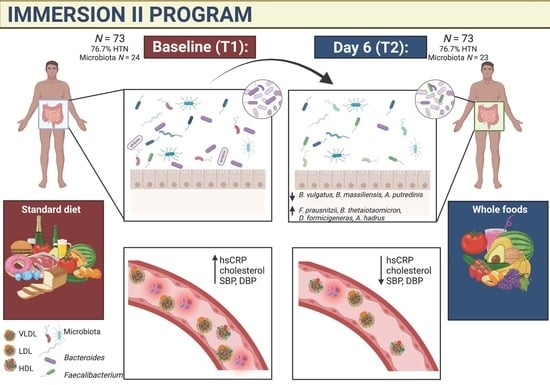
A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Anthropometric and Cardiovascular Measures
2.2. Gut Microbiome
2.3. Data Analyses
3. Results
3.1. Anthropometric and Cardiovascular Measures
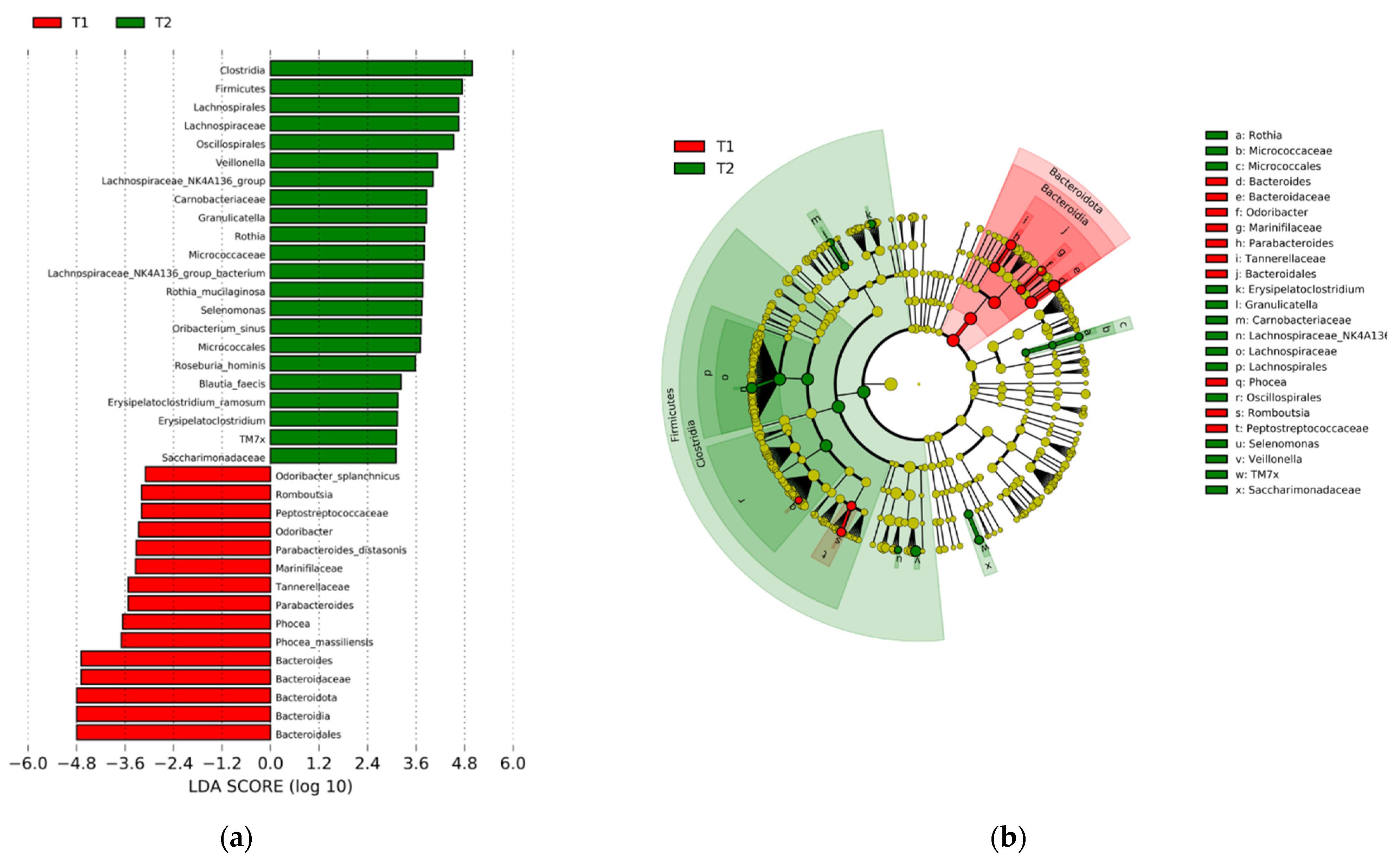
3.2. Gut Microbiome
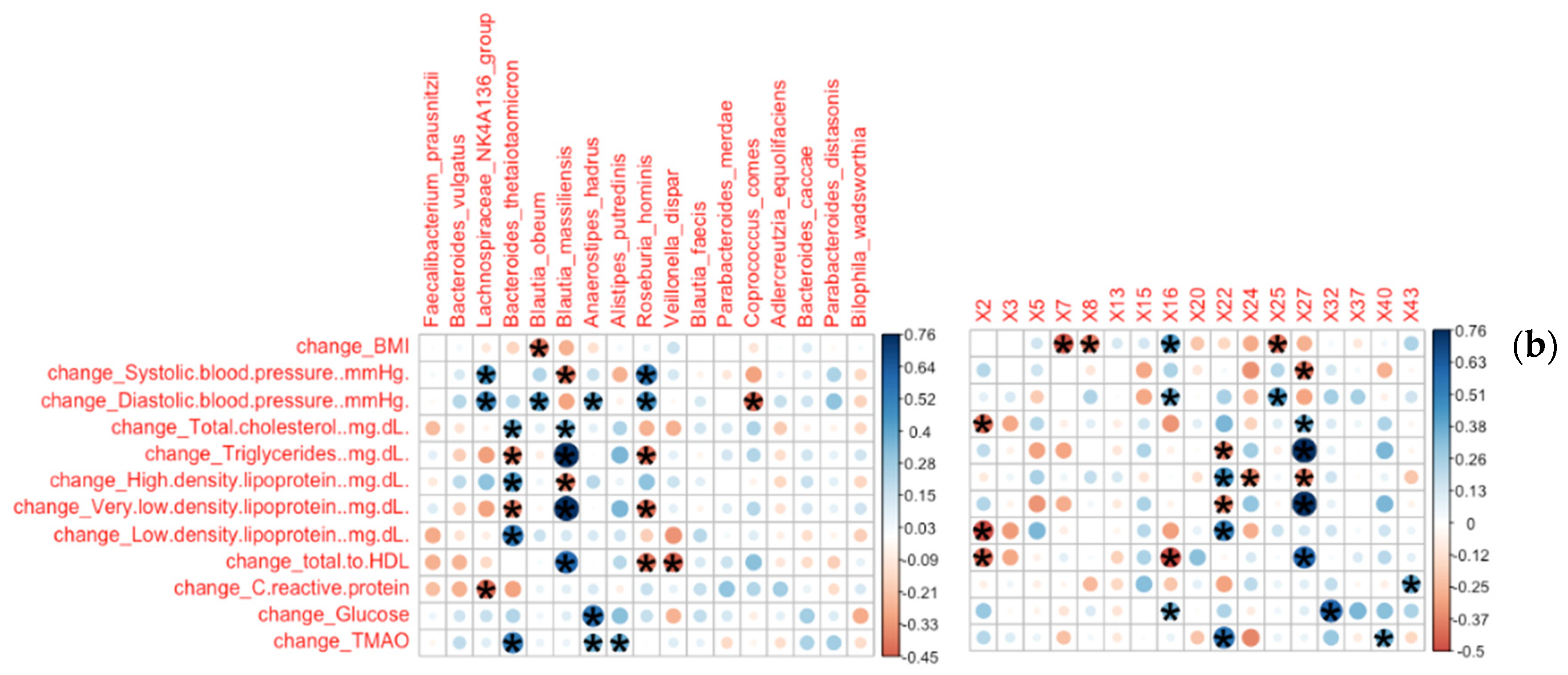
3.2.1. Bacterial Families and Genera Correlated with Clinical Measures
3.2.2. Bacterial Species and ASVs Correlated with Clinical Measures
3.2.3. Microbiota Are Differentially Abundant across Clinical Subgroups of Hypertension, LDL, and hsCRP at T2
3.2.4. Functional Predictions from the 16S Data Suggest a Downregulation of Bacterial Genes Implicated in Metabolism of Amino Acids, Carbohydrates, and Glycans, as Well as Human Cancer and Metabolic Disease
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart Disease and Stroke Statistics-2019 Update: A Report From the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E.; Collins, K.J.; Dennison, H.C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018, 71, e13–e115. [Google Scholar] [CrossRef]
- CDC Hypertension Prevalence in the U.S. Million Hearts®. Available online: https://millionhearts.hhs.gov/data-reports/hypertension-prevalence.html (accessed on 12 March 2021).
- Carroll, M.D.; Fryar, C.D. Total and High-Density Lipoprotein Cholesterol in Adults: United States, 2015–2018. NCHS Data Brief 2020, 8, 1–8. [Google Scholar]
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics—2020 Update: A Report From the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- Fryar, C.D.; Chen, T.-C.; Li, X. Prevalence of Uncontrolled Risk Factors for Cardiovascular Disease: United States, 1999–2010. NCHS Data Brief 2012, 103, 1–8. [Google Scholar]
- Frayr, C.; Carroll, M.; Afful, J. Prevalence of Overweight, Obesity, and Extreme Obesity among Adults Aged 20 and Over: United States, 1960–1962 through 2017–2018. Available online: https://www.cdc.gov/nchs/data/hestat/obesity-adult-17-18/obesity-adult.htm (accessed on 12 March 2021).
