Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal–Fetal Outcomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Establishment of GDM Animal Model
2.2. Sample Collection and Preparation
2.3. LC-MS/MS Analysis
2.4. Statistical Analysis
3. Results
3.1. GDM Animal Models
3.2. QC Sample Analysis
3.3. Multivariate Analysis of LC-MS/MS Data
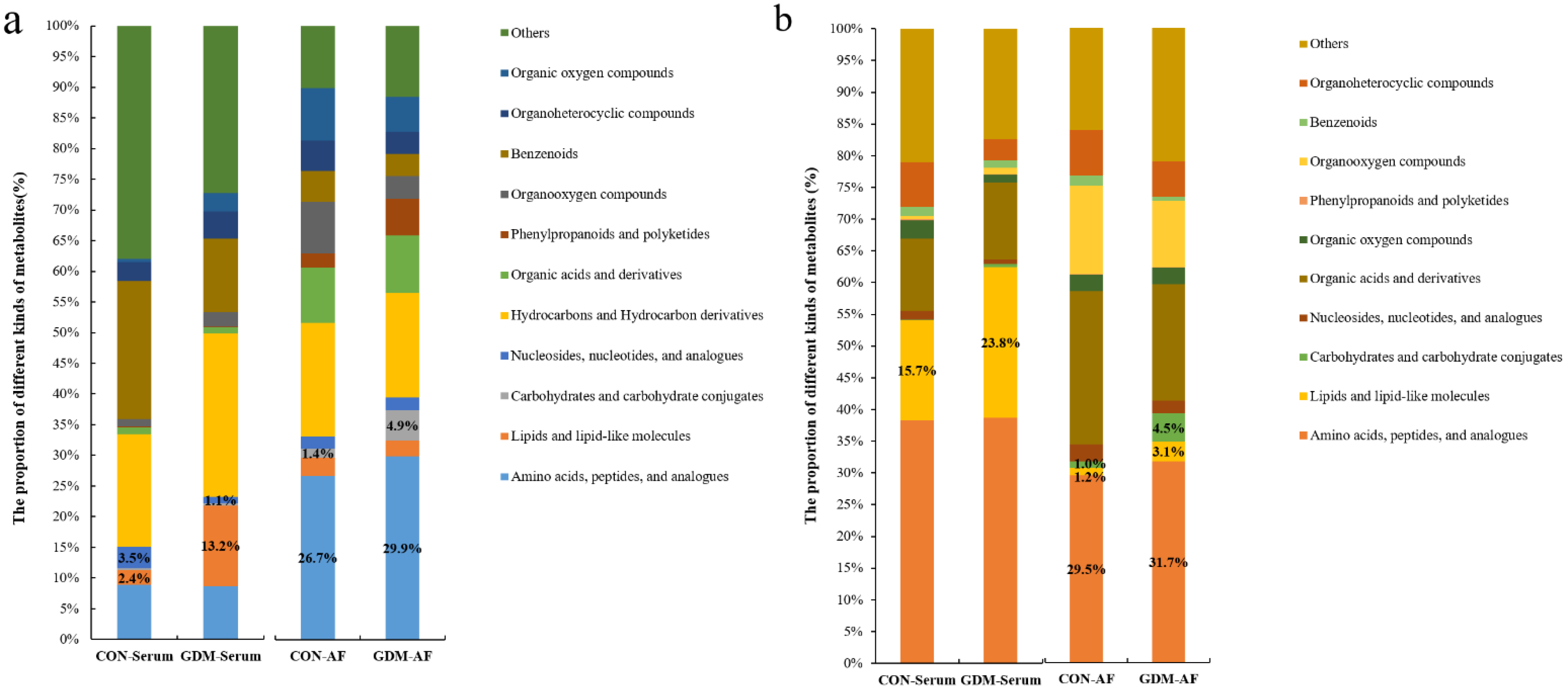
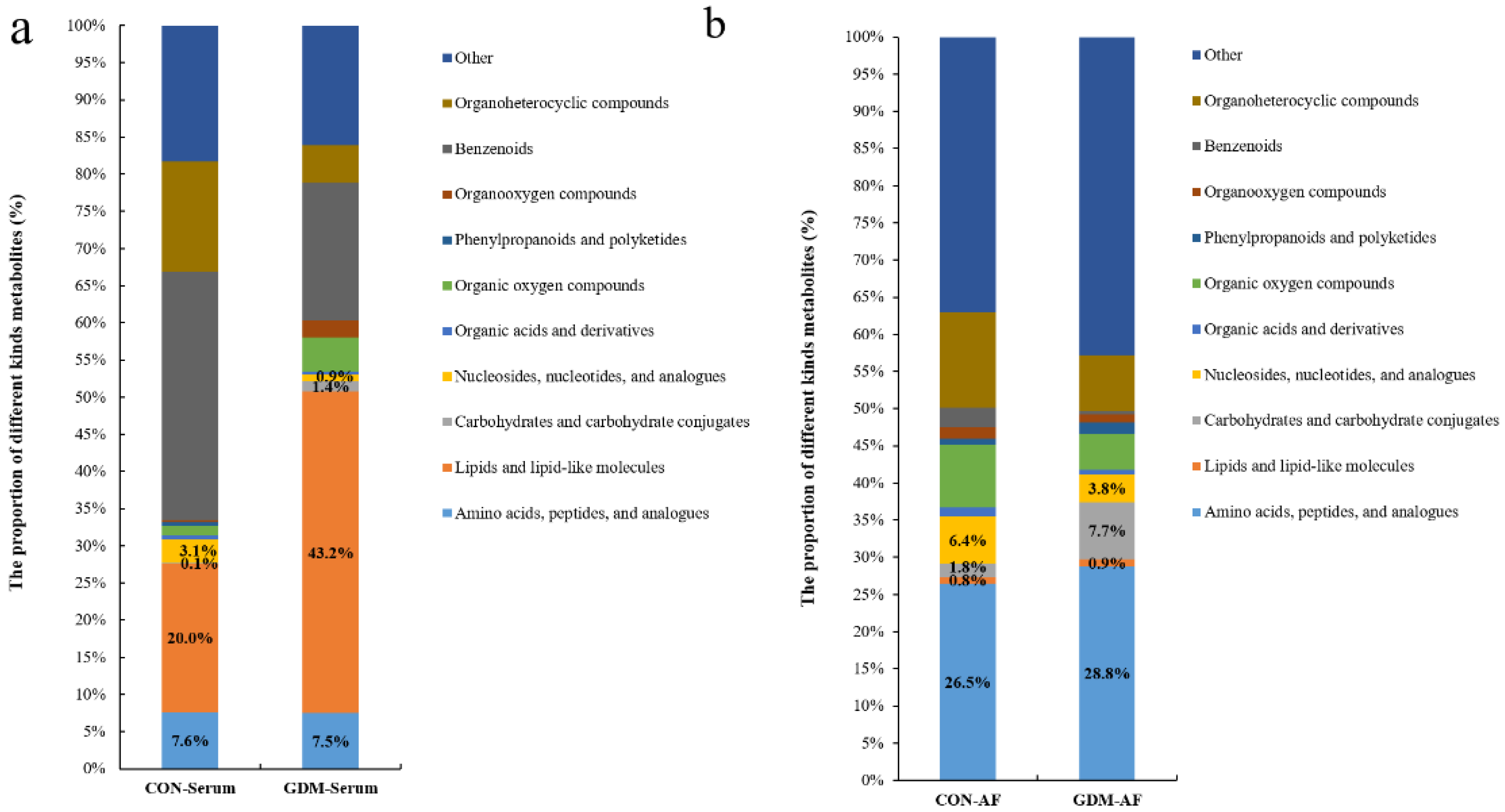
3.4. Proportion of Identified Metabolites in Serum and Amniotic Fluid
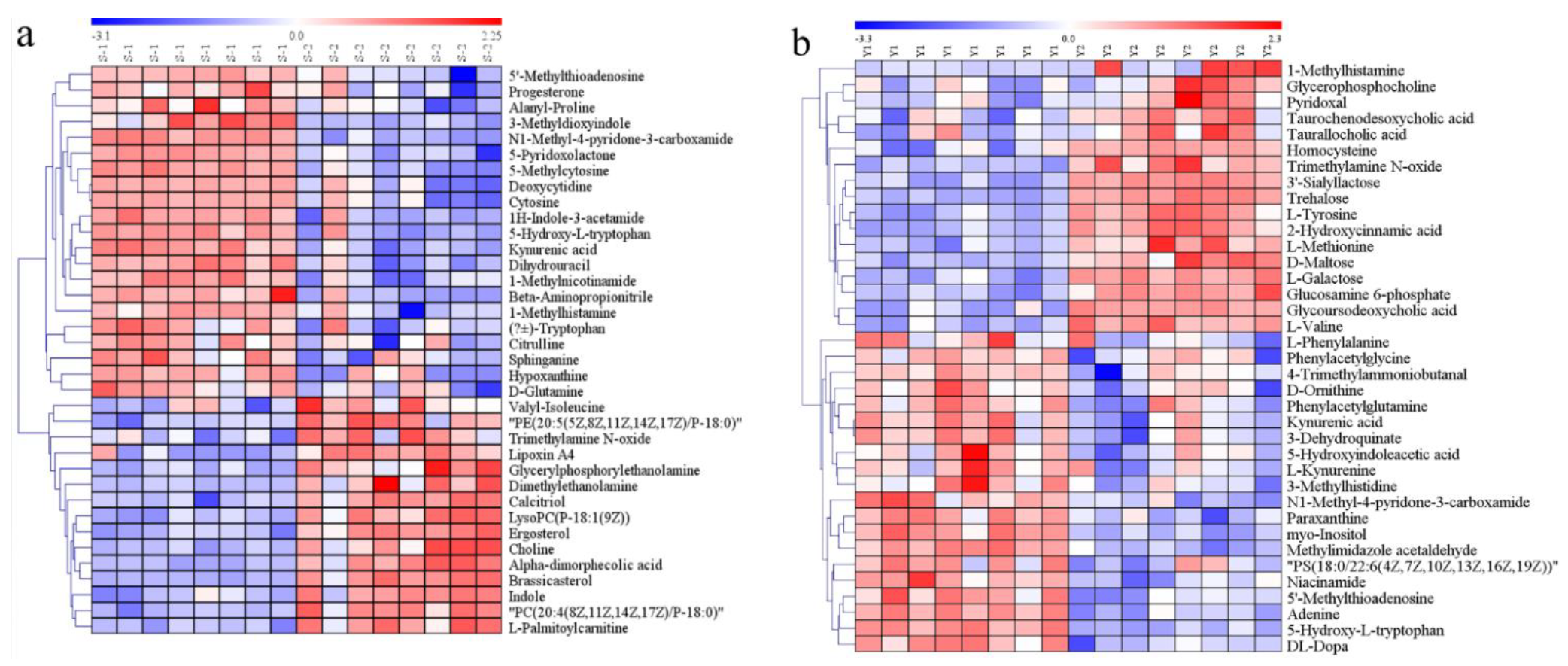
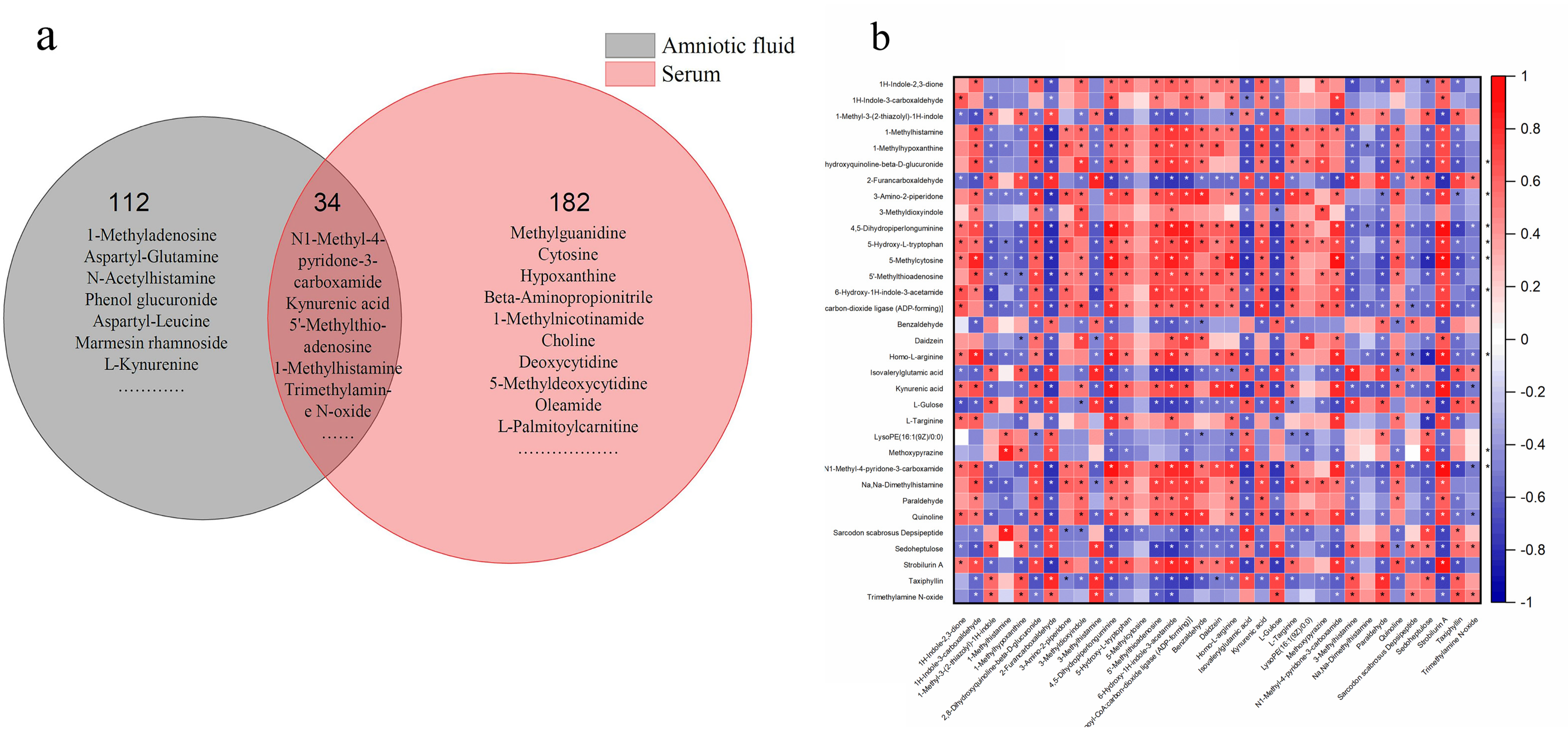
3.5. Identification of Differential Metabolites
3.6. Altered Metabolic Pathways Induced by Differential Metabolites
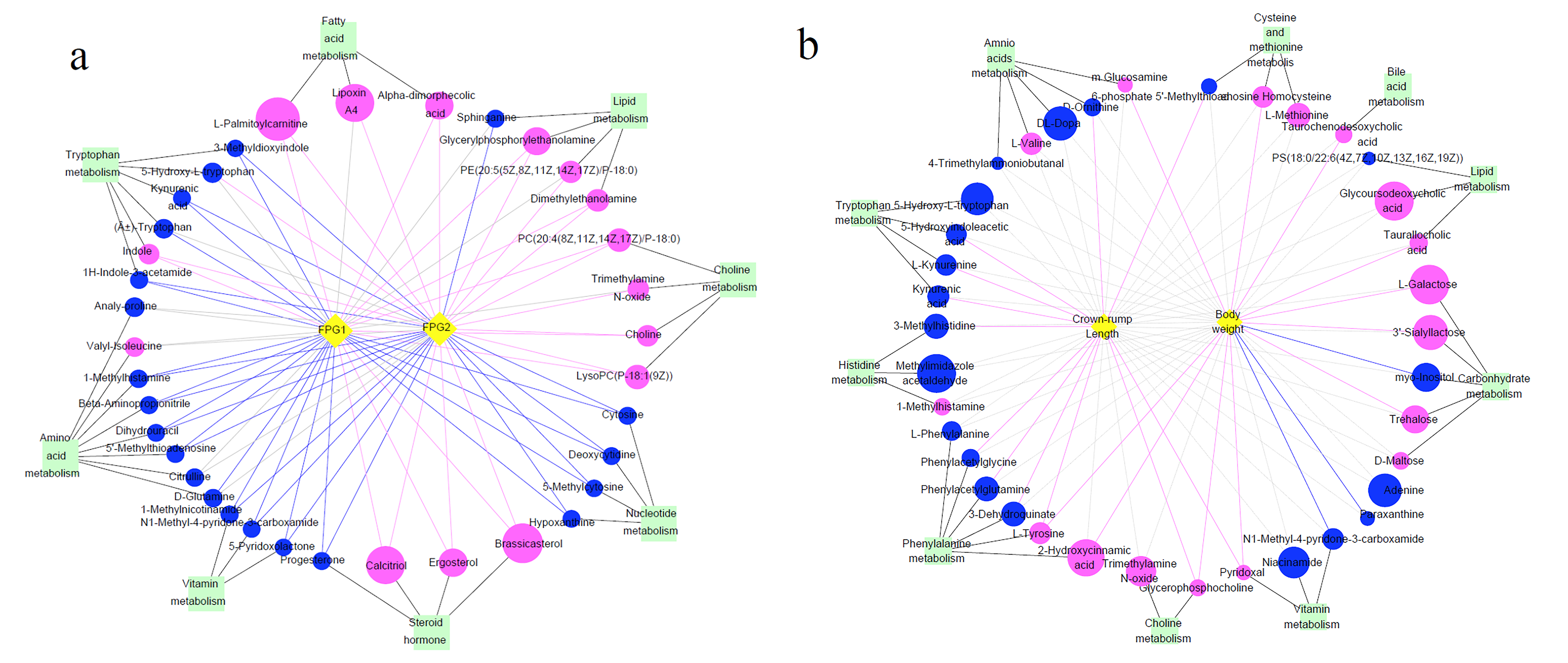
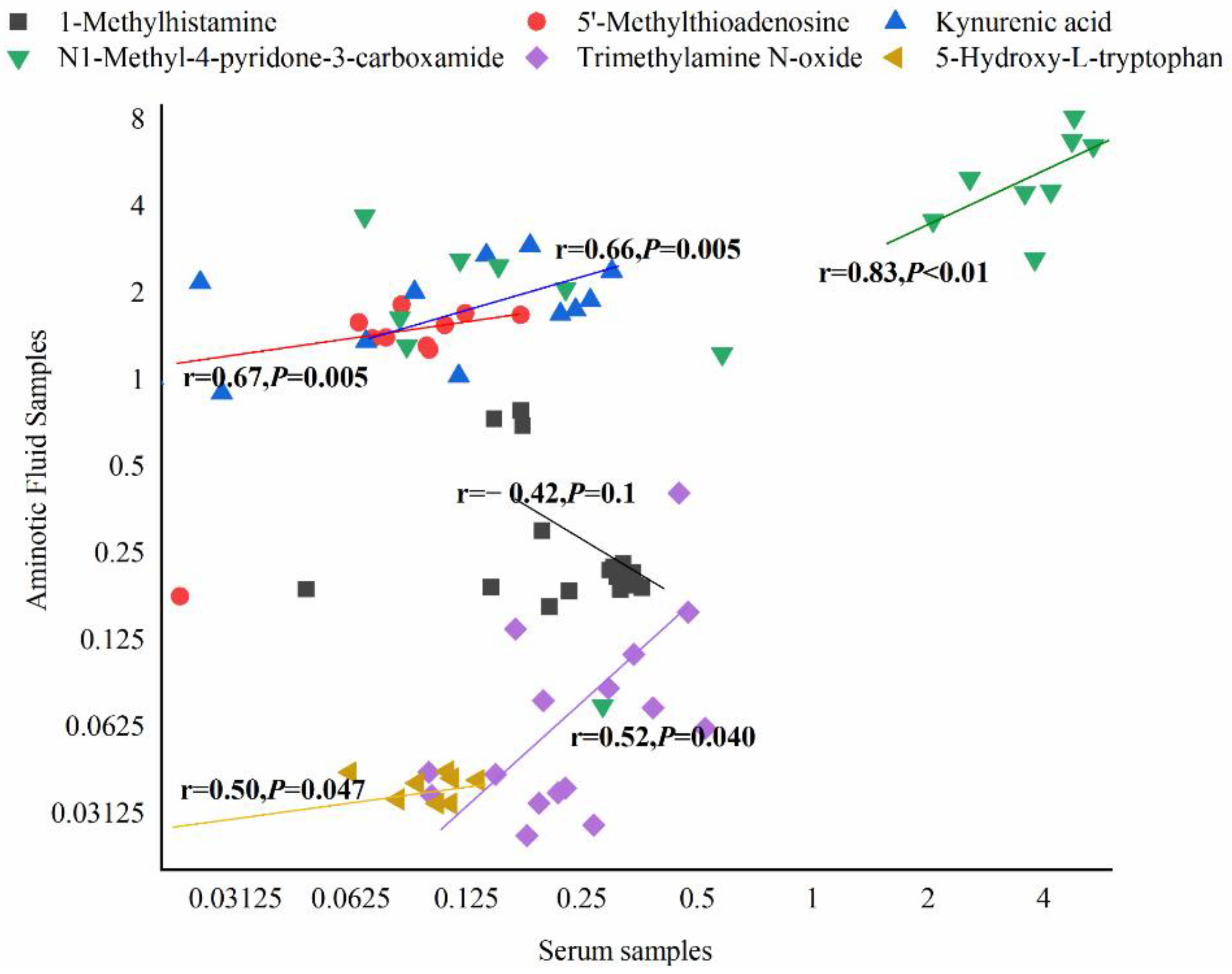
3.7. Differential Metabolites and Their Maternal–Fetal Repercussions
4. Discussion
4.1. Differential Metabolites Related to Amino Acid Metabolism in Serum Exposed to GDM
4.1.1. Differential Metabolites Related to Amino Acid Metabolism and Its Relationship with Maternal FPG
4.1.2. Changes in Amino Acid Metabolism Profiles and Their Relationship with Fetal Weight and Crown–Rump Length
4.2. Lipid Metabolism
4.3. Carbohydrate Metabolism
4.4. Common Metabolism in Serum and AF Exposed to GDM
4.4.1. Choline Metabolism
4.4.2. Tryptophan Metabolism
4.4.3. Histidine Metabolism
4.4.4. Nicotinate and Nicotinamide Metabolism
4.4.5. Methionine Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Lee, K.W.; Ching, S.M.; Ramachandran, V.; Yee, A.; Hoo, F.K.; Chia, Y.C.; Wan Sulaiman, W.A.; Suppiah, S.; Mohamed, M.H.; Veettil, S.K. Prevalence and risk factors of gestational diabetes mellitus in Asia: A systematic review and meta-analysis. BMC Pregnancy Childbirth 2018, 18, 494. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Liu, Q.; Zhang, Y.; Li, J.; Yang, J.; Wang, L. Prevalence and Epidemiologic Characteristics of Gestational Diabetes Mellitus in Hebei Province, 2016. Chin. Gen. Pract. 2018, 21, 2592–2596. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, Q.; Sheng, W.; Wei, M.; Yang, N.; Li, Y. Investigation and analysis of risk factors and pregnancy outcome of gestational diabetes in China. Chin. Med. J. 2019, 54, 1014–1019. [Google Scholar]
- Yu, Y.; Arah, O.A.; Liew, Z.; Cnattingius, S.; Olsen, J.; Sørensen, H.T.; Qin, G.; Li, J. Maternal diabetes during pregnancy and early onset of cardiovascular disease in offspring: Population based cohort study with 40 years of follow-up. BMJ 2019, 367, l6398. [Google Scholar] [CrossRef] [Green Version]
