DEG, significant decrease (1.18 fold) |
| Spermidine/spermine-N1-acetyltransferase 1 (Sat1) | Sat1 overexpression is correlated with poor clinical outcomes [45] | Sat1 is involved in acetylation of histone H3, resulting in chromatin remodeling and regulation of gene expression [45] | DEG, significant decrease (0.82 fold) |
| Interferon-Induced Protein 44 (Ifi44) | Oncogene. Overexpressed Ifi44 is found in head and neck squamous cell carcinoma and functioned heterogeneously in tumor formation and progression [46] | Iifi44 promoter hypomethylation can distinguish systemic lupus erythematosus patients from healthy persons, promising to be first novel epigenetic diagnostic marker [47] | DEG, significant decrease (2.19 fold) |
| WW and C2 domain containing 1 (Wwc1) | Tumor-suppressor gene. Low Wwc1 expression is associated with aggressive breast cancer and poor survival outcomes [48] | DNA methylation is negatively correlated with Wwc1 expression [48] | DEG, significant increase (0.90 fold) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, M.; Arora, I.; Chen, M.; Wu, H.; Crowley, M.R.; Tollefsbol, T.O.; Li, Y. Therapeutic Effects of Dietary Soybean Genistein on Triple-Negative Breast Cancer via Regulation of Epigenetic Mechanisms. Nutrients 2021, 13, 3944. https://doi.org/10.3390/nu13113944
Sharma M, Arora I, Chen M, Wu H, Crowley MR, Tollefsbol TO, Li Y. Therapeutic Effects of Dietary Soybean Genistein on Triple-Negative Breast Cancer via Regulation of Epigenetic Mechanisms. Nutrients. 2021; 13(11):3944. https://doi.org/10.3390/nu13113944
Chicago/Turabian StyleSharma, Manvi, Itika Arora, Min Chen, Huixin Wu, Michael R. Crowley, Trygve O. Tollefsbol, and Yuanyuan Li. 2021. "Therapeutic Effects of Dietary Soybean Genistein on Triple-Negative Breast Cancer via Regulation of Epigenetic Mechanisms" Nutrients 13, no. 11: 3944. https://doi.org/10.3390/nu13113944
APA StyleSharma, M., Arora, I., Chen, M., Wu, H., Crowley, M. R., Tollefsbol, T. O., & Li, Y. (2021). Therapeutic Effects of Dietary Soybean Genistein on Triple-Negative Breast Cancer via Regulation of Epigenetic Mechanisms. Nutrients, 13(11), 3944. https://doi.org/10.3390/nu13113944