- Hajar, R. Risk Factors for Coronary Artery Disease: Historical Perspectives. Heart Views 2017, 18, 109–114. [Google Scholar] [CrossRef]
- Yu, E.; Malik, V.S.; Hu, F.B. Cardiovascular Disease Prevention by Diet Modification: JACC Health Promotion Series. J. Am. Coll. Cardiol. 2018, 72, 914–926. [Google Scholar] [CrossRef]
- Mozaffarian, D. Dietary and Policy Priorities for Cardiovascular Disease, Diabetes, and Obesity—A Comprehensive Review. Circulation 2016, 133, 187–225. [Google Scholar] [CrossRef]
- Zomer, E.; Gurusamy, K.; Leach, R.; Trimmer, C.; Lobstein, T.; Morris, S.; James, W.P.T.; Finer, N. Interventions That Cause Weight Loss and the Impact on Cardiovascular Risk Factors: A Systematic Review and Meta-Analysis. Obes. Rev. 2016, 17, 1001–1011. [Google Scholar] [CrossRef]
- Danielsen, K.K.; Svendsen, M.; Mæhlum, S.; Sundgot-Borgen, J. Changes in Body Composition, Cardiovascular Disease Risk Factors, and Eating Behavior after an Intensive Lifestyle Intervention with High Volume of Physical Activity in Severely Obese Subjects: A Prospective Clinical Controlled Trial. J. Obes. 2013, 2013, 325464. [Google Scholar] [CrossRef] [Green Version]
- López-Domènech, S.; Martínez-Herrera, M.; Abad-Jiménez, Z.; Morillas, C.; Escribano-López, I.; Díaz-Morales, N.; Bañuls, C.; Víctor, V.M.; Rocha, M. Dietary Weight Loss Intervention Improves Subclinical Atherosclerosis and Oxidative Stress Markers in Leukocytes of Obese Humans. Int. J. Obes. 2019, 43, 2200–2209. [Google Scholar] [CrossRef] [PubMed]
- Kwaifa, I.K.; Bahari, H.; Yong, Y.K.; Noor, S.M. Endothelial Dysfunction in Obesity-Induced Inflammation: Molecular Mechanisms and Clinical Implications. Biomolecules 2020, 10, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Caulfield, L.E.; Garcia-Larsen, V.; Steffen, L.M.; Coresh, J.; Rebholz, C.M. Plant-Based Diets Are Associated With a Lower Risk of Incident Cardiovascular Disease, Cardiovascular Disease Mortality, and All-Cause Mortality in a General Population of Middle-Aged Adults. J. Am. Heart Assoc. 2019, 8, e012865. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.M.; Pepine, C.J.; Raizada, M.K.; Kim, S. The Gut, Its Microbiome, and Hypertension. Curr. Hypertens. Rep. 2017, 19, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Li, H. The Role of Gut Microbiota in Atherosclerosis and Hypertension. Front. Pharmacol. 2018, 9, 1082. [Google Scholar] [CrossRef]
- Maslowski, K.M.; Mackay, C.R. Diet, Gut Microbiota and Immune Responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Jie, Z.; Xia, H.; Zhong, S.-L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The Gut Microbiome in Atherosclerotic Cardiovascular Disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [Green Version]
- Cho, C.E.; Taesuwan, S.; Malysheva, O.V.; Bender, E.; Tulchinsky, N.F.; Yan, J.; Sutter, J.L.; Caudill, M.A. Trimethylamine-N-Oxide (TMAO) Response to Animal Source Foods Varies among Healthy Young Men and Is Influenced by Their Gut Microbiota Composition: A Randomized Controlled Trial. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randrianarisoa, E.; Lehn-Stefan, A.; Wang, X.; Hoene, M.; Peter, A.; Heinzmann, S.S.; Zhao, X.; Königsrainer, I.; Königsrainer, A.; Balletshofer, B.; et al. Relationship of Serum Trimethylamine N-Oxide (TMAO) Levels with Early Atherosclerosis in Humans. Sci. Rep. 2016, 6, 26745. [Google Scholar] [CrossRef] [Green Version]
- Ros, E.; Martínez-González, M.A.; Estruch, R.; Salas-Salvadó, J.; Fitó, M.; Martínez, J.A.; Corella, D. Mediterranean Diet and Cardiovascular Health: Teachings of the PREDIMED Study. Adv. Nutr. 2014, 5, 330S–336S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salas-Salvadó, J.; Becerra-Tomás, N.; García-Gavilán, J.F.; Bulló, M.; Barrubés, L. Mediterranean Diet and Cardiovascular Disease Prevention: What Do We Know? Prog. Cardiovasc. Dis. 2018, 61, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Jaworska, K.; Bielinska, K.; Gawrys-Kopczynska, M.; Ufnal, M. TMA (Trimethylamine), but Not Its Oxide TMAO (Trimethylamine-Oxide), Exerts Haemodynamic Effects: Implications for Interpretation of Cardiovascular Actions of Gut Microbiome. Cardiovasc. Res. 2019, 115, 1948–1949. [Google Scholar] [CrossRef] [PubMed]