- Lowe, W.L., Jr.; Scholtens, D.M.; Kuang, A.; Linder, B.; Lawrence, J.M.; Lebenthal, Y.; McCance, D.; Hamilton, J.; Nodzenski, M.; Talbot, O.; et al. Hyperglycemia and Adverse Pregnancy Outcome Follow-up Study (HAPO FUS): Maternal Gestational Diabetes Mellitus and Childhood Glucose Metabolism. Diabetes Care 2019, 42, 372–380. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Zhang, C. Prevalence of Gestational Diabetes and Risk of Progression to Type 2 Diabetes: A Global Perspective. Curr. Diabetes Rep. 2016, 16, 7. [Google Scholar] [CrossRef]
- Kong, L.; Nilsson, I.A.; Gissler, M.; Lavebratt, C. Associations of Maternal Diabetes and Body Mass Index with Offspring Birth Weight and Prematurity. JAMA Pediatr. 2019, 173, 371–378. [Google Scholar] [CrossRef]
- Tan, B.; Ma, Y.; Zhang, L.; Li, N.; Zhang, J. The application of metabolomics analysis in the research of gestational diabetes mellitus and preeclampsia. J. Obstet. Gynaecol. Res. 2020, 46, 1310–1318. [Google Scholar] [CrossRef]
- O’Neill, K.; Alexander, J.; Azuma, R.; Xiao, R.; Snyder, N.W.; Mesaros, C.A.; Blair, I.A.; Pinney, S.E. Gestational Diabetes Alters the Metabolomic Profile in 2nd Trimester Amniotic Fluid in a Sex-Specific Manner. Int. J. Mol. Sci. 2018, 19, 2696. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.-Z.; Gerszten, R.E. Metabolomics and Proteomics in Type 2 Diabetes. Circ. Res. 2020, 126, 1613–1627. [Google Scholar] [CrossRef]
- Guasch-Ferré, M.; Hruby, A.; Toledo, E.; Clish, C.B.; Martínez-González, M.A.; Salas-Salvadó, J.; Hu, F.B. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care 2016, 39, 833–846. [Google Scholar] [CrossRef] [Green Version]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.; Chung, A.C.K.; Xiang, L.; Li, X.; Zheng, Y.; Luan, H.; Zhu, L.; Liu, W.; Peng, Y.; et al. Large-Scale Longitudinal Metabolomics Study Reveals Different Trimester-Specific Alterations of Metabolites in Relation to Gestational Diabetes Mellitus. J. Proteome Res. 2019, 18, 292–300. [Google Scholar] [CrossRef]
- Tian, M.; Ma, S.; You, Y.; Long, S.; Zhang, J.; Guo, C.; Wang, X.; Tan, H. Serum Metabolites as an Indicator of Developing Gestational Diabetes Mellitus Later in the Pregnancy: A Prospective Cohort of a Chinese Population. J. Diabetes Res. 2021, 2021, 8885954. [Google Scholar] [CrossRef]
