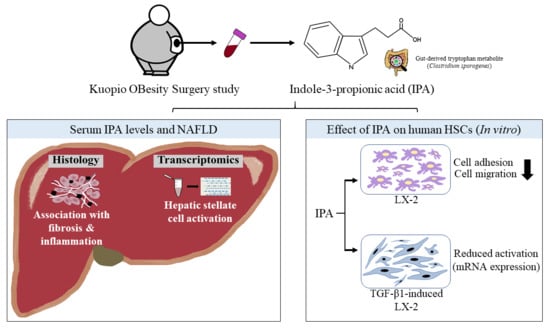
Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Liver Histology
2.3. Measurement of Serum Indole-3-Propionic Acid (IPA)
2.4. Gene Expression in Liver Using RNA-Sequencing
2.5. Validation of Anti-Fibrotic Effects of IPA Using Human Hepatic Stellate Cells (LX-2)
2.5.1. Cytotoxicity Assay
2.5.2. Cell Adhesion Assay
2.5.3. Wound Healing Cell Migration Assay
2.5.4. Activation of LX-2 Cells by TGF-β1 and IPA Treatment
2.5.5. Quantitative RT-PCR
2.6. Statistical Analysis
3. Results
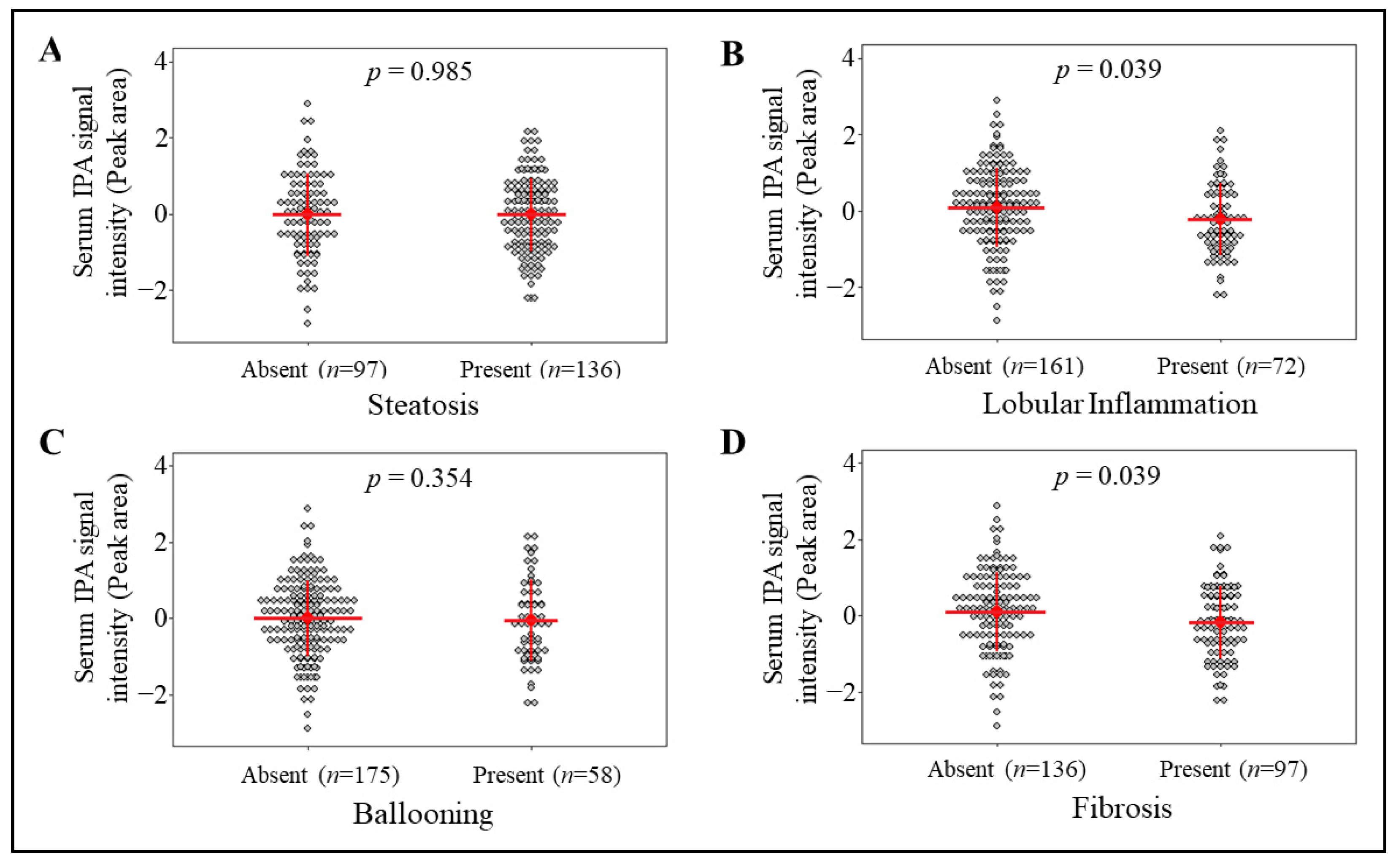
3.1. Circulating IPA Levels Associate with the Lobular Inflammation and Fibrosis
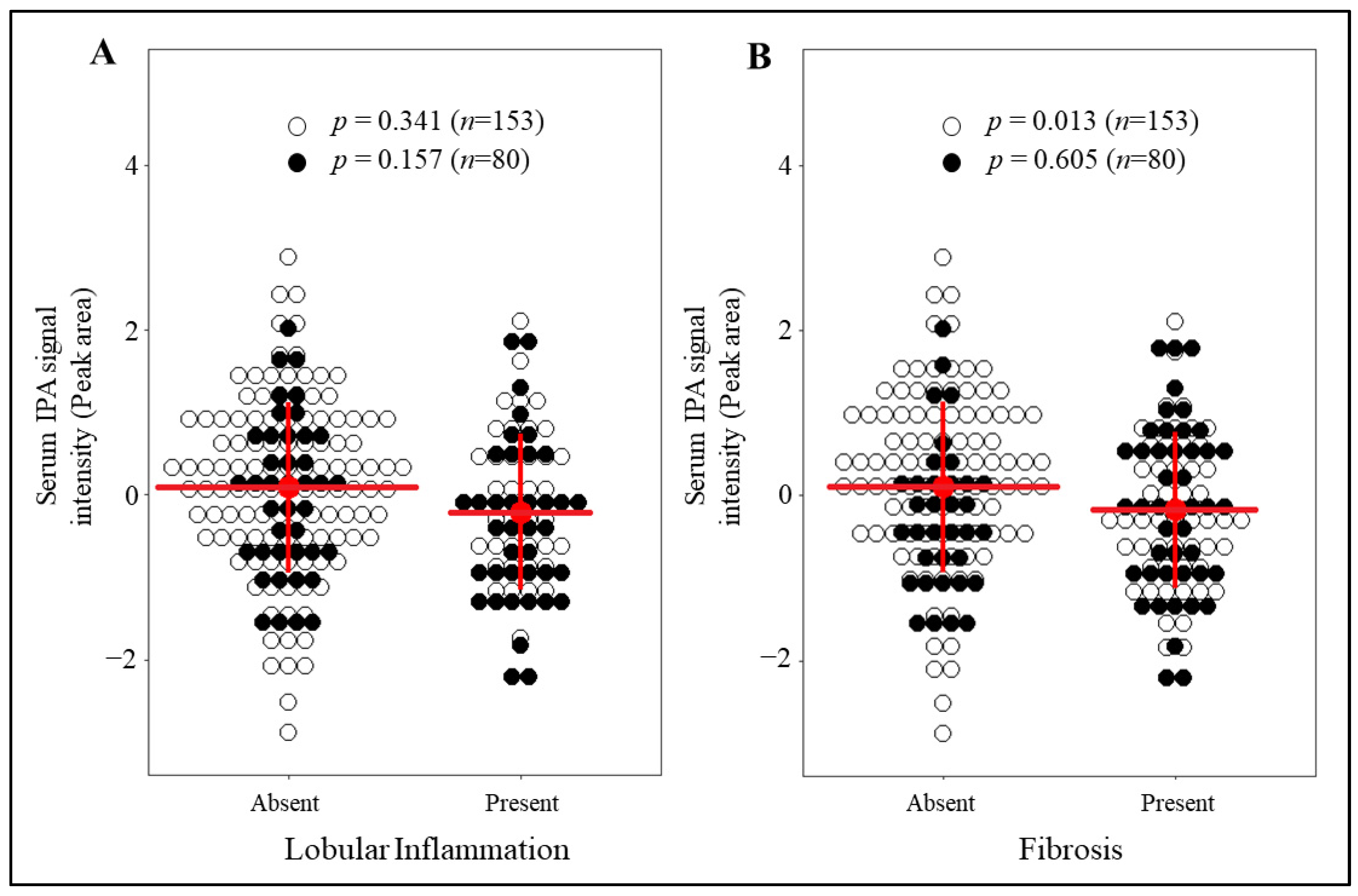
3.2. IPA Levels Are Markedly Reduced in Individuals with Fibrosis and without T2D
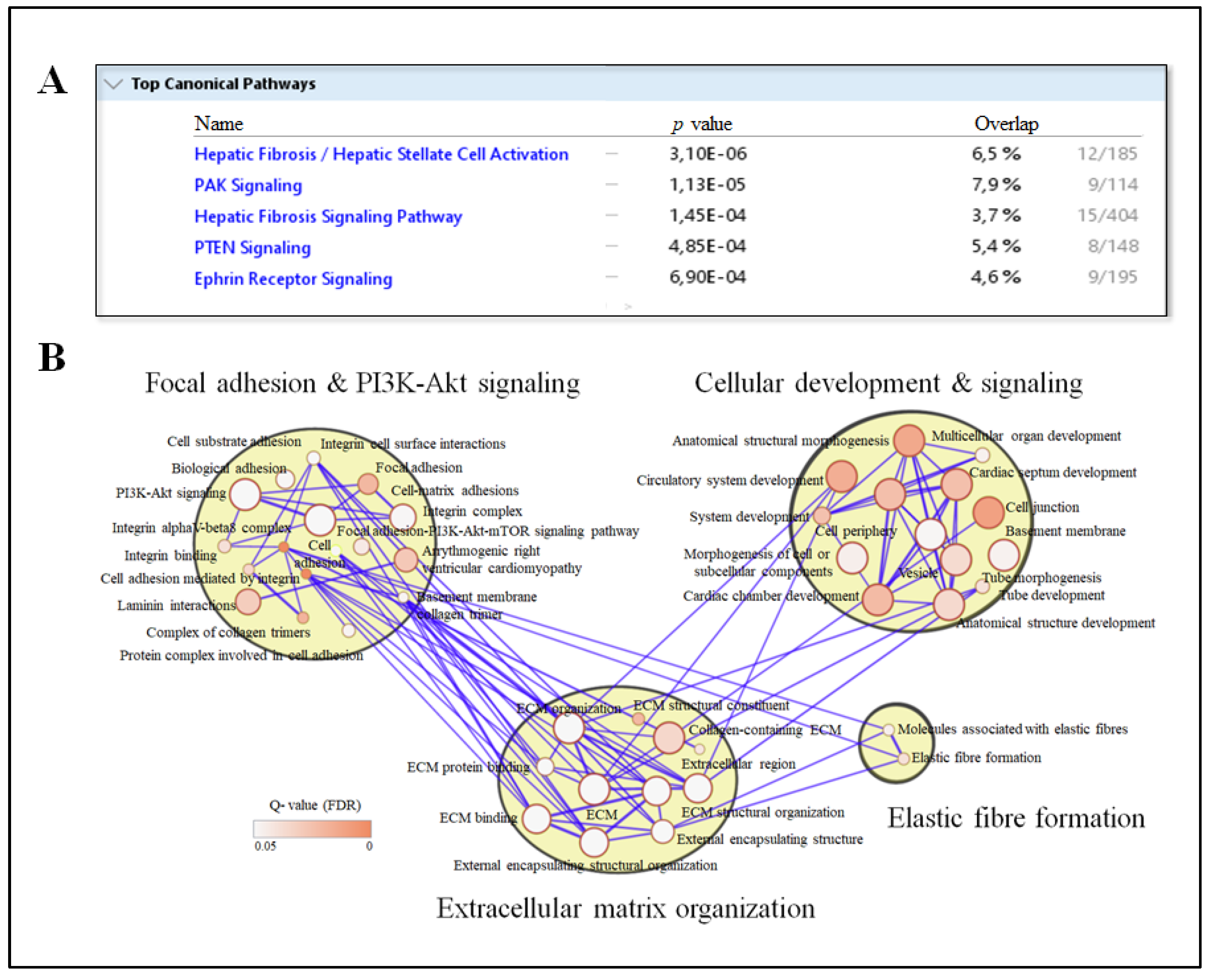
3.3. Circulating IPA Associates with Liver Transcripts Enriched for Pathways Related to Fibrosis
3.4. IPA Reduces Cell Adhesion and Migration of LX-2 Cells
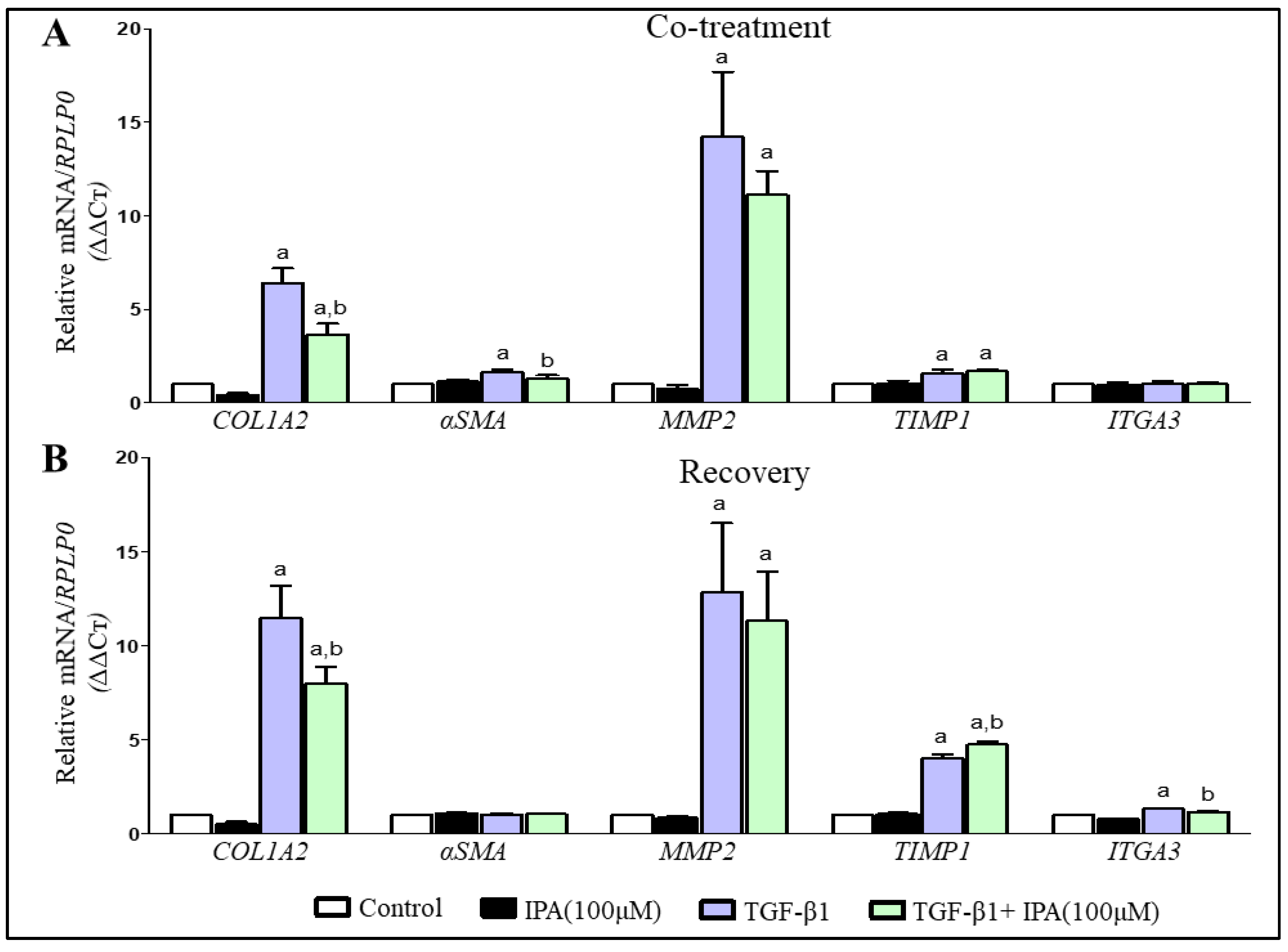
3.5. IPA Subsides Fibrogenesis in Activated LX-2 Cells
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Trefts, E.; Gannon, M.; Wasserman, D.H. The Liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Bechmann, L.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parhofer, K.G. Interaction between Glucose and Lipid Metabolism: More than Diabetic Dyslipidemia. Diabetes Metab. J. 2015, 39, 353–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsarou, A.; Moustakas, I.I.; Pyrina, I.; Lembessis, P.; Koutsilieris, M.; Chatzigeorgiou, A. Metabolic inflammation as an instigator of fibrosis during non-alcoholic fatty liver disease. World J. Gastroenterol. 2020, 26, 1993–2011. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee-Sarwar, K.A.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Metabolome–Microbiome Crosstalk and Human Disease. Metabolites 2020, 10, 181. [Google Scholar] [CrossRef]