- Crimarco, A.; Springfield, S.; Petlura, C.; Streaty, T.; Cunanan, K.; Lee, J.; Fielding-Singh, P.; Carter, M.M.; Topf, M.A.; Wastyk, H.C.; et al. A Randomized Crossover Trial on the Effect of Plant-Based Compared with Animal-Based Meat on Trimethylamine-N-Oxide and Cardiovascular Disease Risk Factors in Generally Healthy Adults: Study with Appetizing Plantfood—Meat Eating Alternative Trial (SWAP-MEAT). Am. J. Clin. Nutr. 2020, 112, 1188–1199. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Handberg, E.; Pepine, C.; George, J.; Jain, A.; Desai, D.; Li, Q.; Stoll, S.; Aggarwal, M. Benefit of One Week Immersion in Lifestyle-Based Program for Sustainable Improvements in Cardiovascular Risk Factors over Time. Int. J. Dis. Reversal Prev. 2020, 2, 10. [Google Scholar] [CrossRef]
- Harris, P.A.; Taylor, R.; Thielke, R.; Payne, J.; Gonzalez, N.; Conde, J.G. Research Electronic Data Capture (REDCap)—A Metadata-Driven Methodology and Workflow Process for Providing Translational Research Informatics Support. J. Biomed. Inform. 2009, 42, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Roesch, L.F.W.; Casella, G.; Simell, O.; Krischer, J.; Wasserfall, C.H.; Schatz, D.; Atkinson, M.A.; Neu, J.; Triplett, E.W. Influence of Fecal Sample Storage on Bacterial Community Diversity. Open Microbiol. J. 2009, 3, 40–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, J.T.; Roesch, L.F.W.; Ördberg, M.; Ilonen, J.; Atkinson, M.A.; Schatz, D.A.; Triplett, E.W.; Ludvigsson, J. Genetic Risk for Autoimmunity Is Associated with Distinct Changes in the Human Gut Microbiome. Nat. Commun. 2019, 10, 3621. [Google Scholar] [CrossRef] [Green Version]
- Davis-Richardson, A.G.; Ardissone, A.N.; Dias, R.; Simell, V.; Leonard, M.T.; Kemppainen, K.M.; Drew, J.C.; Schatz, D.; Atkinson, M.A.; Kolaczkowski, B.; et al. Bacteroides Dorei Dominates Gut Microbiome Prior to Autoimmunity in Finnish Children at High Risk for Type 1 Diabetes. Front. Microbiol. 2014, 5, 678. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S RRNA Gene Sequences from Microbial Communities. Curr. Protoc. Bioinform. 2011, 10, Unit10.7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Weber, N.; Liou, D.; Dommer, J.; MacMenamin, P.; Quiñones, M.; Misner, I.; Oler, A.J.; Wan, J.; Kim, L.; Coakley McCarthy, M.; et al. Nephele: A Cloud Platform for Simplified, Standardized and Reproducible Microbiome Data Analysis. Bioinformatics 2018, 34, 1411–1413. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Roesch, L.F.W.; Dobbler, P.T.; Pylro, V.S.; Kolaczkowski, B.; Drew, J.C.; Triplett, E.W. Pime: A Package for Discovery of Novel Differences among Microbial Communities. Mol. Ecol. Resour. 2020, 20, 415–428. [Google Scholar] [CrossRef]
- Carrero, J.J.; Andersson Franko, M.; Obergfell, A.; Gabrielsen, A.; Jernberg, T. HsCRP Level and the Risk of Death or Recurrent Cardiovascular Events in Patients With Myocardial Infarction: A Healthcare-Based Study. J. Am. Heart Assoc. 2019, 8, e012638. [Google Scholar] [CrossRef]
- Bundy, J.D.; Li, C.; Stuchlik, P.; Bu, X.; Kelly, T.N.; Mills, K.T.; He, H.; Chen, J.; Whelton, P.K.; He, J. Systolic Blood Pressure Reduction and Risk of Cardiovascular Disease and Mortality: A Systematic Review and Network Meta-Analysis. JAMA Cardiol. 2017, 2, 775–781. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Yokoyama, Y.; Levin, S.M.; Barnard, N.D. Association between Plant-Based Diets and Plasma Lipids: A Systematic Review and Meta-Analysis. Nutr. Rev. 2017, 75, 683–698. [Google Scholar] [CrossRef]
- Kim, H.; Caulfield, L.E.; Rebholz, C.M. Healthy Plant-Based Diets Are Associated with Lower Risk of All-Cause Mortality in US Adults. J. Nutr. 2018, 148, 624–631. [Google Scholar] [CrossRef] [Green Version]
- Ornish, D.; Scherwitz, L.W.; Billings, J.H.; Brown, S.E.; Gould, K.L.; Merritt, T.A.; Sparler, S.; Armstrong, W.T.; Ports, T.A.; Kirkeeide, R.L.; et al. Intensive Lifestyle Changes for Reversal of Coronary Heart Disease. JAMA 1998, 280, 2001–2007. [Google Scholar] [CrossRef]