- Mokkala, K.; Vahlberg, T.; Pellonperä, O.; Houttu, N.; Koivuniemi, E.; Laitinen, K. Distinct Metabolic Profile in Early Pregnancy of Overweight and Obese Women Developing Gestational Diabetes. J. Nutr. 2020, 150, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Graça, G.; Goodfellow, B.J.; Barros, A.S.; Diaz, S.; Duarte, I.F.; Spagou, K.; Veselkov, K.; Want, E.J.; Lindon, J.C.; Carreira, I.M.; et al. UPLC-MS metabolic profiling of second trimester amniotic fluid and maternal urine and comparison with NMR spectral profiling for the identification of pregnancy disorder biomarkers. Mol. BioSyst. 2012, 8, 1243–1254. [Google Scholar] [CrossRef]
- Damasceno, D.C.; Sinzato, Y.K.; Bueno, A.; Netto, A.O.; Dallaqua, B.; Gallego, F.Q.; Iessi, I.L.; Corvino, S.B.; Serrano, R.G.; Marini, G.; et al. Mild Diabetes Models and Their Maternal-Fetal Repercussions. J. Diabetes Res. 2013, 2013, 473575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.T.; Lili, L.N.; Li, S.; Tran, V.T.; Stewart, K.B.; Schwartz, C.E.; Jones, D.P.; Sherman, S.L.; Fridovich-Keil, J.L. Low-level maternal exposure to nicotine associates with significant metabolic perturbations in second-trimester amniotic fluid. Environ. Int. 2017, 107, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Hajar, A.A.S.; Mathew, J.C.; Saidaah, M.Y.N.I.; Massita, N.; Rajesh, R.; Aishah, A.; Fazlin, M.F. Animal Model of Gestational Diabetes Mellitus with Pathophysiological Resemblance to the Human Condition Induced by Multiple Factors (Nutritional, Pharmacological, and Stress) in Rats. Biomed Res. Int. 2016, 2016, 9704607. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Castrejon, M.; Powell, T.L. Placental Nutrient Transport in Gestational Diabetic Pregnancies. Front. Endocrinol. 2017, 8, 306. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Pérez, A.; Vilariño-García, T.; Guadix, P.; Dueñas, J.L.; Sánchez-Margalet, V. Leptin and Nutrition in Gestational Diabetes. Nutrients 2020, 12, 1970. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhu, B.; Liu, Y.; Fang, L.; Yin, B.; Sun, Y.; Ma, M.; Huang, Y.; Zhu, Y.; Zhang, Y. Unique Biomarker Characteristics in Gestational Diabetes Mellitus Identified by LC-MS-Based Metabolic Profiling. J. Diabetes Res. 2021, 2021, 6689414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kuang, A.; Bain, J.R.; Muehlbauer, M.J.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; Scholtens, D.M.; Lowe, W.L. Maternal Metabolites Associated with Gestational Diabetes Mellitus and a Postpartum Disorder of Glucose Metabolism. J. Clin. Endocrinol. Metab. 2021. [Google Scholar] [CrossRef]