- Menni, C.; Hernandez, M.M.; Vital, M.; Mohney, R.P.; Spector, T.D.; Valdes, A.M. Circulating levels of the anti-oxidant indoleproprionic acid are associated with higher gut microbiome diversity. Gut Microbes 2019, 10, 688–695. [Google Scholar] [CrossRef] [Green Version]
- de Mello, V.D.; Paananen, J.; Lindström, J.; Lankinen, M.A.; Shi, L.; Kuusisto, J.; Uusitupa, M. Faculty Opinions recommendation of Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Cason, C.A.; Dolan, K.T.; Sharma, G.; Tao, M.; Kulkarni, R.; Helenowski, I.B.; Doane, B.M.; Avram, M.J.; McDermott, M.M.; Chang, E.B.; et al. Plasma Microbiome-Modulated Indole-and Phenyl-Derived Metabolites Associate with Advanced Atherosclerosis and Postoperative Outcomes. J. Vasc. Surg. 2018, 68, 1552–1562.e7. [Google Scholar] [CrossRef]
- Cussotto, S.; Delgado, I.; Anesi, A.; Dexpert, S.; Aubert, A.; Beau, C.; Forestier, D.; Ledaguenel, P.; Magne, E.; Mattivi, F.; et al. Tryptophan Metabolic Pathways Are Altered in Obesity and Are Associated With Systemic Inflammation. Front. Immunol. 2020, 11, 557. [Google Scholar] [CrossRef]
- Jennis, M.; Cavanaugh, C.R.; Leo, G.C.; Mabus, J.R.; Lenhard, J.; Hornby, P.J. Microbiota-Derived Tryptophan Indoles Increase After Gastric Bypass Surgery and Reduce Intestinal Permeability in Vitro and in Vivo. Neurogastroenterol. Motil. 2018, 30, e13178. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-Y.; Lin, C.-J.; Pan, H.-C.; Lee, C.-C.; Lu, S.-C.; Hsieh, Y.-T.; Huang, S.-Y.; Huang, H.-Y. Clinical association between the metabolite of healthy gut microbiota, 3-indolepropionic acid and chronic kidney disease. Clin. Nutr. 2018, 38, 2945–2948. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and Gut Microbial Tryptophan Metabolism and Type 2 Diabetes: An Integrative Analysis of Host Genetics, Diet, Gut Microbiome and Circulating Metabolites in Cohort Studies. Gut 2021, 0, 1–11. [Google Scholar]