- Lopetuso, L.R.; Scaldaferri, F.; Petito, V.; Gasbarrini, A. Commensal Clostridia: Leading Players in the Maintenance of Gut Homeostasis. Gut Pathog. 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bultman, S.J. Bacterial Butyrate Prevents Atherosclerosis. Nat. Microbiol. 2018, 3, 1332–1333. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Y.; Yang, M.; Zhang, M.; Xiao, M.; Li, X. Butyrate Mitigates TNF-α-Induced Attachment of Monocytes to Endothelial Cells. J. Bioenergy Biomembr. 2020, 52, 247–256. [Google Scholar] [CrossRef]
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio 2019, 10, e02566-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toya, T.; Corban, M.T.; Marrietta, E.; Horwath, I.E.; Lerman, L.O.; Murray, J.A.; Lerman, A. Coronary Artery Disease Is Associated with an Altered Gut Microbiome Composition. PLoS ONE 2020, 15, e0227147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, L.; Wang, Y.; Dorfman, R.G.; Tang, D.; Xu, L.; Pan, Y.; Zhou, Q.; Li, Y.; Yin, Y.; et al. Faecalibacterium Prausnitzii Produces Butyrate to Decrease C-Myc-Related Metabolism and Th17 Differentiation by Inhibiting Histone Deacetylase 3. Int. Immunol. 2019, 31, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean Diet Intervention in Overweight and Obese Subjects Lowers Plasma Cholesterol and Causes Changes in the Gut Microbiome and Metabolome Independently of Energy Intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marungruang, N.; Tovar, J.; Björck, I.; Hållenius, F.F. Improvement in Cardiometabolic Risk Markers Following a Multifunctional Diet Is Associated with Gut Microbial Taxa in Healthy Overweight and Obese Subjects. Eur. J. Nutr. 2018, 57, 2927–2936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reunanen, J.; Kainulainen, V.; Huuskonen, L.; Ottman, N.; Belzer, C.; Huhtinen, H.; de Vos, W.M.; Satokari, R. Akkermansia Muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl. Environ. Microbiol. 2015, 81, 3655–3662. [Google Scholar] [CrossRef] [Green Version]
- Pascale, A.; Marchesi, N.; Govoni, S.; Coppola, A.; Gazzaruso, C. The Role of Gut Microbiota in Obesity, Diabetes Mellitus, and Effect of Metformin: New Insights into Old Diseases. Curr. Opin. Pharmacol. 2019, 49, 1–5. [Google Scholar] [CrossRef]
- Remely, M.; Hippe, B.; Zanner, J.; Aumueller, E.; Brath, H.; Haslberger, A.G. Gut Microbiota of Obese, Type 2 Diabetic Individuals Is Enriched in Faecalibacterium Prausnitzii, Akkermansia Muciniphila and Peptostreptococcus Anaerobius after Weight Loss. Endocr. Metab. Immune Disord. Drug Targets 2016, 16, 99–106. [Google Scholar] [CrossRef]
- Zamakhchari, M.; Wei, G.; Dewhirst, F.; Lee, J.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. Identification of Rothia Bacteria as Gluten-Degrading Natural Colonizers of the Upper Gastro-Intestinal Tract. PLoS ONE 2011, 6, e24455. [Google Scholar] [CrossRef] [Green Version]
- Romano, K.A.; Vivas, E.I.; Amador-Noguez, D.; Rey, F.E. Intestinal Microbiota Composition Modulates Choline Bioavailability from Diet and Accumulation of the Proatherogenic Metabolite Trimethylamine-N-Oxide. mBio 2015, 6, e02481. [Google Scholar] [CrossRef] [Green Version]
- Iyer, N. Methods in Microbiome Research. Lab Animal 2016, 45, 323–326. [Google Scholar] [CrossRef] [Green Version]


| Baseline 1 | Day 6 1 | Difference | ||||||
|---|---|---|---|---|---|---|---|---|
| Mean | Std | Mean | Std | Mean | Std | p | BH | |
| Age (years) | 46.89 | 12.38 | - | - | - | - | - | - |
| BMI | 31.14 | 8.83 | 31.21 | 9.75 | −0.47 | 0.39 | <0.0001 | 0.0125 |
| Height (cm) | 169.19 | 8.13 | - | - | - | - | - | - |
| Weight (kg) | 89.59 | 27.58 | 90.24 | 31.86 | −1.35 | 1.15 | <0.0001 | 0.025 |
| Waist (cm) | 104.90 | 23.32 | 102.21 | 21.06 | −2.69 | 10.95 | 0.0408 | 0.1500 |
| Hip (cm) | 114.33 | 19.56 | 113.23 | 18.54 | −1.09 | 4.37 | 0.0372 | 0.1375 |
| SBP (mmHg) | 129.92 | 17.52 | 125.46 | 14.82 | −4.46 | 15.63 | 0.0181 | 0.1250 |
| DBP (mmHg) | 84.76 | 10.36 | 81.39 | 8.27 | −3.38 | 10.17 | 0.0063 | 0.1125 |
| TC (mg/dL) | 184.15 | 33.36 | 167.26 | 32.86 | −16.89 | 16.39 | <0.0001 | 0.0375 |
| TRG (mg/dL) | 132.18 | 76.12 | 107.38 | 51.89 | −24.81 | 46.04 | <0.0001 | 0.0750 |
| HDL (mg/dL) | 52.90 | 15.25 | 51.07 | 14.75 | −1.83 | 4.98 | 0.0026 | 0.1000 |
| LDL (mg/dL) | 104.81 | 28.48 | 94.99 | 27.53 | −9.82 | 15.61 | <0.0001 | 0.0500 |
| VLDL (mg/dL) | 26.46 | 15.21 | 21.47 | 10.38 | −4.99 | 9.17 | <0.0001 | 0.0625 |