- Huynh, J.; Xiong, G.; Bentley-Lewis, R. A systematic review of metabolite profiling in gestational diabetes mellitus. Diabetologia 2014, 57, 2453–2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Kuang, A.; Talbot, O.; Bain, J.R.; Muehlbauer, M.J.; Hayes, M.G.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Metabolomic and genetic associations with insulin resistance in pregnancy. Diabetologia 2020, 63, 1783–1795. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Wu, S.; Fang, C.; Wang, C.; Yang, Y.; Liu, C.; Hu, J.; Huang, Y. Amino acids levels in early pregnancy predict subsequent gestational diabetes. J. Diabetes 2020, 12, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Kadakia, R.; Talbot, O.; Kuang, A.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; Lowe, L.P.; Metzger, B.E.; Newgard, C.B.; et al. Cord Blood Metabolomics: Association With Newborn Anthropometrics and C-Peptide Across Ancestries. J. Clin. Endocrinol. Metab. 2019, 104, 4459–4472. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Duparc, T.; Plovier, H.; Marrachelli, V.G.; Van Hul, M.; Essaghir, A.; Ståhlman, M.; Matamoros, S.; Geurts, L.; Pardo-Tendero, M.M.; Druart, C.; et al. Hepatocyte MyD88 affects bile acids, gut microbiota and metabolome contributing to regulate glucose and lipid metabolism. Gut 2017, 66, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Molinaro, A.; Wahlström, A.; Marschall, H.-U. Role of Bile Acids in Metabolic Control. Trends Endocrinol. Metab. 2018, 29, 31–41. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E.; et al. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef] [PubMed]
- Maessen, S.E.; Derraik, J.G.B.; Binia, A.; Cutfield, W.S. Perspective: Human Milk Oligosaccharides: Fuel for Childhood Obesity Prevention? Adv. Nutr. 2020, 11, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Jantscher-Krenn, E.; Treichler, C.; Brandl, W.; Schönbacher, L.; Kofeler, H.; Van Poppel, M.N.M. The association of human milk oligosaccharides with glucose metabolism in overweight and obese pregnant women. Am. J. Clin. Nutr. 2019, 110, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Wise, A.; Robertson, B.; Choudhury, B.; Rautava, S.; Isolauri, E.; Salminen, S.; Bode, L. Infants Are Exposed to Human Milk Oligosaccharides Already in utero. Front. Pediatr. 2018, 6, 270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wang, Y.; Fan, Q.; Liu, Y.; Liu, H.; Yan, J.; Li, M.; Dong, W.; Li, W. High levels of fucosylation and sialylation of milk N-glycans from mothers with gestational diabetes mellitus alter the offspring gut microbiome and immune balance in mice. FASEB J. 2020, 34, 3715–3731. [Google Scholar] [CrossRef]
