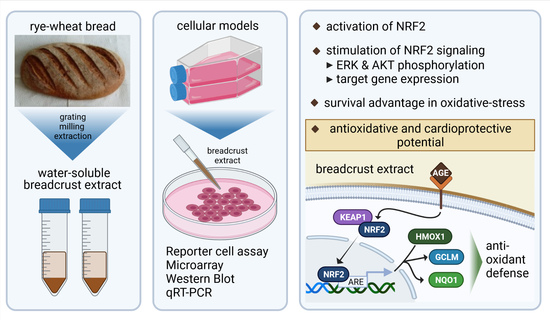
AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway
Abstract
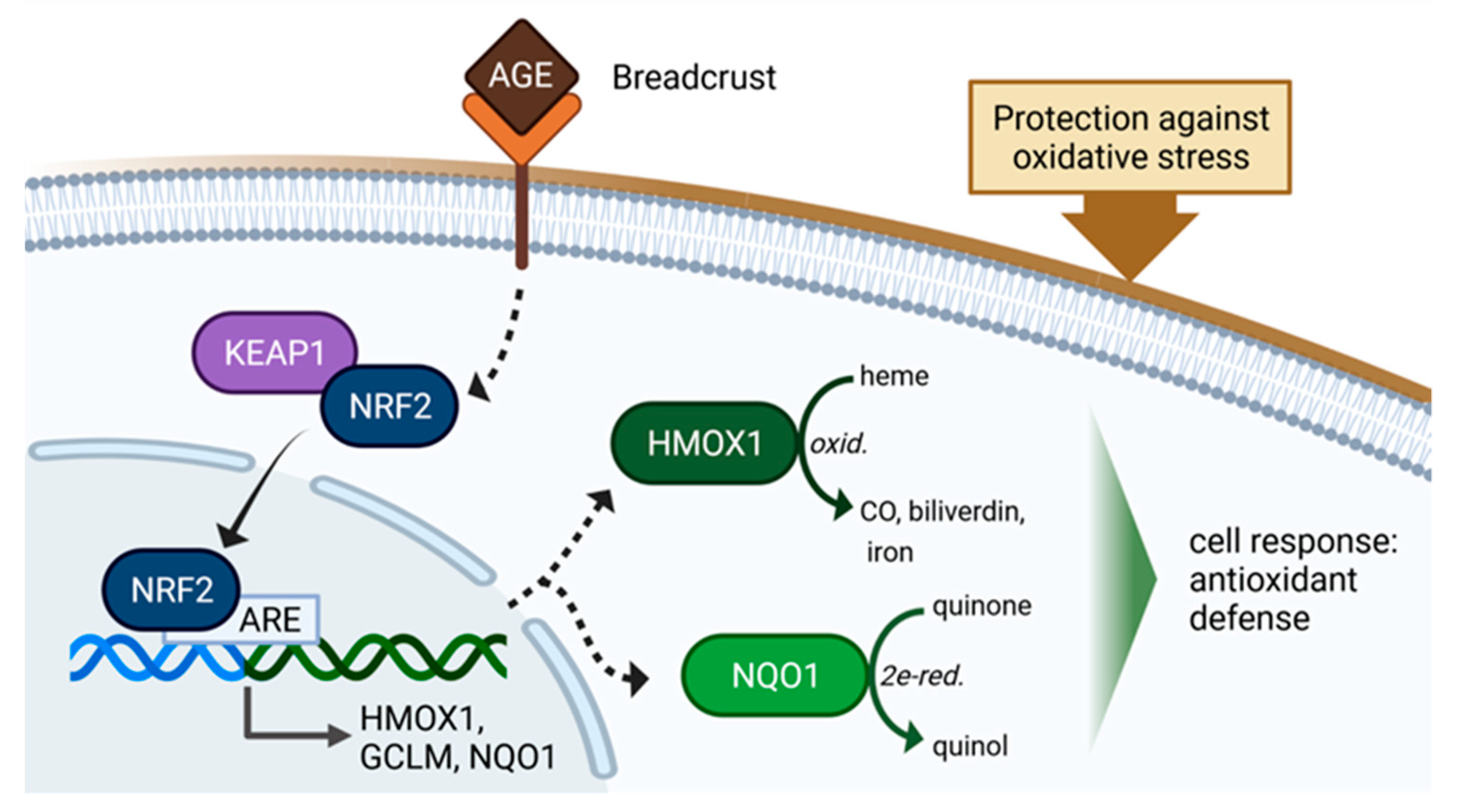
:1. Introduction
2. Materials and Methods
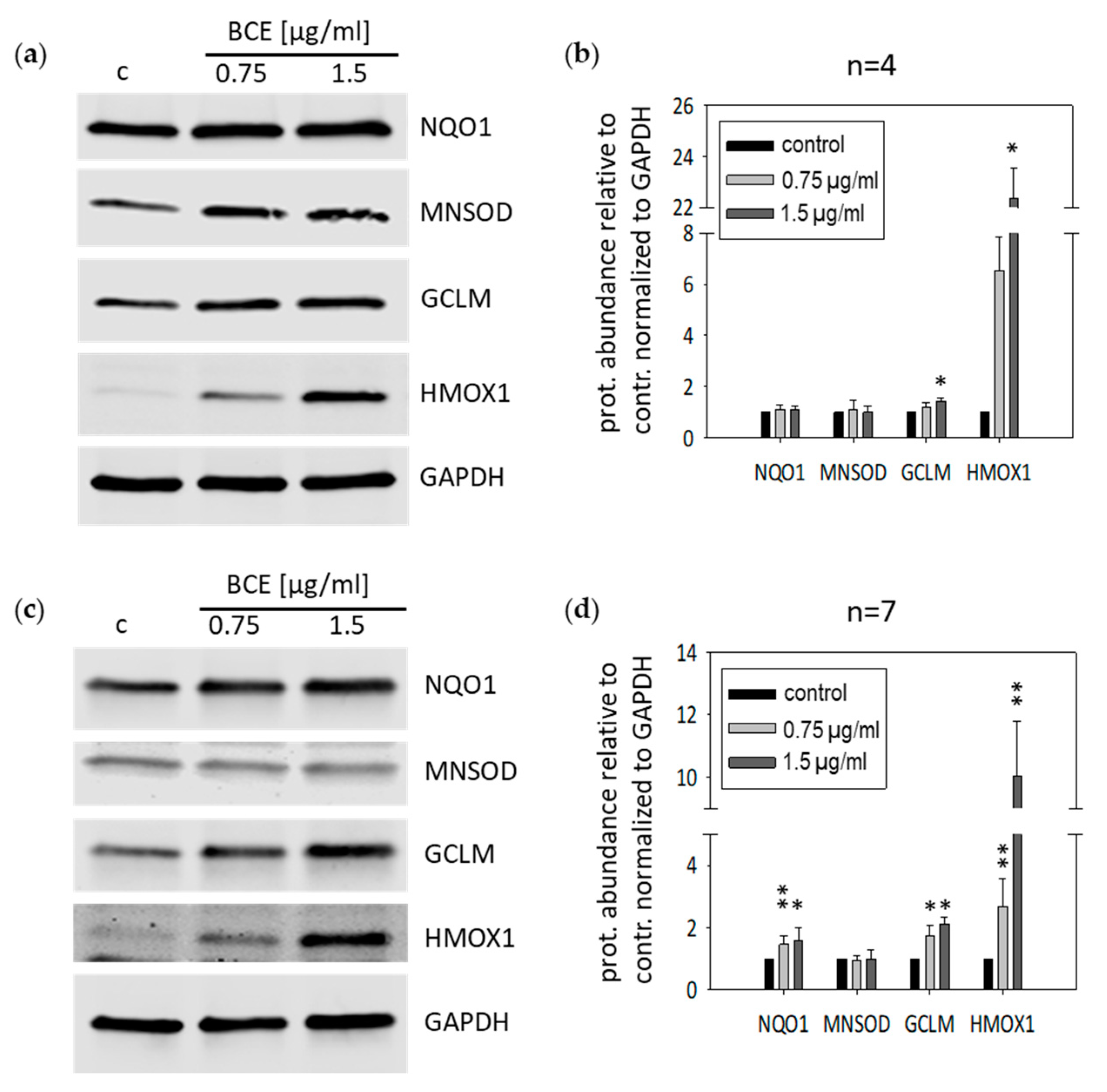
2.1. Western Blot Analysis
2.2. RNA Extraction, Reverse-Transcription, and quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.3. Microarray Analyses
2.4. Cell Culture, Transfection, Inducers and Inhibitor
2.5. Hydrogen Peroxide Stress Assay
2.6. Statistical Analysis
2.7. Production of Bread Crust (BC) and Preparation of Bread Crust Extract (BCE)
2.8. TCA-Precipitation, Acidic Hydrolysis and Mass Spectrometric Identification of Advanced Glycation End Products in BCE
3. Results
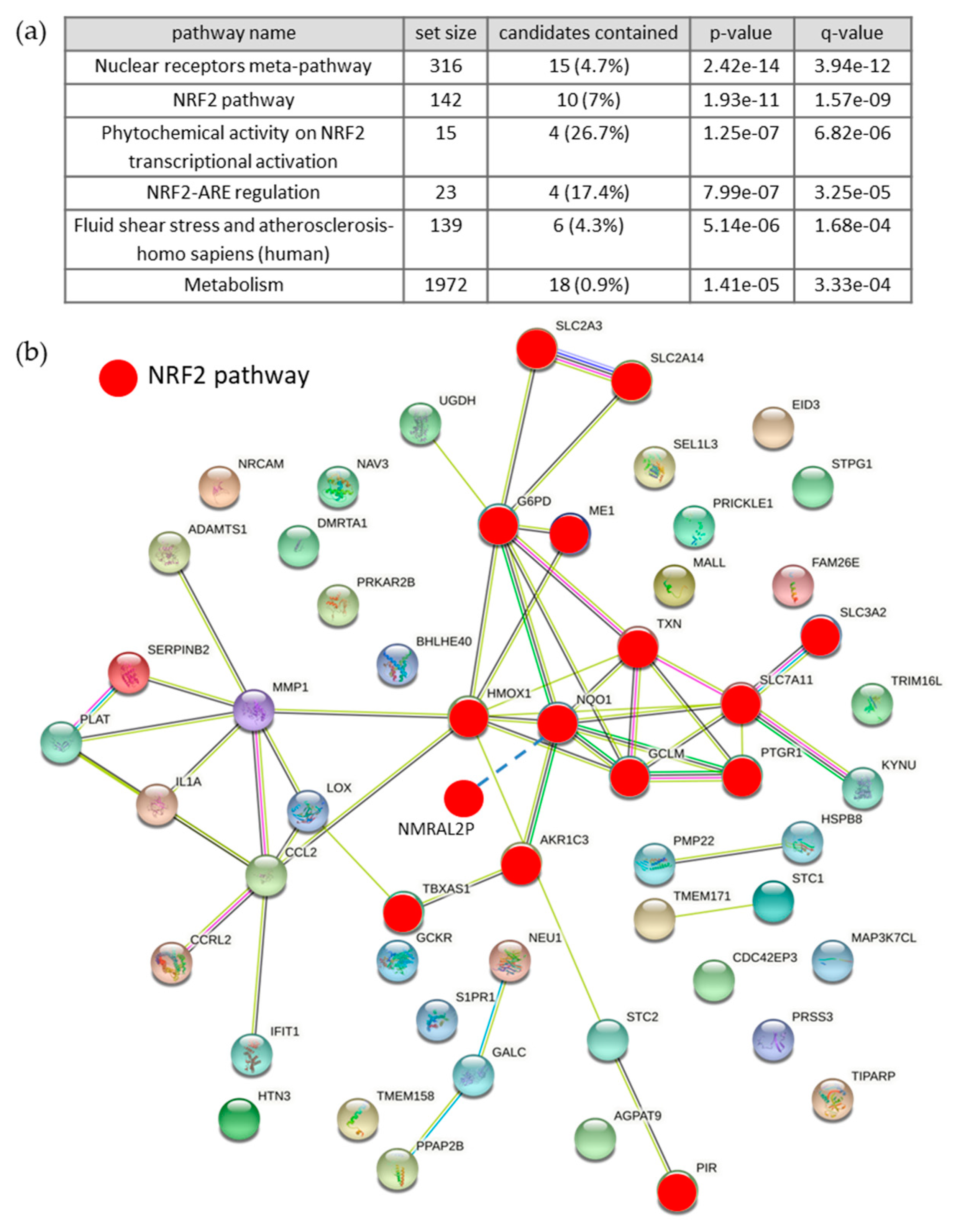
3.1. Differentially Expressed RNAs of Control vs. BCE Treated EA.hy926 Cells
3.1.1. Microarray Analysis
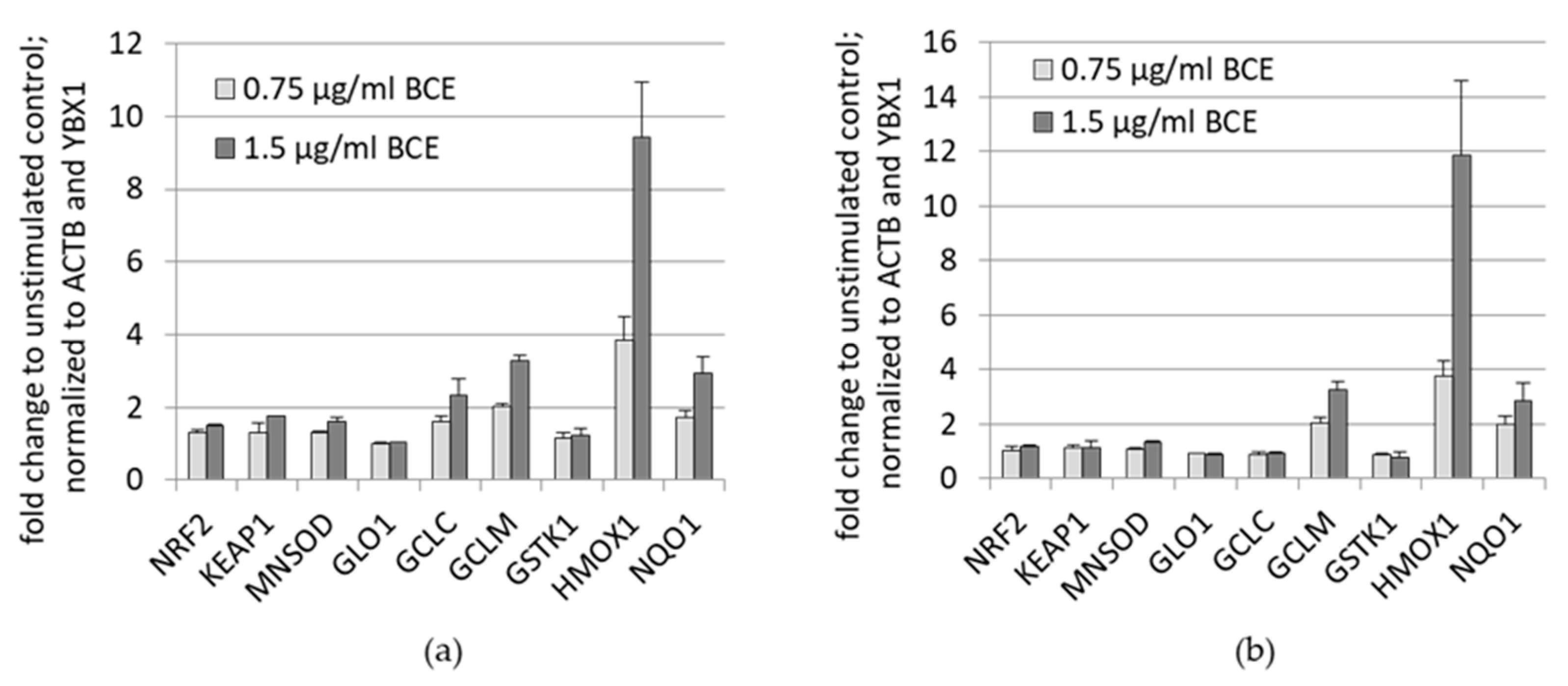
3.1.2. Effects of BCE on mRNA Expression Levels in EA.hy926 and HeLa Cells
3.2. Effects of BCE on Protein Expression Levels in EA.hy926 and HeLa Cells
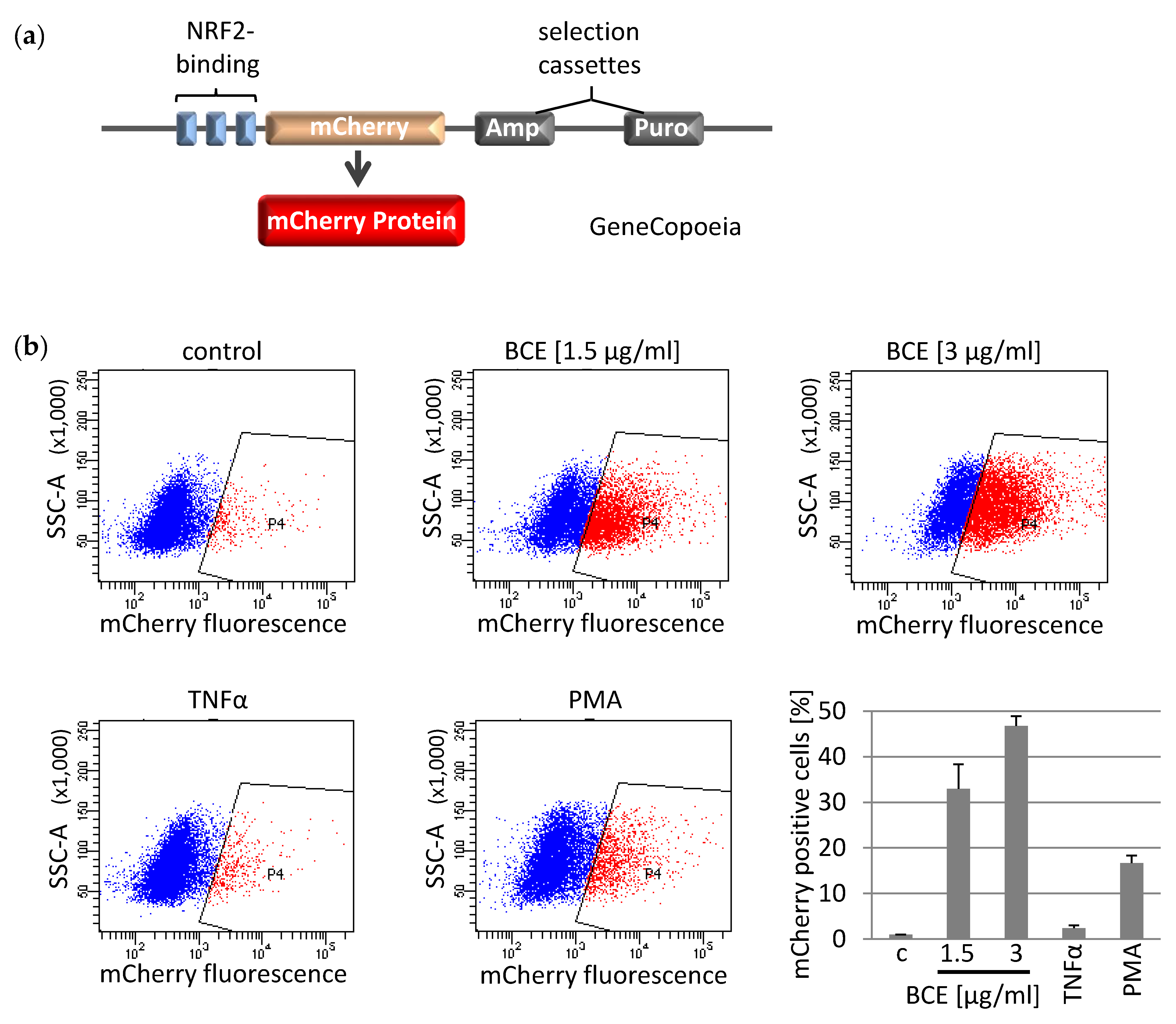
3.3. Effects of BCE on NRF2 Induced Gene Expression in EA.hy926 Reporter Cells
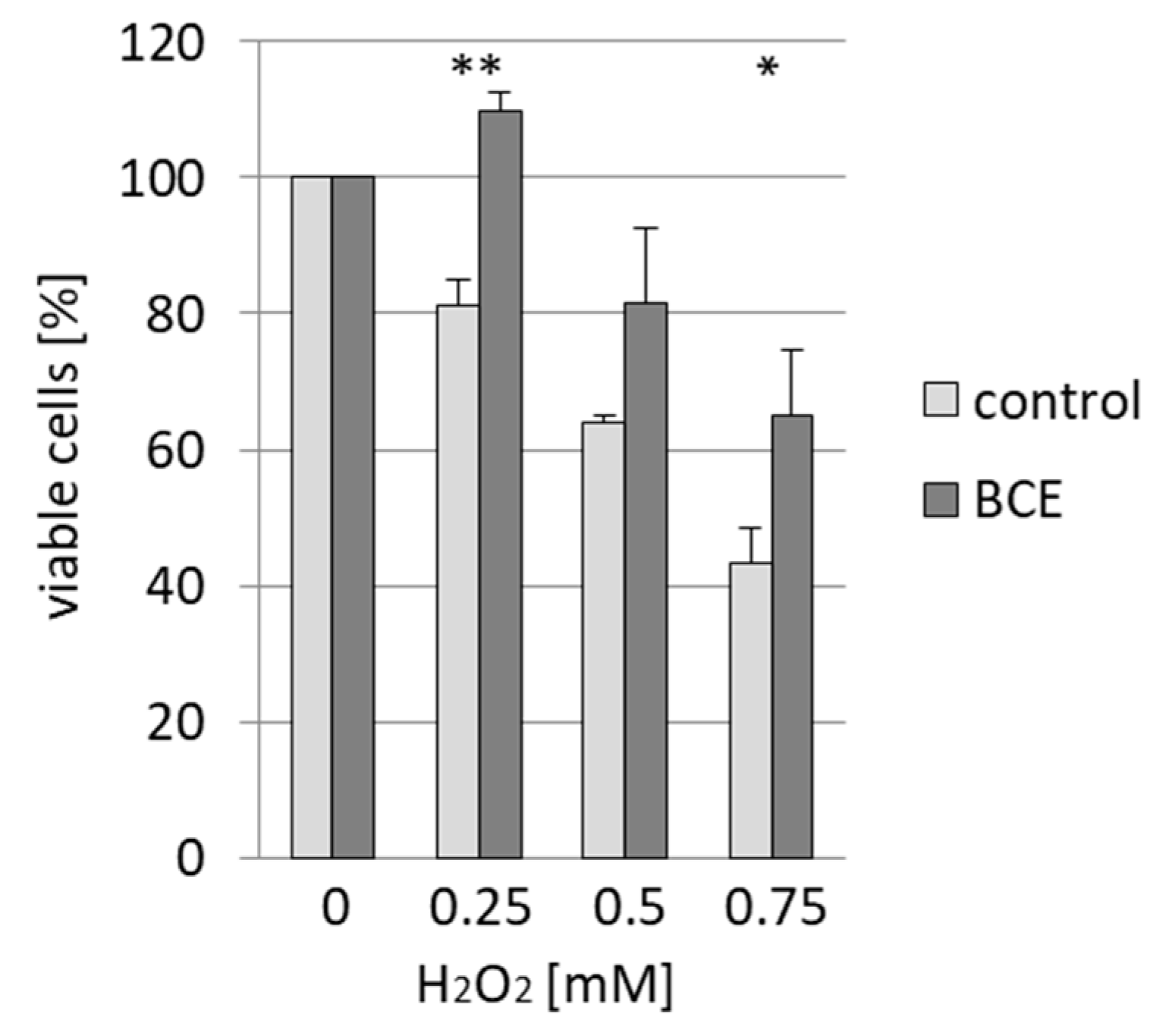
3.4. Effects of BCE on H2O2-Induced Cell Death in EA.hy926 Cells
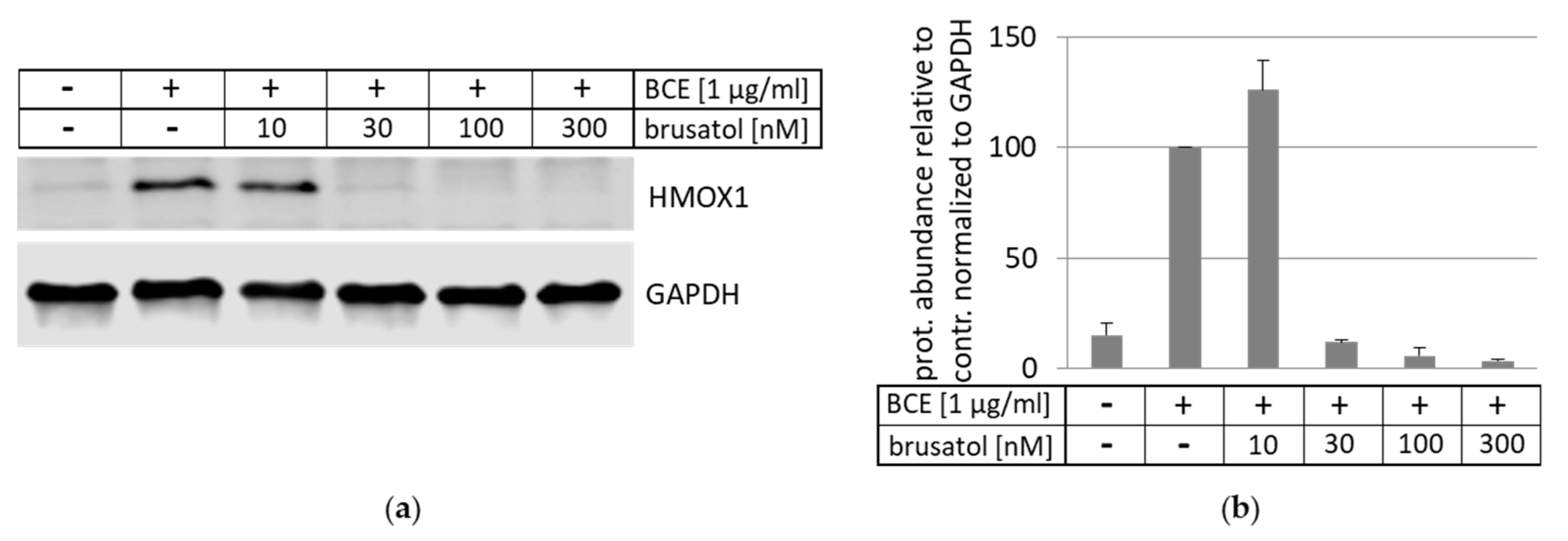
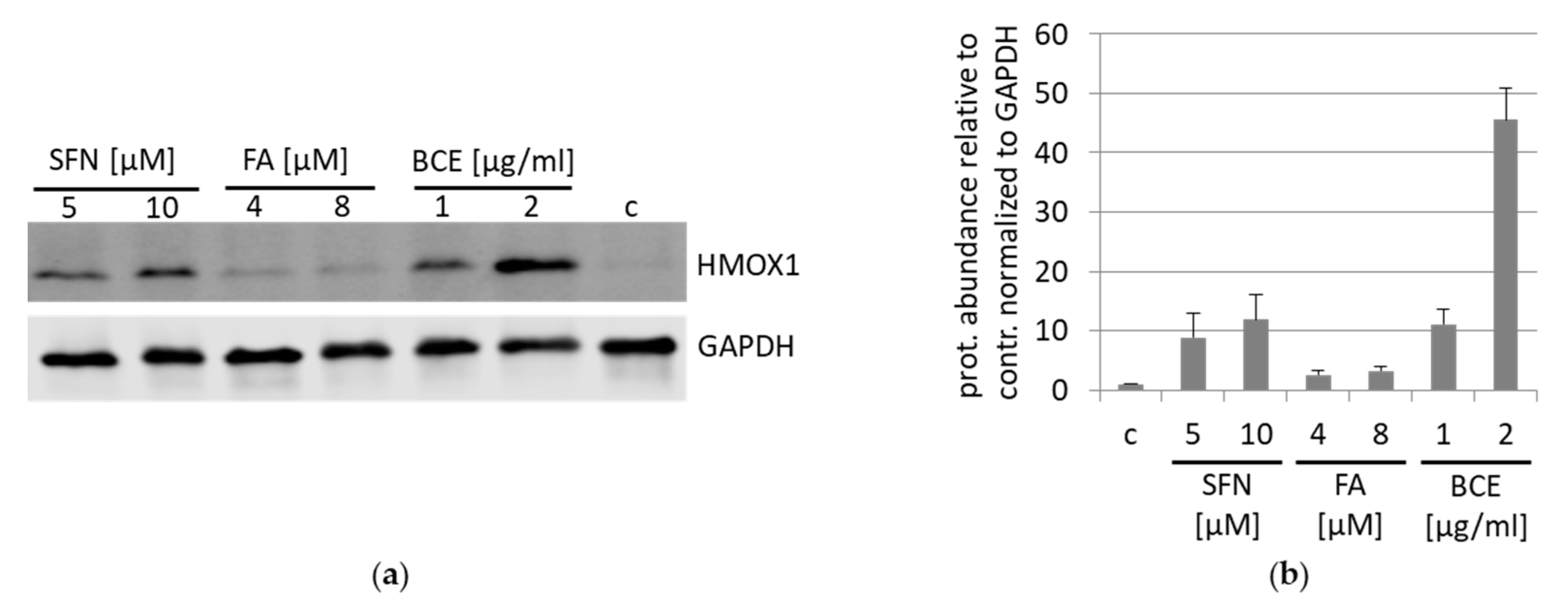
3.5. Inhibition and Induction of NRF2-Pathway
3.6. Effects of BCE on Signaling Pathways
3.7. Identification of AGE-Modifications in BCE by Mass Spectrometry
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | actin beta |
| AGEs | advanced glycation end products |
| AGE-R1-3 | AGE receptors 1-3 |
| AKT | AKT serine/threonine kinase |
| AVG | average |
| ARE | antioxidant response element |
| Arg | arginine |
| BCE | bread crust extract |
| Caco | colorectal adenocarcinoma (human) |
| DME’s | drug-metabolizing enzymes |
| EA.hy926 | immortalized human vascular endothelial cells |
| EMT | epithelial-mesenchymal transition |
| ERK | mitogen-activated protein kinase |
| FA | falcarinol |
| FCS | fetal calf serum |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GCLC | glutamate-cysteine ligase catalytic subunit |
| GCLM | glutamate-cysteine ligase modifier subunit |
| GLO1 | glyoxalase I |
| GSH | glutathione |
| GSTK1 | glutathione S-transferase kappa 1 |
| H2O2 | hydrogen peroxide |
| HMOX1 | heme oxygenase 1 |
| KEAP1 | kelch-like ECH associated protein 1 |
| Lys | lysine |
| MNSOD | superoxide dismutase 2 |
| MRPs | Maillard reaction products |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NQO1 | NAD(P)H quinone dehydrogenase 1 |
| NRF2 | nuclear factor E2-related factor 2 |
| NF-κB | nuclear factor kappa B |
| P | phosphorylation |
| PBS | phosphate-buffered saline |
| PE | phycoerythrin |
| PI3K | phosphoinositide 3-kinase |
| PMA | phorbol 12-myristate 13-acetate |
| qRT-PCR | quantitative Real time-PCR |
| RAGE | advanced glycosylation end product receptor |
| RIN | RNA integrity number |
| ROS | reactive oxygen species |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SFN | sulforaphane |
| TNFα | tumor necrosis factor a |
| Trp | tryptophan |
| Tyr | tyrosine |