- Pallister, T.; Jennings, A.; Mohney, R.P.; Yarand, D.; Mangino, M.; Cassidy, A.; MacGregor, A.; Spector, T.D.; Menni, C. Characterizing Blood Metabolomics Profiles Associated with Self-Reported Food Intakes in Female Twins. PLoS ONE 2016, 11, e0158568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Coker, O.O.; Chu, E.S.H.; Fu, K.; Lau, H.C.H.; Wang, Y.; Chan, A.W.H.; Wei, H.; Yang, X.; Sung, J.J.Y.; et al. Dietary Cholesterol Drives Fatty Liver-Associated Liver Cancer by Modulating Gut Microbiota and Metabolites. Gut 2021, 70, 761. [Google Scholar] [CrossRef] [PubMed]
- Abildgaard, A.; Elfving, B.; Hokland, M.; Wegener, G.; Lund, S. The microbial metabolite indole-3-propionic acid improves glucose metabolism in rats, but does not affect behaviour. Arch. Physiol. Biochem. 2017, 124, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Bendheim, P.E.; Poeggeler, B.; Neria, E.; Ziv, V.; Pappolla, M.A.; Chain, D.G. Development of Indole-3-Propionic Acid (OXIGON™) for Alzheimer’s Disease. J. Mol. Neurosci. 2002, 19, 213–217. [Google Scholar] [CrossRef]
- Hwang, I.K.; Yoo, K.-Y.; Li, H.; Park, O.K.; Lee, C.H.; Choi, J.H.; Jeong, Y.-G.; Lee, Y.L.; Kim, Y.-M.; Kwon, Y.-G.; et al. Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J. Neurosci. Res. 2009, 87, 2126–2137. [Google Scholar] [CrossRef] [PubMed]
- Negatu, D.A.; Gengenbacher, M.; Dartois, V.; Dick, T. Indole Propionic Acid, an Unusual Antibiotic Produced by the Gut Microbiota, With Anti-inflammatory and Antioxidant Properties. Front. Microbiol. 2020, 11, 575586. [Google Scholar] [CrossRef]
- Yisireyili, M.; Takeshita, K.; Saito, S.; Murohara, T.; Niwa, T. Indole-3-propionic acid suppresses indoxyl sulfate-induced expression of fibrotic and inflammatory genes in proximal tubular cells. Nagoya J. Med Sci. 2017, 79, 477–486. [Google Scholar] [CrossRef]
- Karbownik, M.; Reiter, R.J.; Garcia, J.J.; Cabrera, J.; Burkhardt, S.; Osuna, C.; Lewiński, A. Indole-3-Propionic Acid, a Melatonin-Related Molecule, Protects Hepatic Microsomal Membranes from Iron-Induced Oxidative Damage: Relevance to Cancer Reduction. J. Cell. Biochem. 2001, 81, 507–513. [Google Scholar] [CrossRef]