- Tarr, A.J.; Galley, J.D.; Fisher, S.E.; Chichlowski, M.; Berg, B.M.; Bailey, M.T. The prebiotics 3′Sialyllactose and 6′Sialyllactose diminish stressor-induced anxiety-like behavior and colonic microbiota alterations: Evidence for effects on the gut–brain axis. Brain Behav. Immun. 2015, 50, 166–177. [Google Scholar] [CrossRef] [Green Version]
- Raubenheimer, P.J.; Nyirenda, M.J.; Walker, B.R. A Choline-Deficient Diet Exacerbates Fatty Liver but Attenuates Insulin Resistance and Glucose Intolerance in Mice Fed a High-Fat Diet. Diabetes 2006, 55, 2015–2020. [Google Scholar] [CrossRef] [Green Version]
- Musso, G.; Gambino, R.; Cassader, M. Obesity, Diabetes, and Gut Microbiota: The hygiene hypothesis expanded? Diabetes Care 2010, 33, 2277–2284. [Google Scholar] [CrossRef] [Green Version]
- Heianza, Y.; Sun, D.; Li, X.; DiDonato, J.A.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: The POUNDS Lost trial. Gut 2019, 68, 263–270. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef]
- Galligan, J.J. Beneficial actions of microbiota-derived tryptophan metabolites. Neurogastroenterol Motil. 2018, 30, e13283. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, H.; Li, Y.; Huang, S.; Zhang, L.; Cao, C.; Baker, P.N.; Tong, C.; Zheng, P.; Qi, H. Altered gut bacterial and metabolic signatures and their interaction in gestational diabetes mellitus. Gut Microbes 2020, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Liu, Y.; Ronnett, G.V.; Wu, A.; Cox, B.J.; Dai, F.F.; Röst, H.L.; Gunderson, E.P.; Wheeler, M.B. Amino acid and lipid metabolism in post-gestational diabetes and progression to type 2 diabetes: A metabolic profiling study. PLoS Med. 2020, 17, e1003112. [Google Scholar] [CrossRef]
- Maintz, L.; Schwarzer, V.; Bieber, T.; van der Ven, K.; Novak, N. Effects of histamine and diamine oxidase activities on pregnancy: A critical review. Hum. Reprod. Updat. 2008, 14, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, L.L.; Sun, Q.H.; Liu, G.R.; Guo, J.Y. Urinary Metabolomics Study of the Intervention Effect of Hypoglycemic Decoction on Type 2 Diabetes Mellitus Rats Model. Evid.-Based Complement. Altern. Med. 2019, 2019, 1394641. [Google Scholar] [CrossRef]
- HoleČek, M. Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients 2020, 12, 848. [Google Scholar] [CrossRef] [Green Version]
- Jutel, M.; Blaser, K.; Akdis, C.A. The role of histamine in regulation of immune responses. Chem. Immunol. Allergy 2006, 91, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, M.; Maeyama, K. The metabolism of histamine in rat hypothalamus and cortex after reserpine treatment. Neurochem. Int. 2015, 85–86, 31–39. [Google Scholar] [CrossRef]