| YBX1 | Y-box binding protein 1 |
Appendix A
| Primary antibodies | |||
| antibody | Company (order number) | dilution | isotype |
| GCLM | Abcam (ab126704) | 1:1500 | rabbit monoclonal IgG |
| HMOX1 | Abcam (ab68477) | 1:2500 | rabbit monoclonal |
| HMOX1 | Enzo Life Sciences (ADI-SPA-896) | 1:1000 | rabbit polyclonal |
| NQO1 | Santa Cruz (sc-32793) | 1:500 | mouse monoclonal |
| GAPDH | Cell Signaling (#5174) | 1:1000 | rabbit monoclonal IgG |
| GAPDH | Sigma-Aldrich (G8795) | 1:20000 | mouse monoclonal IgM |
| MNSOD | Biomol (600-401-G13) | 1:1000 | rabbit |
| Phospho-AKT (Ser473) | Cell Signaling (#4060) | 1:2000 | rabbit polyclonal IgG |
| AKT-pan | Cell Signaling (#2920) | 1:2000 | mouse monoclonal IgG1 |
| Phospho-ERK1/2 (Thr202/Tyr204) | Cell Signaling (#4370) | 1:2000 | rabbit monoclonal IgG |
| ERK1/2 | Abcam (ab184699) | 1:10000 | rabbit monoclonal |
| RAGE | Abcam (ab65965) | 1:3000 | rabbit polyclonal |
| AGE-R1 | Santa Cruz (sc-74408) | 1:1000 | mouse monoclonal IgG1 |
| AGE-R2 | Abcam (ab134071) | 1:2000 | rabbit monoclonal IgG |
| AGE-R3 | Santa Cruz (sc-19283) | 1:1000 | goat polyclonal IgG |
| Secondary antibodies | |||
| antibody | Company (order number) | dilution | isotype |
| IRDye 800CW | Li-Cor (926-32211) | 1:15000 | goat anti-rabbit |
| IRDye 680LT | Li-Cor (926-68020) | 1:15000 | goat anti-mouse |
| Genes | Forward Primers (5′–3′) | Reverse Primers (5′–3′) | Source |
|---|---|---|---|
| NRF2 | 5′-CAGCGACGGAAAGAGTATGA-3′ | 5′-TGGGCAACCTGGGAGTAG-3′ | [83] |
| KEAP1 | 5′-GGCTGTCCTCAATCGTCTCC-3′ | 5′-TCTGTTTCCACATCGTAGCG-3 | [83] |
| HMOX1 | 5′-CAACATCCAGCTCTTTGAGG-3′ | 5′-GGCAGAATCTTGCACTTTG-3′ | [83] |
| ACTB | 5′ -CTGGAACGGTGAAGGTGACA-3′ | 5′-AAGGGACTTCCTGTAACAATGCA-3′ | [83] |
| MNSOD | 5′-CACCGAGGAGAAGTACCAGG-3′ | 5′-TAGGGCTGAGGTTTGTCCAG-3′ | [84] |
| GCLC | 5′-GTCCTTTCCCCCTTCTCTTG-3′ | 5′-AGGACGTTCTCAAGTGGGG-3′ | [84] |
| GSTK1 | 5′-TGGTCTCCTTGAGCTGGTTC-3′ | 5′-AATGAAGACATCACCGAGCC-3′ | [84] |
| GLO1 | 5′-TGGATTAGCGTCATTCCAAG-3′ | 5′-CAGTTGCTGCTCCGACG-3′ | [84] |
| YBX1 | 5′-ACTGCGAAGGTACTTCCTGG-3′ | 5′-TGGTTCAATGTAAGGAACGGA-3′ | [84] |
| GCLM | 5′-ACTCGTGCGCTTGAATGTC-3′ | 5′-CTGTGTGATGCCACCAGATT-3′ | [84] |
| NQO1 | 5′-GCATAGAGGTCCGACTCCAC-3′ | 5′-GGACTGCACCAGAGCCAT-3′ | [84] |
References
- Poulsen, M.W.; Hedegaard, R.V.; Andersen, J.M.; de Courten, B.; Bügel, S.; Nielsen, J.; Skibsted, L.H.; Dragsted, L.O. Advanced glycation endproducts in food and their effects on health. Food Chem. Toxicol. 2013, 60, 10–37. [Google Scholar] [CrossRef] [PubMed]
- Ruhs, S.; Nass, N.; Somoza, V.; Friess, U.; Schinzel, R.; Silber, R.-E.; Simm, A. Maillard reaction products enriched food extract reduce the expression of myofibroblast phenotype markers. Mol. Nutr. Food Res. 2007, 51, 488–495. [Google Scholar] [CrossRef]
- Jost, T.; Henning, C.; Heymann, T.; Glomb, M.A. Comprehensive Analyses of Carbohydrates, 1,2-Dicarbonyl Compounds, and Advanced Glycation End Products in Industrial Bread Making. J. Agric. Food Chem. 2021, 69, 3720–3731. [Google Scholar] [CrossRef]
- Lindenmeier, M.; Faist, A.V.; Hofmann, T. Structural and Functional Characterization of Pronyl-lysine, a Novel Protein Modification in Bread Crust Melanoidins Showing in Vitro Antioxidative and Phase I/II Enzyme Modulating Activity. J. Agric. Food Chem. 2002, 50, 6997–7006. [Google Scholar] [CrossRef] [PubMed]
