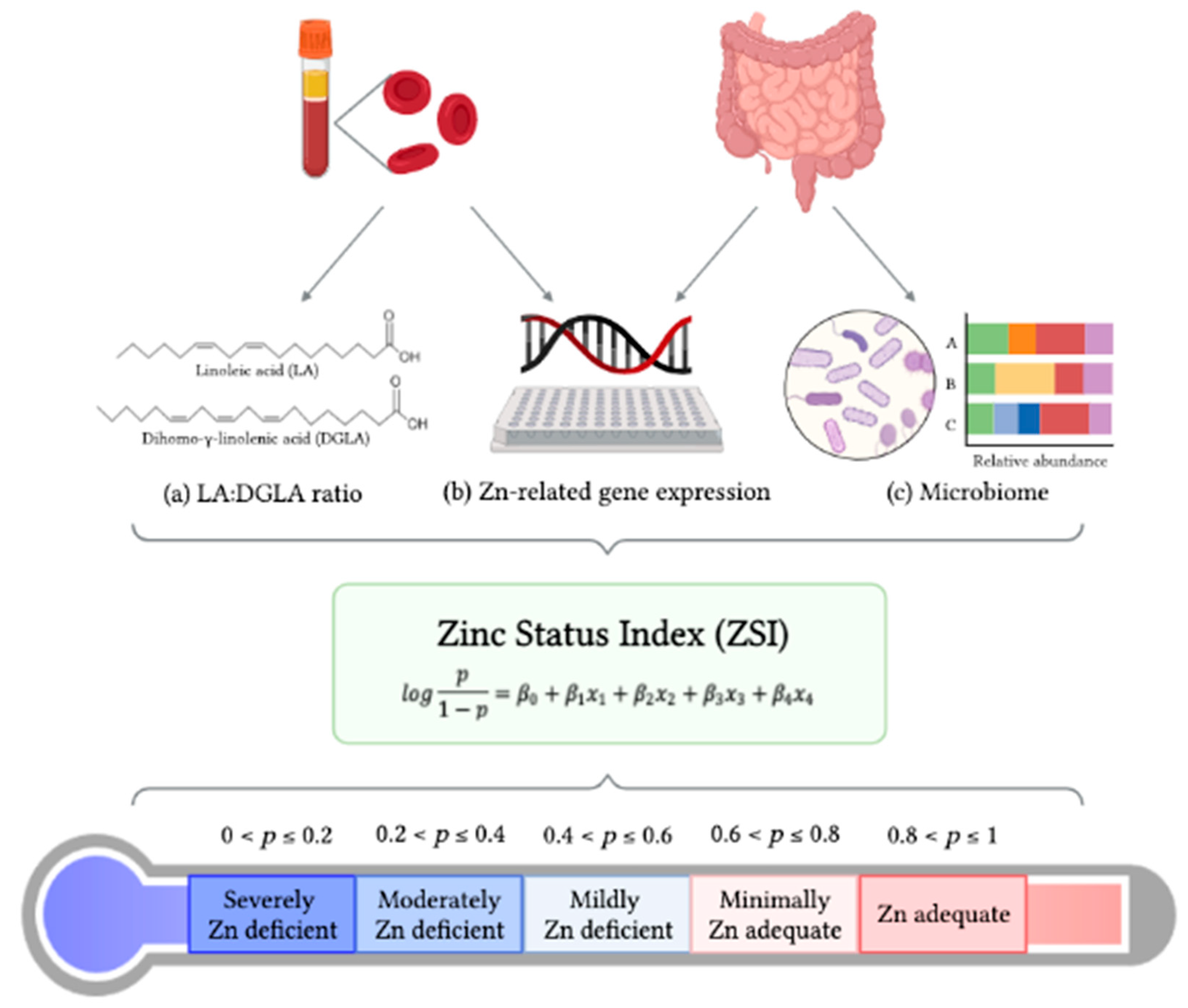
Zinc Status Index (ZSI) for Quantification of Zinc Physiological Status
Abstract
:1. Introduction
2. Review of Literature on the Three Pillars of the ZSI: LA:DGLA Ratio, Zn-Related Gene Expression, and Gut Microbiome Modulation
2.1. Materials and Methods
2.1.1. Animal Model, Study Design, and Experimental Diets
2.1.2. Blood Collection and Erythrocyte Fatty Acid Analysis
2.1.3. Determination of Serum, Nail, Feather, and Liver Zn Content
2.1.4. Isolation of Total RNA
2.1.5. Real-Time Polymerase Chain Reaction (RT-PCR)
2.1.6. 16S rRNA Gene Amplification, Sequencing, and Analysis
2.1.7. Statistical Analysis
2.2. Results
2.2.1. Zn Consumption
2.2.2. LA:DGLA Ratio
2.2.3. Zn-Related Gene Expression
2.2.4. Analysis of the Gut Microbiota
2.2.5. Additional Biomarkers of Zn Physiological Status
2.3. Discussion
2.3.1. The LA:DGLA Ratio as a Potential Reactive Biomarker of Zn Physiological Status
2.3.2. Zn-Related Gene Expression in Relation to Zn Dietary Intake In Vivo
2.3.3. Gut Microbiome as a Potential Indicator of Zn Status
3. Development of the ZSI
3.1. Statistical Strategy for Creating the Zinc Status Index (ZSI)
3.2. Examples of the ZSI as a Predictor of Zn Status
3.3. Zinc Status Index as an Accurate Predictor of Zn Physiological Status
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wessells, K.R.; Brown, K.H. Estimating the global prevalence of zinc deficiency: Results based on zinc availability in national food supplies and the prevalence of stunting. PLoS ONE 2012, 7, e50568. [Google Scholar] [CrossRef] [Green Version]
- Maret, W.; Sandstead, H.H. Zinc requirements and the risks and benefits of zinc supplementation. J. Trace Elem. Med. Biol. 2006, 20, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Andreini, C.; Banci, L.; Bertini, I.; Rosato, A. Counting the Zinc-Proteins Encoded in the Human Genome. J. Proteome Res. 2005, 5, 196–291. [Google Scholar] [CrossRef] [PubMed]
- Gaither, L.; Eide, D.J. Eukaryotic zinc transporters and their regulation. BioMetals 2001, 14, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Frederickson, C.J.; Koh, J.Y.; Bush, A.I. The neurobiology of zinc in health and disease. Nat. Rev. Neurosci 2005, 6, 449–462. [Google Scholar] [CrossRef]
- Maares, M.; Haase, H. A Guide to Human Zinc Absorption: General Overview and Recent Advances of In Vitro Intestinal Models. Nutrients 2020, 12, 762. [Google Scholar] [CrossRef] [Green Version]