- Kimura, N.; Fukuwatari, T.; Sasaki, R.; Shibata, K. Comparison of Metabolic Fates of Nicotinamide, NAD+ and NADH Administered Orally and Intraperitoneally; Characterization of Oral NADH. J. Nutr. Sci. Vitaminol. 2006, 52, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Salek, R.M.; Maguire, M.L.; Bentley, E.; Rubtsov, D.V.; Hough, T.; Cheeseman, M.; Nunez, D.; Sweatman, B.C.; Haselden, J.N.; Cox, R.D.; et al. A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol. Genom. 2007, 29, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, Y.; Wu, P. 5’-Methylthioadenosine and Cancer: Old molecules, new understanding. J. Cancer 2019, 10, 927–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadakia, R.; Nodzenski, M.; Talbot, O.; Kuang, A.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Ilkayeva, O.R.; O’Neal, S.K.; Lowe, L.P.; et al. Maternal metabolites during pregnancy are associated with newborn outcomes and hyperinsulinaemia across ancestries. Diabetologia 2019, 62, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
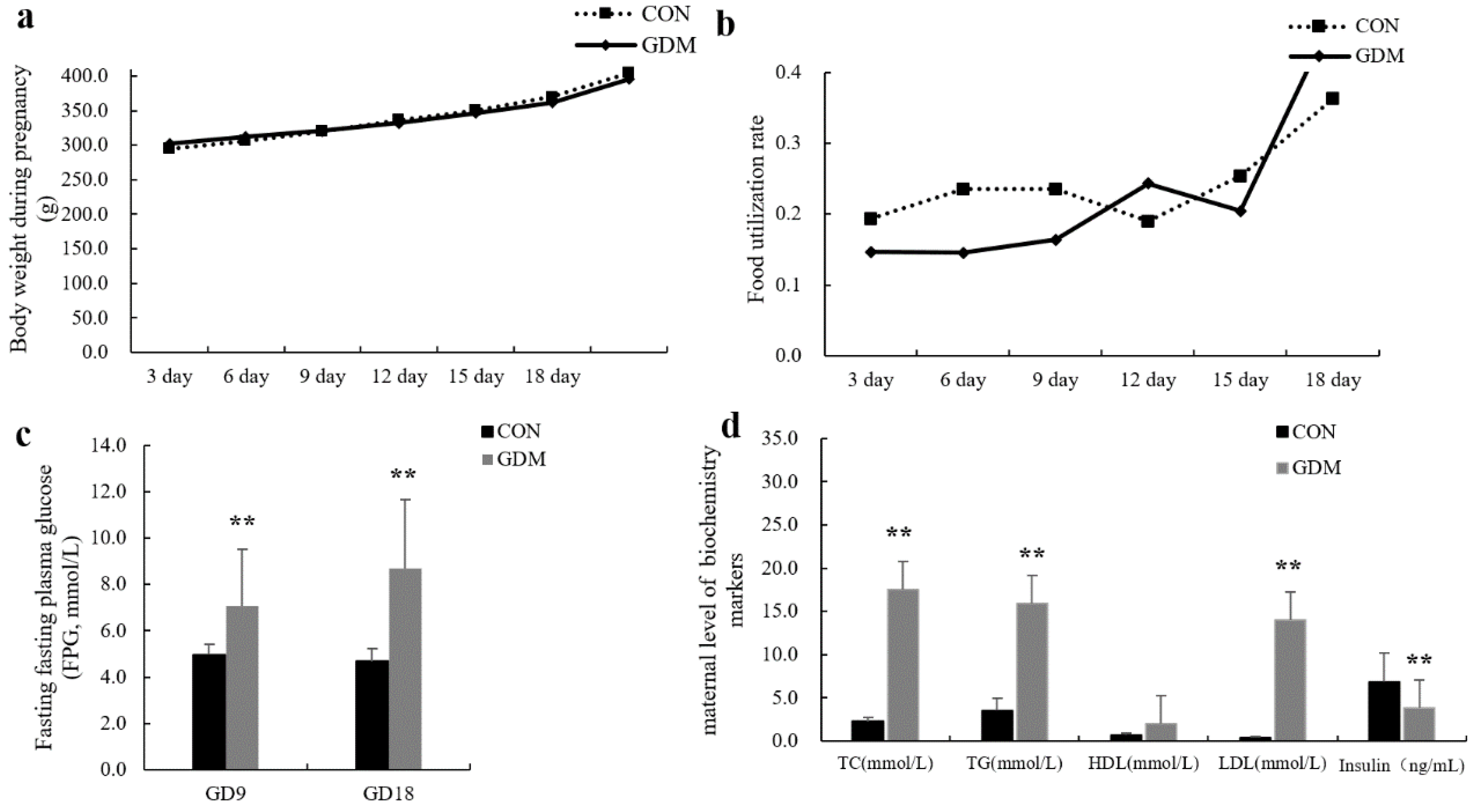
| Group | Live Fetus | Resorption Rate | Stillborn Fetus Rate | Plateau Weight (g) | Fetus Weight (g) | CRL (mm) |
|---|---|---|---|---|---|---|
| CON (n = 8) | 13.7 ± 3.43 | 1.80% (2/111) | 0 | 0.51 ± 0.07 | 4.29 ± 0.68 | 29.01 ± 2.79 |
| GDM (n = 8) | 10.07 ± 4.58 * | 8.49% (9/106) * | 1.89% (2/106) | 0.72 ± 0.15 ** | 4.64 ± 0.35 * | 29.34 ± 1.48 |
| Pathway | Metabolites | p-Value | FC |
|---|---|---|---|
| Amino Acid Metabolism | |||
| D-Glutamine and D-glutamate metabolism | D-Glutamine | 0.010 | 0.61 |
| Histidine metabolism | 1-Methylhistamine | 0.001 | 0.57 |
| β-Alanine metabolism | β-Aminopropionitrile | 0.001 | 0.40 |
| Dihydrouracil | <0.001 | 0.27 | |
| Cysteine and methionine metabolism | 5’-Methylthioadenosine | <0.001 | 0.20 |
| Arginine and proline metabolism | Citrulline | 0.009 | 0.59 |
| Cyanoamino acid metabolism | β-Aminopropionitrile | 0.001 | 0.40 |
| Isoleucine metabolism | Valyl-isoleucine | 0.024 | 2.13 |
| Proline | Analy-proline | 0.0151 | 0.43 |
| Tryptophan metabolism | (±)-Tryptophan | 0.0150 | 0.54 |
| Kynurenic acid | <0.001 | 0.12 | |
| 3-Methyldioxyindole | 0.029 | 0.02 | |
| 1H-Indole−3-acetamide | <0.001 | 0.22 | |
| Indole | <0.001 | 1.65 | |
| 5-Hydroxy-L-tryptophan | <0.001 | 0.24 | |
| Lipid metabolism | |||
| Glycerophospholipid metabolism | Choline | 0.002 | 2.19 |
| Dimethylethanolamine | 0.016 | 2.77 | |
| LysoPC(P-18:1(9Z)) | 0.001 | 3.76 | |