- Pastoriza, S.; Roncero-Ramos, I.; Rufian-Henares, J.A.; Delgado-Andrade, C. Antioxidant balance after long-term consumption of standard diets including bread crust glycated compounds by adult rats. Food Res. Int. 2014, 64, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Somoza, V.; Wenzel, E.; Lindenmeier, M.; Grothe, D.; Erbersdobler, H.F.; Hofmann, T. Influence of Feeding Malt, Bread Crust, and a Pronylated Protein on the Activity of Chemopreventive Enzymes and Antioxidative Defense Parameters in Vivo. J. Agric. Food Chem. 2005, 53, 8176–8182. [Google Scholar] [CrossRef] [PubMed]
- Ruhs, S.; Nass, N.; Bartling, B.; Brömme, H.-J.; Leuner, B.; Somoza, V.; Friess, U.; Silber, R.-E.; Simm, A. Preconditioning with Maillard reaction products improves antioxidant defence leading to increased stress tolerance in cardiac cells. Exp. Gerontol. 2010, 45, 752–762. [Google Scholar] [CrossRef] [Green Version]
- Bartling, B.; Hofmann, H.-S.; Sohst, A.; Hatzky, Y.; Somoza, V.; Silber, R.-E.; Simm, A. Prognostic Potential and Tumor Growth-Inhibiting Effect of Plasma Advanced Glycation End Products in Non-Small Cell Lung Carcinoma. Mol. Med. 2011, 17, 980–989. [Google Scholar] [CrossRef]
- Potzsch, S.; Blankenhorn, A.; Navarrete Santos, A.; Silber, R.-E.; Somoza, V.; Simm, A. The effect of an AGE-rich dietary extract on the activation of NF-kappaB depends on the cell model used. Food Funct. 2013, 4, 1023–1031. [Google Scholar] [CrossRef]
- Sauer, T.; Raithel, M.; Kressel, J.; Münch, G.; Pischetsrieder, M. Activation of the transcription factor Nrf2 in macrophages, Caco-2 cells and intact human gut tissue by Maillard reaction products and coffee. Amino Acids 2012, 44, 1427–1439. [Google Scholar] [CrossRef]
- He, M.; Siow, R.C.M.; Sugden, D.; Gao, L.; Cheng, X.; Mann, G.E. Induction of HO-1 and redox signaling in endothelial cells by advanced glycation end products: A role for Nrf2 in vascular protection in diabetes. Nutr. Metab. Cardiovasc. Dis. 2011, 21, 277–285. [Google Scholar] [CrossRef]
- Pall, M.L.; Levine, S. Nrf2, a master regulator of detoxification and also antioxidant, anti-inflammatory and other cytoprotective mechanisms, is raised by health promoting factors. Sheng Li Xue Bao Acta Physiol. Sin. 2015, 67, 1–18. [Google Scholar]
- Fuse, Y.; Kobayashi, M. Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time. Molecules 2017, 22, 436. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [Green Version]
- Alam, J.; Cook, J.L. Transcriptional regulation of the heme oxygenase-1 gene via the stress response element pathway. Curr. Pharm. Des. 2003, 9, 2499–2511. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, R.; Jaiswal, A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. USA 1996, 93, 14960–14965. [Google Scholar] [CrossRef] [Green Version]