- Zhao, Z.-H.; Xin, F.-Z.; Xue, Y.; Hu, Z.; Han, Y.; Ma, F.; Zhou, D.; Liu, X.-L.; Cui, A.; Liu, Z.; et al. Indole-3-propionic acid inhibits gut dysbiosis and endotoxin leakage to attenuate steatohepatitis in rats. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Aron-Wisnewsky, J.; Warmbrunn, M.V.; Nieuwdorp, M.; Clément, K. Nonalcoholic Fatty Liver Disease: Modulating Gut Microbiota to Improve Severity? Gastroenterology 2020, 158, 1881–1898. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A Gut Bacterial Pathway Metabolizes Aromatic Amino Acids into Nine Circulating Metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Fan, J.-G. Microbial metabolites in non-alcoholic fatty liver disease. World J. Gastroenterol. 2019, 25, 2019–2028. [Google Scholar] [CrossRef]
- Kirpich, I.A.; Marsano, L.S.; McClain, C.J. Gut-Liver Axis, Nutrition, and Non-Alcoholic Fatty Liver Disease. Clin. Biochem. 2015, 48, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aragonès, G.; Colom-Pellicer, M.; Aguilar, C.; Guiu-Jurado, E.; Martínez, S.; Sabench, F.; Antonio Porras, J.; Riesco, D.; Del Castillo, D.; Richart, C.; et al. Circulating Microbiota-Derived Metabolites: A "Liquid Biopsy? Int. J. Obes. 2020, 44, 875–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohtani, N.; Kawada, N. Role of the Gut-Liver Axis in Liver Inflammation, Fibrosis, and Cancer: A Special Focus on the Gut Microbiota Relationship. Hepatol. Commun. 2019, 3, 456–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazagova, M.; Wang, L.; Anfora, A.T.; Wissmueller, M.; Lesley, S.A.; Miyamoto, Y.; Eckmann, L.; Dhungana, S.; Pathmasiri, W.; Sumner, S.; et al. Commensal Microbiota is Hepatoprotective and Prevents Liver Fibrosis in Mice. Fed. Am Soc. Exp. Biol. 2015, 29, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Ni, Y.; Ni, L.; Zhuge, F.; Fu, Z. The Gut Microbiota and its Metabolites, Novel Targets for Treating and Preventing Non-Alcoholic Fatty Liver Disease. Mol. Nutr. Food Res. 2020, 64, 2000375. [Google Scholar] [CrossRef]