| PC(20:4(8Z,11Z,14Z,17Z)/P-18:0) | <0.001 | 3.27 | |
| Glycerylphosphorylethanolamine | 0.022 | 5.60 | |
| PE(20:5(5Z,8Z,11Z,14Z,17Z)/P-18:0) | 0.001 | 2.58 | |
| Sphingolipid metabolism | Sphinganine | 0.004 | 0.39 |
| Fatty acid metabolism | |||
| Fatty acid degradation metabolism | L-Palmitoylcarnitine | 0.004 | 13.69 |
| Arachidonic acid metabolism | Lipoxin A4 | 0.012 | 10.87 |
| PC(20:4(8Z,11Z,14Z,17Z)/P-18:0) | <0.001 | 3.27 | |
| Linoleic acid metabolism | α-Dimorphecolic acid | 0.001 | 5.51 |
| PC(20:4(8Z,11Z,14Z,17Z)/P-18:0) | <0.001 | 3.27 | |
| α-Linolenic acid metabolism | PC(20:4(8Z,11Z,14Z,17Z)/P-18:0) | <0.001 | 3.27 |
| Steroid hormone | |||
| Steroid biosynthesis | Brassicasterol | <0.001 | 11.81 |
| Calcitriol | 0.001 | 10.85 | |
| Ergosterol | <0.001 | 5.83 | |
| Steroid hormone biosynthesis | Progesterone | 0.035 | 0.36 |
| Nucleotide metabolism | |||
| Pyrimidine metabolism | Cytosine | <0.001 | 0.32 |
| Deoxycytidine | <0.001 | 0.30 | |
| 5-Methylcytosine | <0.001 | 0.12 | |
| Dihydrouracil | <0.001 | 0.27 | |
| Purine metabolism | Hypoxanthine | 0.007 | 0.21 |
| Vitamin metabolism | |||
| Nicotinate and nicotinamide metabolism | 1-Methylnicotinamide N1-Methyl-4-pyridone-3-carboxamide | <0.001 <0.001 | 0.28 0.05 |
| Vitamin B6 metabolism | 5-Pyridoxolactone | <0.001 | 0.04 |
| Pantothenate and CoA biosynthesis | Dihydrouracil | <0.001 | 0.27 |
| Choline metabolism | Choline | 0.002 | 2.19 |
| Trimethylamine N-oxide | 0.004 | 1.97 | |
| LysoPC(P-18:1(9Z)) | 0.001 | 3.76 | |
| PC(20:4(8Z,11Z,14Z,17Z)/P-18:0) | <0.001 | 3.27 | |
| Pathway | Metabolites | p-Value | FC |
|---|---|---|---|
| Amino acids metabolism | |||
| Phenylalanine, tyrosine, and tryptophan biosynthesis | L-Phenylalanine | 0.047 | 0.67 |
| L-Tyrosine | 0.002 | 2.97 | |
| 3-Dehydroquinate | 0.002 | 0.45 | |
| Phenylalanine metabolism | L-Phenylalanine | 0.047 | 0.67 |
| L-Tyrosine | 0.002 | 2.97 | |
| Phenylacetylglutamine | 0.022 | 0.53 | |
| Phenylacetylglycine | 0.050 | 0.36 | |
| 2-Hydroxycinnamic acid | <0.001 | 3.07 | |
| Cysteine and methionine metabolism | 5’-Methylthioadenosine | 0.002 | 0.66 |
| Homocysteine | 0.001 | 11.46 | |
| L-Methionine | 0.008 | 2.61 | |
| Histidine metabolism | 1-Methylhistamine | 0.031 | 2.23 |
| Methylimidazole acetaldehyde | 0.000 | 0.40 | |
| 3-Methylhistidine | 0.037 | 0.37 | |
| D-Arginine and D-ornithine metabolism | D-Ornithine | 0.015 | 0.67 |
| Valine, leucine, and isoleucine biosynthesis | L-Valine | 0.044 | 1.42 |
| Tryptophan metabolism | 5-Hydroxyindoleacetic acid | 0.021 | 0.64 |