- Solis, W.A.; Dalton, T.P.; Dieter, M.Z.; Freshwater, S.; Harrer, J.M.; He, L.; Shertzer, H.G.; Nebert, D.W. Glutamate–cysteine ligase modifier subunit: Mouse Gclm gene structure and regulation by agents that cause oxidative stress. Biochem. Pharmacol. 2002, 63, 1739–1754. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Angeli, J.P.F.; Bayir, H.; Bush, A.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Baldensperger, T.; Eggen, M.; Kappen, J.; Winterhalter, P.R.; Pfirrmann, T.; Glomb, M.A. Comprehensive analysis of posttranslational protein modifications in aging of subcellular compart-ments. Sci. Rep. 2020, 10, 7596. [Google Scholar] [CrossRef] [PubMed]
- Smuda, M.; Henning, C.; Raghavan, C.T.; Johar, K.; Vasavada, A.R.; Nagaraj, R.H.; Glomb, M.A. Comprehensive Analysis of Maillard Protein Modifications in Human Lenses: Effect of Age and Cataract. Biochemistry 2015, 54, 2500–2507. [Google Scholar] [CrossRef]
- Glomb, M.A.; Lang, G. Isolation and Characterization of Glyoxal−Arginine Modifications. J. Agric. Food Chem. 2001, 49, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Klopfer, A.; Spanneberg, R.; Glomb, M.A. Formation of arginine modifications in a model system of Nal-pha-tert-butoxycarbonyl (Boc)-arginine with methylglyoxal. J. Agric. Food Chem. 2011, 59, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Glomb, M.A.; Pfahler, C. Amides Are Novel Protein Modifications Formed by Physiological Sugars. J. Biol. Chem. 2001, 276, 41638–41647. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Tamba, M.; Ishii, T.; Bannai, S. Cloning and Expression of a Plasma Membrane Cystine/Glutamate Exchange Transporter Composed of Two Distinct Proteins. J. Biol. Chem. 1999, 274, 11455–11458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, P.; Wang, Q.; Sun, F.; Liu, F. Sulforaphane Attenuates H(2)O(2)-induced Oxidant Stress in Human Trabecular Meshwork Cells (HTMCs) via the Phosphatidylinositol 3-Kinase (PI3K)/Serine/Threonine Kinase (Akt)-Mediated Factor-E2-Related Factor 2 (Nrf2) Signal-ing Activation. Med. Sci. Monit. 2019, 25, 811–818. [Google Scholar] [CrossRef]
- Assar, S.H.; Moloney, C.; Lima, M.; Magee, R.; Ames, J.M. Determination of Nepsilon-(carboxymethyl)lysine in food systems by ultra performance liquid chromatog-raphy-mass spectrometry. Amino Acids 2009, 36, 317–326. [Google Scholar] [CrossRef]
- Treibmann, S.; Hellwig, A.; Hellwig, M.; Henle, T. Lysine-Derived Protein-Bound Heyns Compounds in Bakery Products. J. Agric. Food Chem. 2017, 65, 10562–10570. [Google Scholar] [CrossRef]
- van der Lugt, T.; Venema, K.; van Leeuwen, S.; Vrolijk, M.F.; Opperhuizen, A.; Bast, A. Gastrointestinal digestion of dietary advanced glycation endproducts using an in vitro model of the gastrointestinal tract (TIM-1). Food Funct. 2020, 11, 6297–6307. [Google Scholar] [CrossRef] [PubMed]
- Michalska, A.; Amigo-Benavent, M.; Zielinski, H.; Castillo, D. Effect of bread making on formation of Maillard reaction products contributing to the overall antioxidant activity of rye bread. J. Cereal Sci. 2008, 48, 123–132. [Google Scholar] [CrossRef]
- Scheijen, J.L.; Clevers, E.; Engelen, L.; Dagnelie, P.C.; Brouns, F.; Stehouwer, C.D.; Schalkwijk, C.G. Analysis of advanced glycation endproducts in selected food items by ultra-performance liquid chromatography tandem mass spectrometry: Presentation of a dietary AGE database. Food Chem. 2016, 190, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Satoh, K.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Moreno-Murciano, P.; Pedraza-Chaverri, J. The transcription factor Nrf2 as a new therapeutic target in Parkinson’s disease. Expert Opin. Ther. Targets 2009, 13, 319–329. [Google Scholar] [CrossRef]