- Karsdal, M.A.; Manon-Jensen, T.; Genovese, F.; Kristensen, J.H.; Nielsen, M.J.; Sand, J.M.B.; Hansen, N.-U.B.; Bay-Jensen, A.-C.; Bager, C.L.; Krag, A.; et al. Novel insights into the function and dynamics of extracellular matrix in liver fibrosis. Am. J. Physiol. Liver Physiol. 2015, 308, G807–G830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marra, F. Chemokines in Liver Inflammation and Fibrosis. Front. Biosci. 2002, 7, 1899. [Google Scholar] [CrossRef] [PubMed]
- Elsharkawy, A.M.; Oakley, F.; Mann, D.A. The role and regulation of hepatic stellate cell apoptosis in reversal of liver fibrosis. Apoptosis 2005, 10, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Pihlajamäki, J.; Grönlund, S.; Simonen, M.; Käkelä, P.; Moilanen, L.; Pääkkönen, M.; Pirinen, E.; Kolehmainen, M.; Kärjä, V.; Kainulainen, S.; et al. Cholesterol Absorption Decreases After Roux-En-Y Gastric Bypass but Not After Gastric Banding. Metabolism 2010, 59, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Männistö, V.T.; Simonen, M.; Soininen, P.; Tiainen, M.; Kangas, A.J.; Kaminska, D.; Venesmaa, S.; Käkelä, P.; Kärjä, V.; Gylling, H.; et al. Lipoprotein Subclass Metabolism in Nonalcoholic Steatohepatitis. J. Lipid Res. 2014, 55, 2676–2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunt, E.M.; Janney, C.G.; Di Bisceglie, A.M.; Neuschwander-Tetri, B.A.; Bacon, B.R. Nonalcoholic Steatohepatitis: A Proposal for Grading and Staging the Histological Lesions. Am. J. Gastroenterol. 1999, 94, 2467–2474. [Google Scholar] [CrossRef]
- de Mello, V.D.; Sehgal, R.; Männistö, V.; Klåvus, A.; Nilsson, E.; Perfilyev, A.; Kaminska, D.; Miao, Z.; Pajukanta, P.; Ling, C.; et al. Serum aromatic and branched-chain amino acids associated with NASH demonstrate divergent associations with serum lipids. Liver Int. 2020, 41, 754–763. [Google Scholar] [CrossRef]