| LDL/HDL | 2.16 | 0.91 | 2.02 | 0.87 | −0.14 | 0.35 | 0.0016 | 0.0875 |
| hsCRP | 2.32 | 2.16 | 2.51 | 2.82 | −0.23 | 1.66 | 0.2905 | 0.1625 |
| Glucose | 99.81 | 31.94 | 97.90 | 23.87 | −1.90 | 16.09 | 0.3191 | 0.1750 |
| TMAO | 4.58 | 8.12 | 4.73 | 8.34 | 0.14 | 3.39 | 0.7488 | 0.1875 |
| Hypertensive (SBP ≥ 130/80 mmHg) | <130/80 mmHg | |
|---|---|---|
| No. of participants (% of total) | 56 (76.7%) | 17 (23.3%) |
| Average age (years) | 48.41 | 41.75 |
| Male (% of total) | 22 (39.3%) | 15 (88.2%) |
| Female (% of total) | 34 (60.7%) | 2 (11.8%) |
| No. Taking antihypertensive medications (% of total) | 17 (21.25%) | 2 (2.5%) |
| Mean BMI (kg/m2) | 31.96 | 28.33 |
| Mean height (cm) | 170.51 | 164.49 |
| Mean weight (kg) | 93.35 | 76.65 |
| Mean waist size (cm) | 108.43 | 92.51 |
| Mean hip size (cm) | 116.33 | 107.24 |
| Mean waist to hip ratio | 0.93 | 0.86 |
| Mean total cholesterol (mg/dL) | 184 | 185 |
| Mean triglycerides (mg/dL) | 131 | 138 |
| Mean HDL (mg/dL) | 51 | 59 |
| Mean LDL (mg/dL) | 107 | 98 |
| Mean LDL/HDL Ratio | 2.25 | 1.83 |
| Mean VLDL (mg/dL) | 26 | 28 |
| High sensitivity C-reactive protein | 3.74 | 4.39 |
| Glucose | 103 | 87 |
| TMAO | 4.03 | 6.16 |
| Phylum | Class | Order | Family | Genus | Species | ASV | T1 | T2 | %Diff | p |
|---|---|---|---|---|---|---|---|---|---|---|
| Family-level | ||||||||||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | 17.53% | 27.83% | 58.78% | 0.0002 | |||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | 9.32% | 16.97% | 82.05% | 0.0003 | |||
| Firmicutes | Clostridia | Monoglobales | Monoglobaceae | 0.53% | 1.15% | 117.39% | 0.0004 | |||
| Actinobacteriota | Coriobacteriia | Coriobacteriales | Eggerthellaceae | 0.74% | 1.28% | 73.96% | 0.002 | |||
| Firmicutes | Clostridia | Christensenellales | Christensenellaceae | 0.27% | 0.70% | 157.75% | 0.001 | |||
| Firmicutes | Clostridia | Oscillospirales | Butyricicoccaceae | 0.39% | 0.75% | 92.08% | 0.023 | |||
| Firmicutes | Bacilli | Erysipelotrichales | Erysipelatoclostridiaceae | 0.15% | 0.38% | 157.89% | 0.054 | |||
| Proteobacteria | Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | 0.22% | 0.37% | 66.67% | 0.095 | |||
| Bacteroidota | Bacteroidia | Bacteroidales | Barnesiellaceae | 0.16% | 0.01% | −92.86% | 0.001 | |||
| Proteobacteria | Gammaproteobacteria | Burkholderiales | Sutterellaceae | 0.36% | 0.21% | −42.55% | 0.0003 | |||
| Bacteroidota | Bacteroidia | Bacteroidales | Marinifilaceae | 0.21% | 0.04% | −79.63% | 0.0001 | |||
| Desulfobacterota | Desulfovibrionia | Desulfovibrionales | Desulfovibrionaceae | 0.21% | 0.05% | −78.18% | 0.001 | |||
| Bacteroidota | Bacteroidia | Bacteroidales | Tannerellaceae | 0.97% | 0.43% | −54.98% | 0.001 | |||
| Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | 1.24% | 0.58% | −53.56% | 0.013 | |||
| Firmicutes | Negativicutes | Acidaminococcales | Acidaminococcaceae | 13.99% | 3.12% | −77.74% | 0.025 | |||
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | 27.58% | 11.83% | −57.09% | 0.005 | |||
| Genus-level | ||||||||||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Faecalibacterium | 6.57% | 10.96% | 66.86% | 0.001 | ||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Roseburia | 0.50% | 2.06% | 308.40% | 0.002 | ||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae NK4A136 group | 0.33% | 1.70% | 418.82% | 0.0004 | ||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Subdoligranulum | 0.60% | 1.91% | 220.65% | 0.001 | ||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Anaerostipes | 0.63% | 1.94% | 205.45% | 0.007 | ||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | 2.18% | 3.30% | 51.06% | 0.011 | ||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Ruminococcus | 0.80% | 1.85% | 130.14% | 0.028 | ||
| Firmicutes | Clostridia | Monoglobales | Monoglobaceae | Monoglobus | 0.53% | 1.15% | 117.39% | 0.0004 | ||
| Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Veillonellaceae | Veillonella | 0.20% | 0.76% | 271.70% | 0.018 | ||
| Firmicutes | Clostridia | Christensenellales | Christensenellaceae | Christensenellaceae R-7 group | 0.23% | 0.69% | 198.33% | 0.001 | ||