- Cheng, X.; Siow, R.C.M.; Mann, G.E. Impaired Redox Signaling and Antioxidant Gene Expression in Endothelial Cells in Diabetes: A Role for Mitochondria and the Nuclear Factor-E2-Related Factor 2-Kelch-Like ECH-Associated Protein 1 Defense Pathway. Antioxid. Redox Signal. 2011, 14, 469–487. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Ji, Y.; Hu, Q.; Yan, W.; Chen, G.; Yin, H. Increased intestinal inflammatory response and gut barrier dysfunction in Nrf2-deficient mice after traumatic brain injury. Cytokine 2008, 44, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ichikawa, T.; Janicki, J.S.; Cui, T. Targeting the Nrf2 pathway against cardiovascular disease. Expert Opin. Ther. Targets 2009, 13, 785–794. [Google Scholar] [CrossRef]
- Yet, S.-F.; Tian, R.; Layne, M.; Wang, Z.Y.; Maemura, K.; Solovyeva, M.; Ith, B.; Melo, L.G.; Zhang, L.; Ingwall, J.S.; et al. Cardiac-Specific Expression of Heme Oxygenase-1 Protects Against Ischemia and Reperfusion Injury in Transgenic Mice. Circ. Res. 2001, 89, 168–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrella, M.A.; Yet, S.F. Role of heme oxygenase-1 in cardiovascular function. Curr. Pharm. Des. 2003, 9, 2479–2487. [Google Scholar] [CrossRef]
- Marelli, G.; Erreni, M.; Anselmo, A.; Taverniti, V.; Guglielmetti, S.; Mantovani, A.; Allavena, P. Heme-oxygenase-1 Production by Intestinal CX3CR1+ Macrophages Helps to Resolve Inflammation and Prevents Carcinogenesis. Cancer Res. 2017, 77, 4472–4485. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Huang, S.; Zeng, L.; Ma, J.; Sun, S.; Zeng, F.; Kong, F.; Cheng, X. HMOX-1 inhibits TGF-beta-induced epithelial-mesenchymal transition in the MCF-7 breast cancer cell line. Int. J. Mol. Med. 2017, 40, 411–417. [Google Scholar] [CrossRef] [Green Version]
- Ross, D.; Kepa, J.K.; Winski, S.L.; Beall, H.D.; Anwar, A.; Siegel, D. NAD(P)H:quinone oxidoreductase 1 (NQO1): Chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem. Interact. 2000, 129, 77–97. [Google Scholar] [CrossRef]
- Landi, L.; Fiorentini, D.; Galli, M.; Segura-Aguilar, J.; Beyer, R. DT-Diaphorase Maintains the Reduced State of Ubiquinones in Lipid Vesicles thereby Promoting their Antioxidant Function. Free Radic. Biol. Med. 1997, 22, 329–335. [Google Scholar] [CrossRef]
- Siegel, D.; Bolton, E.M.; Burr, J.A.; Liebler, D.; Ross, D. The Reduction of α-Tocopherolquinone by Human NAD(P)H:Quinone Oxidoreductase: The Role of α-Tocopherolhydroquinone as a Cellular Antioxidant. Mol. Pharmacol. 1997, 52, 300–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinkova-Kostova, A.T.; Talalay, P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikami, K.; Naito, M.; Tomida, A.; Yamada, M.; Sirakusa, T.; Tsuruo, T. DT-diaphorase as a critical determinant of sensitivity to mitomycin C in human colon and gastric carcinoma cell lines. Cancer Res. 1996, 56, 2823–2826. [Google Scholar] [PubMed]