- Tuomainen, M.; Lindström, J.; Lehtonen, M.; Auriola, S.; Pihlajamäki, J.; Peltonen, M.; Tuomilehto, J.; Uusitupa, M.; de Mello, V.D.; Hanhineva, K. Associations of Serum Indolepropionic Acid, a Gut Microbiota Metabolite, with Type 2 Diabetes and Low-Grade Inflammation in High-Risk Individuals. Nutr. Diabetes 2018, 8, 35. [Google Scholar] [CrossRef]
- Männistö, V.; Kaminska, D.; Käkelä, P.; Neuvonen, M.; Niemi, M.; Alvarez, M.; Pajukanta, P.; Romeo, S.; Nieuwdorp, M.; Groen, A.; et al. Protein Phosphatase 1 Regulatory Subunit 3B Genotype at rs4240624 Has a Major Effect on Gallbladder Bile Composition. Hepatol. Commun. 2020, 5, 244–257. [Google Scholar] [CrossRef]
- Gesper, M.; Nonnast, A.B.H.; Kumowski, N.; Stoehr, R.; Schuett, K.; Marx, N.; Kappel, B.A. Gut-Derived Metabolite Indole-3-Propionic Acid Modulates Mitochondrial Function in Cardiomyocytes and Alters Cardiac Function. Front. Med. 2021, 8, 264. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; D’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewidar, B.; Meyer, C.; Dooley, S.; Meindl-Beinker, A. TGF-Β in Hepatic Stellate Cell Activation and Liver Fibrogenesis-Updated 2019. Cells 2019, 8, 1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Wang, S.; Friedman, S.L. The Power of Plasticity—Metabolic Regulation of Hepatic Stellate Cells. Cell Metab. 2020, 33, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.; Gicquel, T.; Bodin, A.; Lagente, V.; Boichot, E. Characterization of the MMP/TIMP Imbalance and Collagen Production Induced by IL-1β Or TNF-A Release from Human Hepatic Stellate Cells. PLoS ONE 2016, 11, e0153118. [Google Scholar] [CrossRef] [Green Version]
- Hendrikx, T.; Schnabl, B. Indoles: Metabolites Produced by Intestinal Bacteria Capable of Controlling Liver Disease Manifestation. J. Intern. Med. 2019, 286, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Li, H.; Hu, J.; Zheng, J.; Zhou, J.; Botchlett, R.; Matthews, D.; Zeng, T.; Chen, L.; Xiao, X.; et al. Indole Alleviates Diet-Induced Hepatic Steatosis and Inflammation in a Manner Involving Myeloid Cell 6-Phosphofructo-2-Kinase/Fructose-2,6-Biphosphatase 3. Hepatology 2020, 72, 1191–1203. [Google Scholar] [CrossRef]