| L-Kynurenine | 0.026 | 0.64 | |
| Kynurenic acid | 0.002 | 0.46 | |
| 5-Hydroxy-L-tryptophan | 0.001 | 0.56 | |
| Lysine degradation | 4-Trimethylammoniobutanal | 0.028 | 0.65 |
| Valine, leucine, and isoleucine degradation | L-Valine | 0.044 | 1.42 |
| Tyrosine metabolism | L-Tyrosine | 0.002 | 2.97 |
| DL-Dopa | 0.06 | ||
| Carbohydrate metabolism | |||
| Starch and sucrose metabolism | Trehalose | <0.000 | 8.34 |
| D-Maltose | 0.010 | 5.40 | |
| Amino sugar and nucleotide sugar metabolism | Glucosamine 6-phosphate | 0.004 | 2.00 |
| Galactose metabolism | myo-Inositol | 0.000 | 0.40 |
| 3’-Sialyllactose | 0.000 | 18.10 | |
| L-Galactose | 12.95 | ||
| Nucleotides metabolism | |||
| Aminoacyl-tRNA biosynthesis | L-Phenylalanine | 0.047 | 0.67 |
| L-Methionine | 0.008 | 2.61 | |
| L-Valine | 0.044 | 1.42 | |
| L-Tyrosine | 0.002 | 2.97 | |
| Purine metabolism | Adenine | 0.000 | 0.44 |
| Lipids metabolism | |||
| Glycerophospholipid metabolism | Glycerophosphocholine | 0.022 | 1.77 |
| PS(18:0/22:6(4Z,7Z,10Z,13Z,16Z,19Z)) | 0.010 | 0.53 | |
| Vitamin metabolism | |||
| Nicotinate and nicotinamide metabolism | Niacinamide | 0.003 | 0.35 |
| N1-Methyl-4-pyridone-3-carboxamide | <0.001 | 0.37 | |
| Vitamin B6 metabolism | Pyridoxal | 0.042 | 1.57 |
| Pantothenate and CoA biosynthesis | L-Valine | 0.044 | 1.42 |
| Choline | Trimethylamine N-oxide | 0.031 | 3.64 |
| Bile acid metabolism | |||
| Primary bile acid biosynthesis | Taurochenodesoxycholic acid | 0.043 | 6.67 |
| Others | |||
| Caffeine metabolism | Paraxanthine | <0.001 | 0.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Zhao, R.; Lyu, Y.; Shi, H.; Ye, W.; Tan, Y.; Li, R.; Xu, Y. Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal–Fetal Outcomes. Nutrients 2021, 13, 3644. https://doi.org/10.3390/nu13103644
Zhou Y, Zhao R, Lyu Y, Shi H, Ye W, Tan Y, Li R, Xu Y. Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal–Fetal Outcomes. Nutrients. 2021; 13(10):3644. https://doi.org/10.3390/nu13103644
Chicago/Turabian StyleZhou, Yalin, Runlong Zhao, Ying Lyu, Hanxu Shi, Wanyun Ye, Yuwei Tan, Rui Li, and Yajun Xu. 2021. "Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal–Fetal Outcomes" Nutrients 13, no. 10: 3644. https://doi.org/10.3390/nu13103644
APA StyleZhou, Y., Zhao, R., Lyu, Y., Shi, H., Ye, W., Tan, Y., Li, R., & Xu, Y. (2021). Serum and Amniotic Fluid Metabolic Profile Changes in Response to Gestational Diabetes Mellitus and the Association with Maternal–Fetal Outcomes. Nutrients, 13(10), 3644. https://doi.org/10.3390/nu13103644