- Oh, E.-T.; Park, H.J. Implications of NQO1 in cancer therapy. BMB Rep. 2015, 48, 609–617. [Google Scholar] [CrossRef] [Green Version]
- Ott, C.; Jacobs, K.; Haucke, E.; Santos, A.N.; Grune, T.; Simm, A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014, 2, 411–429. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.-Y.; Lu, C.-H.; Wu, C.-H.; Li, K.-J.; Kuo, Y.-M.; Hsieh, S.-C.; Yu, C.-L. The Development of Maillard Reaction, and Advanced Glycation End Product (AGE)-Receptor for AGE (RAGE) Signaling Inhibitors as Novel Therapeutic Strategies for Patients with AGE-Related Diseases. Molecules 2020, 25, 5591. [Google Scholar] [CrossRef]
- Del Turco, S.; Basta, G. An update on advanced glycation endproducts and atherosclerosis. BioFactors 2012, 38, 266–274. [Google Scholar] [CrossRef]
- Lu, C.; He, J.C.; Cai, W.; Liu, H.; Zhu, L.; Vlassara, H. Advanced glycation endproduct (AGE) receptor 1 is a negative regulator of the inflammatory response to AGE in mesangial cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11767–11772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, W.; He, J.C.; Zhu, L.; Lu, C.; Vlassara, H. Advanced glycation end product (AGE) receptor 1 suppresses cell oxidant stress and activation signaling via EGF receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 13801–13806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.M.; Mitsuhashi, T.; Wojciechowicz, D.; Shimizu, N.; Li, J.; Stitt, A.; He, C.; Banerjee, D.; Vlassara, H. Molecular identity and cellular distribution of advanced glycation endproduct receptors: Relationship of p60 to OST-48 and p90 to 80K-H membrane proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 11047–11052. [Google Scholar] [CrossRef] [Green Version]
- Kanai, M.; Göke, M.; Tsunekawa, S.; Podolsky, D.K. Signal Transduction Pathway of Human Fibroblast Growth Factor Receptor 3. J. Biol. Chem. 1997, 272, 6621–6628. [Google Scholar] [CrossRef] [Green Version]
- Goh, K.C.; Lim, Y.P.; Ong, S.H.; Bin Siak, C.; Cao, X.; Tan, Y.H.; Guy, G.R. Identification of p90, a Prominent Tyrosine-phosphorylated Protein in Fibroblast Growth Factor-stimulated Cells, as 80K-H. J. Biol. Chem. 1996, 271, 5832–5838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkinson, C.P.; Mander, A.; Sale, G.J. Identification of 80K-H as a protein involved in GLUT4 vesicle trafficking. Biochem. J. 2005, 388, 785–793. [Google Scholar] [CrossRef]
- Vlassara, H.; Li, Y.M.; Imani, F.; Wojciechowicz, D.; Yang, Z.; Liu, F.T.; Cerami, A. Identification of galectin-3 as a high-affinity binding protein for advanced glycation end products (AGE): A new member of the AGE-receptor complex. In Proceedings of the 2009 IEEE 3rd International Conference on Nano/Molecular Medicine and Engineering, Tainan, Taiwan, 18–21 October 2009. [Google Scholar]
- Yamaoka, A.; Kuwabara, I.; Frigeri, L.G.; Liu, F.T. A human lectin, galectin-3 (epsilon bp/Mac-2), stimulates superoxide production by neutrophils. J. Immunol. 1995, 154, 3479–3487. [Google Scholar]
- Liu, F.T.; Hsu, D.K.; Zuberi, R.I.; Kuwabara, I.; Chi, E.Y.; Henderson, W.R. Expression and function of galectin-3, a beta-galactoside-binding lectin, in human monocytes and macrophages. Am. J. Pathol. 1995, 147, 1016–1028. [Google Scholar]