- Gao, J.; Guo, X.; Wei, W.; Li, R.; Hu, K.; Liu, X.; Jiang, W.; Liu, S.; Wang, W.; Sun, H.; et al. The Association of Fried Meat Consumption With the Gut Microbiota and Fecal Metabolites and Its Impact on Glucose Homoeostasis, Intestinal Endotoxin Levels, and Systemic Inflammation: A Randomized Controlled-Feeding Trial. Diabetes Care 2021, 44, 1970–1979. [Google Scholar] [CrossRef]
- Ji, Y.; Yin, Y.; Li, Z.; Zhang, W. Gut Microbiota-Derived Components and Metabolites in the Progression of Non-Alcoholic Fatty Liver Disease (NAFLD). Nutrients 2019, 11, 1712. [Google Scholar] [CrossRef] [Green Version]
- Gastaldelli, A.; Cusi, K. From NASH to diabetes and from diabetes to NASH: Mechanisms and treatment options. JHEP Rep. 2019, 1, 312–328. [Google Scholar] [CrossRef] [Green Version]
- Tsuchida, T.; Friedman, S.L. Mechanisms of Hepatic Stellate Cell Activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic Bacterial Metabolites Regulate Gastrointestinal Barrier Function Via the Xenobiotic Sensor PXR and Toll-Like Receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sari, Z.; Miko, E.; Kovacs, T.; Janko, L.; Csonka, T.; Lente, G.; Sebo, E.; Toth, J.; Toth, D.; Arkosy, P.; et al. Indolepropionic Acid, a Metabolite of the Microbiome, Has Cytostatic Properties in Breast Cancer by Activating AHR and PXR Receptors and Inducing Oxidative Stress. Cancers 2020, 12, 2411. [Google Scholar] [CrossRef] [PubMed]

| Normal Liver | Simple Steatosis | NASH | p a | |
|---|---|---|---|---|
| Total, N (men/women) | 79 (20/59) | 40 (9/31) | 45 (18/27) | 0.14 |
| Age (years) | 47.4 ± 9.7 | 46.5 ± 8.6 | 49.5 ± 9.6 | 0.31 |
| BMI (kg/m2) | 42.6 ± 5.5 | 43.3 ± 4.8 | 43.4 ± 5.4 | 0.61 |
| fS-Total cholesterol (mmol/L) | 4.2 ± 0.8 | 4.2 ± 0.9 | 4.5 ± 1.1 | 0.27 |
| fS-LDL cholesterol (mmol/L) | 2.4 ± 0.7 | 2.4 ± 0.9 | 2.5 ± 1.0 | 0.80 |
| fS-HDL cholesterol (mmol/L) | 1.2 ± 0.3 | 1.1 ± 0.2 | 1.2 ± 0.4 | 0.27 |