| Firmicutes | Clostridia | Oscillospirales | Butyricicoccaceae | Butyricicoccus | 0.39% | 0.75% | 92.08% | 0.020 | ||
| Actinobacteriota | Coriobacteriia | Coriobacteriales | Eggerthellaceae | Adlercreutzia | 0.40% | 0.65% | 60.00% | 0.004 | ||
| Bacteroidota | Bacteroidia | Bacteroidales | Tannerellaceae | Parabacteroides | 0.97% | 0.43% | −54.98% | 0.001 | ||
| Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | 1.24% | 0.53% | −57.59% | 0.013 | ||
| Firmicutes | Negativicutes | Acidaminococcales | Acidaminococcaceae | Phascolarctobacterium | 13.99% | 3.12% | −77.74% | 0.029 | ||
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 27.58% | 11.83% | −57.09% | 0.005 | ||
| Species-level | ||||||||||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Faecalibacterium | Faecalibacterium prausnitzii | 4.83% | 7.47% | 54.49% | 0.002 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | Blautia obeum | 0.33% | 0.80% | 141.86% | 0.004 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Anaerostipes | Anaerostipes hadrus | 0.35% | 0.68% | 93.48% | 0.013 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Lachnospiraceae NK4A136 group | Lachnospiraceae NK4A136 group bacterium | 0.10% | 0.35% | 260.00% | 0.003 | |
| Actinobacteriota | Coriobacteriia | Coriobacteriales | Eggerthellaceae | Adlercreutzia | Adlercreutzia equolifaciens | 0.11% | 0.29% | 167.86% | 0.020 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Roseburia | Roseburia hominis | 0.07% | 0.22% | 222.22% | 0.0002 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | Blautia faecis | 0.21% | 0.35% | 63.64% | 0.0004 | |
| Firmicutes | Negativicutes | Veillonellales-Selenomonadales | Veillonellaceae | Veillonella | Veillonella dispar | 0.06% | 0.20% | 218.75% | 0.033 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides thetaiotaomicron | 0.35% | 0.46% | 30.77% | 0.038 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Dorea | Dorea formicigenerans | 0.13% | 0.19% | 40.00% | 0.064 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Coprococcus | Coprococcus comes | 0.23% | 0.13% | −40.68% | 0.044 | |
| Desulfobacterota | Desulfovibrionia | Desulfovibrionales | Desulfovibrionaceae | Bilophila | Bilophila wadsworthia | 0.17% | 0.03% | −81.40% | 0.002 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides caccae | 0.32% | 0.17% | −47.56% | 0.004 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Tannerellaceae | Parabacteroides | Parabacteroides merdae | 0.20% | 0.01% | −96.08% | 0.036 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Tannerellaceae | Parabacteroides | Parabacteroides distasonis | 0.37% | 0.12% | −67.37% | 0.002 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | Blautia massiliensis | 0.50% | 0.22% | −56.15% | 0.099 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | Alistipes putredinis | 0.73% | 0.15% | −79.47% | 0.001 | |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides vulgatus | 5.71% | 2.71% | −52.49% | 0.003 | |
| ASV-level | ||||||||||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Faecalibacterium | Faecalibacterium prausnitzii | 13 | 0.50% | 1.49% | 195.42% | 0.001 |
| Firmicutes | Clostridia | Monoglobales | Monoglobaceae | Monoglobus | 16 | 0.37% | 1.08% | 195.79% | 0.0004 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | Blautia obeum | 25 | 0.33% | 0.80% | 141.86% | 0.004 |
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Faecalibacterium | 15 | 0.57% | 1.00% | 76.19% | 0.033 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | 20 | 0.48% | 0.86% | 76.98% | 0.0004 | ||
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Anaerostipes | Anaerostipes hadrus | 32 | 0.21% | 0.53% | 157.41% | 0.017 |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | 37 | 0.24% | 0.52% | 114.29% | 0.004 | ||
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Faecalibacterium | Faecalibacterium prausnitzii | 2 | 1.57% | 1.77% | 12.50% | 0.067 |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides thetaiotaomicron | 22 | 0.29% | 0.46% | 58.67% | 0.046 |
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Subdoligranulum | 24 | 0.33% | 0.50% | 49.43% | 0.027 | |