- Jiang, G.-P.; Liao, Y.-J.; Huang, L.-L.; Zeng, X.-J.; Liao, X.-H. Effects and molecular mechanism of pachymic acid on ferroptosis in renal ischemia reperfusion injury. Mol. Med. Rep. 2020, 23, 1. [Google Scholar] [CrossRef]
- Dong, H.; Qiang, Z.; Chai, D.; Peng, J.; Xia, Y.; Hu, R.; Jiang, H. Nrf2 inhibits ferroptosis and protects against acute lung injury due to intestinal ischemia reperfusion via regulating SLC7A11 and HO-1. Aging 2020, 12, 12943–12959. [Google Scholar] [CrossRef]
- Tian, Y.; Lu, J.; Hao, X.; Li, H.; Zhang, G.; Liu, X.; Li, X.; Zhao, C.; Kuang, W.; Chen, D.; et al. FTH1 Inhibits Ferroptosis Through Ferritinophagy in the 6-OHDA Model of Parkinson’s Disease. Neurotherapeutics 2020, 17, 1796–1812. [Google Scholar] [CrossRef] [PubMed]
- Abdalkader, M.; Lampinen, R.; Kanninen, K.M.; Malm, T.; Liddell, J.R. Targeting Nrf2 to Suppress Ferroptosis and Mitochondrial Dysfunction in Neurodegeneration. Front. Neurosci. 2018, 12, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, H.; Han, D.; Xie, E.; Yang, X.; Wei, J.; Gu, S.; Gao, F.; Zhu, N.; Yin, X.; et al. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. USA 2019, 116, 2672–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Z.; Wirth, A.-K.; Chen, D.; Wruck, C.J.; Rauh, M.; Buchfelder, M.; Savaskan, N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017, 6, e371. [Google Scholar] [CrossRef] [Green Version]
- Dixon, S.J.; Patel, D.N.; Welsch, M.; Skouta, R.; Lee, E.; Hayano, M.; Thomas, A.G.; Gleason, C.; Tatonetti, N.; Slusher, B.S.; et al. Pharmacological inhibition of cystine–glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 2014, 3, e02523. [Google Scholar] [CrossRef] [PubMed]
- Dodson, M.; Castro-Portuguez, R.; Zhang, D.D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019, 23, 101107. [Google Scholar] [CrossRef]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Kasai, S.; Mimura, J.; Ozaki, T.; Itoh, K. Emerging Regulatory Role of Nrf2 in Iron, Heme, and Hemoglobin Metabolism in Physiology and Disease. Front. Veter- Sci. 2018, 5, 242. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.Q.; Feng, Y.L.; Chen, L.; Liu, J.R.; Wang, M.; Vaziri, N.D.; Zhao, Y.Y. Poricoic acid A enhances melatonin inhibition of AKI-to-CKD transition by regulating Gas6/AxlNFkappaB/Nrf2 axis. Free Radic. Biol. Med. 2019, 134, 484–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adedoyin, O.; Boddu, R.; Traylor, A.M.; Lever, J.M.; Bolisetty, S.; George, J.F.; Agarwal, A. Heme oxygenase-1 mitigates ferroptosis in renal proximal tubule cells. Am. J. Physiol. Physiol. 2018, 314, F702–F714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, Q.; Shi, C.; Jiao, F.; Gong, Z. Mechanism of glycyrrhizin on ferroptosis during acute liver failure by inhibiting oxidative stress. Mol. Med. Rep. 2019, 20, 4081–4090. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zou, Y.; Xiang, J.; Fu, Y.-Y.; Wang, K.-Y.; Wan, P.-Z.; Zhai, X.-Y. Pretreatment with Roxadustat (FG-4592) Attenuates Folic Acid-Induced Kidney Injury through Antiferroptosis via Akt/GSK-3beta/Nrf2 Pathway. Oxid. Med. Cell Longev. 2020, 2020, 6286984. [Google Scholar]