| fS-Triglycerides (mmol/L) | 1.3 (1.0–2.3) | 1.4 (0.9–2.0) | 1.6 (1.3–2.2) | 0.60 |
| fP-glucose (mmol/L) | 6.0 ± 1.3 | 6.3 ± 1.9 | 7.3 ± 2.2 * | 0.0005 |
| fS-insulin (mU/L) | 13.8 (7.8–18.6) | 16.0 (11.0–23.4) * | 20.5 (14.4–28.8) * | 0.00006 |
| Type 2 diabetes, N (%) | 15 (18.9) | 11 (27.5) | 26 (57.8) * | 0.00003 |
| Lipid lowering medication, N (%) | 22 (28.6) | 9 (22.5) | 20 (44.4) | 0.07 |
| Glucose lowering medication, N (%) | 14 (17.7) | 11 (25) | 24 (53.3) * | 0.0002 |
| IPA levels (Inverse normalized) | 0.04 ± 1.11 | 0.18 ± 0.93 | −0.04±0.98 | 0.63 |
| Steatosis grade, N | ||||
| <5% | 79 | 0 | 0 | |
| 5–33% | 0 | 32 | 13 | |
| 33–66% | 0 | 5 | 19 | |
| >66% | 0 | 3 | 13 | |
| Lobular inflammation, N | 0 | 0 | 45 | |
| Ballooning, N | 0 | 0 | 37 | |
| Fibrosis, N (stage range) | 0 | 0 | 42 (1–3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sehgal, R.; Ilha, M.; Vaittinen, M.; Kaminska, D.; Männistö, V.; Kärjä, V.; Tuomainen, M.; Hanhineva, K.; Romeo, S.; Pajukanta, P.; et al. Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis. Nutrients 2021, 13, 3509. https://doi.org/10.3390/nu13103509
Sehgal R, Ilha M, Vaittinen M, Kaminska D, Männistö V, Kärjä V, Tuomainen M, Hanhineva K, Romeo S, Pajukanta P, et al. Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis. Nutrients. 2021; 13(10):3509. https://doi.org/10.3390/nu13103509
Chicago/Turabian StyleSehgal, Ratika, Mariana Ilha, Maija Vaittinen, Dorota Kaminska, Ville Männistö, Vesa Kärjä, Marjo Tuomainen, Kati Hanhineva, Stefano Romeo, Päivi Pajukanta, and et al. 2021. "Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis" Nutrients 13, no. 10: 3509. https://doi.org/10.3390/nu13103509
APA StyleSehgal, R., Ilha, M., Vaittinen, M., Kaminska, D., Männistö, V., Kärjä, V., Tuomainen, M., Hanhineva, K., Romeo, S., Pajukanta, P., Pihlajamäki, J., & de Mello, V. D. (2021). Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis. Nutrients, 13(10), 3509. https://doi.org/10.3390/nu13103509