| Firmicutes | Clostridia | Oscillospirales | Ruminococcaceae | Subdoligranulum | 43 | 0.19% | 0.35% | 82.00% | 0.052 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Agathobacter | 5 | 2.07% | 2.11% | 2.04% | 0.059 | |
| Firmicutes | Clostridia | Lachnospirales | Lachnospiraceae | Blautia | Blautia massiliensis | 27 | 0.50% | 0.22% | −56.15% | 0.099 |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides uniformis | 7 | 1.06% | 0.61% | −42.75% | 0.036 |
| Bacteroidota | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | Alistipes putredinis | 40 | 0.73% | 0.15% | −79.47% | 0.001 |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides vulgatus | 8 | 1.28% | 0.68% | −46.69% | 0.005 |
| Bacteroidota | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides vulgatus | 3 | 2.97% | 0.73% | −75.42% | 0.019 |
| Taxon | Taxonomic Level | Measure | Higher Abundance | log2FC | lfcSE | p | FDR |
|---|---|---|---|---|---|---|---|
| Akkermansia | Genus | LDL | Desirable | −8.84 | 2.30 | <0.001 | 0.005 |
| Akkermansia muciniphila | Species | hsCRP | Low | 9.43 | 2.32 | <0.001 | 0.004 |
| Akkermansiaceae | Family | LDL | Desirable | −8.91 | 2.25 | <0.001 | 0.002 |
| Alistipes onderdonkii | Species | hsCRP | High | −4.65 | 1.60 | 0.004 | 0.130 |
| Alistipes onderdonkii | Species | LDL | High | 5.61 | 1.79 | 0.002 | 0.029 |
| ASV1 | ASV | LDL | Desirable | −8.31 | 1.45 | <0.001 | <0.001 |
| ASV837 | ASV | hsCRP | Low | 7.05 | 2.11 | <0.001 | 0.038 |
| Lactobacillaceae | Family | hsCRP | Low | 4.82 | 1.38 | <0.001 | 0.029 |
| Lactobacillus | Genus | hsCRP | Low | 4.76 | 1.42 | 0.001 | 0.129 |
| Lactobacillus paracasei | Species | hsCRP | Low | 7.00 | 1.80 | <0.001 | 0.006 |
| Phascolarctobacterium | Genus | LDL | Desirable | −7.37 | 1.21 | <0.001 | <0.001 |
| Phascolarctobacterium faecium | Species | LDL | Desirable | −8.28 | 1.24 | <0.001 | <0.001 |
| Prevotella copri | Species | LDL | High | 9.56 | 3.52 | 0.007 | 0.093 |
| Prevotella undetermined | Species | LDL | High | 10.22 | 3.18 | 0.001 | 0.025 |
| Romboutsia | Genus | BP | Hypertensive or elevated | −3.28 | 1.26 | 0.009 | 0.117 |
| Romboutsia ilealis | Species | BP | Hypertensive or elevated | −3.43 | 1.31 | 0.009 | 0.110 |
| PICRUSt Annotation | T1 Median | T2 Median | z | p |
|---|---|---|---|---|
| Human Diseases; Cancers | 0.00097 | 0.00092 | −3.62 | 0.0003 |
| Human Diseases; Metabolic Diseases | 0.00106 | 0.00099 | −4.01 | <0.00001 |
| Metabolism; Amino Acid Metabolism | 0.10014 | 0.09853 | 3.43 | 0.0006 |
| Metabolism; Biosynthesis of Other Secondary Metabolites | 0.01037 | 0.00961 | −2.74 | 0.0061 |
| Metabolism; Carbohydrate Metabolism | 0.11120 | 0.10954 | −2.32 | 0.0203 |
| Metabolism; Glycan Biosynthesis and Metabolism | 0.02372 | 0.01953 | −3.75 | 0.0002 |
| Metabolism; Metabolism of Cofactors and Vitamins | 0.04409 | 0.04324 | −3.36 | 0.0008 |
| Metabolism; Metabolism of Other Amino Acids | 0.01515 | 0.01466 | −3.17 | 0.0015 |
| Metabolism; Metabolism of Terpenoids and Polyketides | 0.01645 | 0.01612 | −3.39 | 0.0007 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahrens, A.P.; Culpepper, T.; Saldivar, B.; Anton, S.; Stoll, S.; Handberg, E.M.; Xu, K.; Pepine, C.; Triplett, E.W.; Aggarwal, M. A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study. Nutrients 2021, 13, 3459. https://doi.org/10.3390/nu13103459
Ahrens AP, Culpepper T, Saldivar B, Anton S, Stoll S, Handberg EM, Xu K, Pepine C, Triplett EW, Aggarwal M. A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study. Nutrients. 2021; 13(10):3459. https://doi.org/10.3390/nu13103459
Chicago/Turabian StyleAhrens, Angelica P., Tyler Culpepper, Brittany Saldivar, Stephen Anton, Scott Stoll, Eileen M. Handberg, Ke Xu, Carl Pepine, Eric W. Triplett, and Monica Aggarwal. 2021. "A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study" Nutrients 13, no. 10: 3459. https://doi.org/10.3390/nu13103459
APA StyleAhrens, A. P., Culpepper, T., Saldivar, B., Anton, S., Stoll, S., Handberg, E. M., Xu, K., Pepine, C., Triplett, E. W., & Aggarwal, M. (2021). A Six-Day, Lifestyle-Based Immersion Program Mitigates Cardiovascular Risk Factors and Induces Shifts in Gut Microbiota, Specifically Lachnospiraceae, Ruminococcaceae, Faecalibacterium prausnitzii: A Pilot Study. Nutrients, 13(10), 3459. https://doi.org/10.3390/nu13103459