- Xin, Y.; Bai, Y.; Jiang, X.; Zhou, S.; Wang, Y.; Wintergerst, K.A.; Cui, T.; Ji, H.; Tan, Y.; Cai, L. Sulforaphane prevents angiotensin II-induced cardiomyopathy by activation of Nrf2 via stimulating the Akt/GSK-3ß/Fyn pathway. Redox Biol. 2018, 15, 405–417. [Google Scholar] [CrossRef]
- Chen, Q.M.; Maltagliati, A.J. Nrf2 at the heart of oxidative stress and cardiac protection. Physiol. Genom. 2018, 50, 77–97. [Google Scholar] [CrossRef]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef] [Green Version]
- Salazar, M.; Rojo, A.I.; Velasco, D.; de Sagarra, R.M.; Cuadrado, A. Glycogen synthase kinase-3beta inhibits the xenobiotic and antioxidant cell response by direct phosphorylation and nuclear exclusion of the transcription factor Nrf2. J. Biol. Chem. 2006, 281, 14841–14851. [Google Scholar] [CrossRef] [Green Version]
- Zakkar, M.; van der Heiden, K.; Luong, L.A.; Chaudhury, H.; Cuhlmann, S.; Hamdulay, S.S.; Krams, R.; Edirisinghe, I.; Rahman, I.; Carlsen, H.; et al. Activation of Nrf2 in Endothelial Cells Protects Arteries from Exhibiting a Proinflammatory State. Arter. Thromb. Vasc. Biol. 2009, 29, 1851–1857. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Li, Y.; Ji, Z. Atorvastatin attenuates inflammation and oxidative stress induced by ischemia/reperfusion in rat heart via the Nrf2 transcription factor. Int. J. Clin. Exp. Med. 2015, 8, 14837–14845. [Google Scholar]
- Li, X.-H.; Li, C.-Y.; Xiang, Z.-G.; Hu, J.-J.; Lu, J.-M.; Tian, R.-B.; Jia, W. Allicin Ameliorates Cardiac Hypertrophy and Fibrosis through Enhancing of Nrf2 Antioxidant Signaling Pathways. Cardiovasc. Drugs Ther. 2012, 26, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Xie, J.; Song, C.; Liu, J.; Zheng, J.; Liu, C.; Zhang, X.; Li, P.; Wang, F. The Activation of Nrf2 and Its Downstream Regulated Genes Mediates the Antioxidative Activities of Xueshuan Xinmaining Tablet in Human Umbilical Vein Endothelial Cells. Evid.-Based Complement. Altern. Med. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, W.; Taub, D.D.; Gardner, K. qPrimerDepot: A primer database for quantitative real time PCR. Nucleic Acids Res. 2007, 35, D805–D809. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wächter, K.; Navarrete Santos, A.; Großkopf, A.; Baldensperger, T.; Glomb, M.A.; Szabó, G.; Simm, A. AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway. Nutrients 2021, 13, 3874. https://doi.org/10.3390/nu13113874
Wächter K, Navarrete Santos A, Großkopf A, Baldensperger T, Glomb MA, Szabó G, Simm A. AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway. Nutrients. 2021; 13(11):3874. https://doi.org/10.3390/nu13113874
Chicago/Turabian StyleWächter, Kristin, Alexander Navarrete Santos, Anne Großkopf, Tim Baldensperger, Marcus A. Glomb, Gábor Szabó, and Andreas Simm. 2021. "AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway" Nutrients 13, no. 11: 3874. https://doi.org/10.3390/nu13113874
APA StyleWächter, K., Navarrete Santos, A., Großkopf, A., Baldensperger, T., Glomb, M. A., Szabó, G., & Simm, A. (2021). AGE-Rich Bread Crust Extract Boosts Oxidative Stress Interception via Stimulation of the NRF2 Pathway. Nutrients, 13(11), 3874. https://doi.org/10.3390/nu13113874