- Black, R.E.; Sazawal, S. Zinc and childhood infectious disease morbidity and mortality. Br. J. Nutr. 2001, 85 (Suppl. 2), S125–S129. [Google Scholar] [CrossRef] [Green Version]
- Lowe, N.M.; Fekete, K.; Decsi, T. Methods of assessment of zinc status in humans: A systematic review. Am. J. Clin. Nutr. 2009, 89, 2040S–2051S. [Google Scholar] [CrossRef] [Green Version]
- WHO/FAO. Vitamin and Mineral Requirements in Human Nutrition; WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Ezzati, M.; Lopez, A.D.; Rodgers, A.A.; Murray, C.J.L. Comparative Quantification of Health Risks: Global and Regional Burden of Disease Attributable to Selected Major Risk Factors; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Lowe, N.M. Assessing zinc in humans. Curr. Opin. Clin. Nutr. Metab. Care. 2016, 19, 321–327. [Google Scholar] [CrossRef]
- King, J.C.; Brown, K.H.; Gibson, R.S.; Krebs, N.F.; Lowe, N.M.; Siekmann, J.H.; Raiten, D.J. Biomarkers of Nutrition for Development (BOND)-Zinc Review. J. Nutr. 2015, 146, 858S–885S. [Google Scholar] [CrossRef] [Green Version]
- Reed, S.; Qin, X.; Ran-Ressler, R.; Brenna, J.T.; Glahn, R.P.; Tako, E. Dietary zinc deficiency affects blood linoleic acid: Dihomo-gamma-linolenic acid (LA:DGLA) ratio; a sensitive physiological marker of zinc status in vivo (Gallus gallus). Nutrients 2014, 6, 1164–1180. [Google Scholar] [CrossRef] [PubMed]
- Tako, E.; Ferket, P.R.; Uni, Z. Changes in chicken intestinal zinc exporter mRNA expression and small intestinal functionality following intra-amniotic zinc-methionine administration. J. Nutr. Biochem. 2005, 16, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Andree, K.; Kim, J.; Kirschke, C.P.; J., G.; H., P.; Joung, H.; Woodhouse, L.; King, J.C.; Huang, L. Investigation of Lymphocyte Gene Expression for Use as Biomarkers for Zinc Status in Humans. J. Nutr. 2004, 134, 1716–1723. [Google Scholar] [CrossRef] [Green Version]
- Carboni, J.; Reed, S.; Kolba, N.; Eshel, A.; Koren, O.; Tako, E. Alterations in the Intestinal Morphology, Gut Microbiota, and Trace Mineral Status Following Intra-Amniotic Administration (Gallus gallus) of Teff (Eragrostis tef) Seed Extracts. Nutrients 2020, 12, 3020. [Google Scholar] [CrossRef]
- Reed, S.; Neuman, H.; Moscovich, S.; Glahn, R.P.; Koren, O.; Tako, E. Chronic Zinc Deficiency Alters Chick Gut Microbiota Composition and Function. Nutrients 2015, 7, 9768–9784. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.; Knez, M.; Uzan, A.; Stangoulis, J.C.R.; Glahn, R.P.; Koren, O.; Tako, E. Alterations in the Gut (Gallus gallus) Microbiota Following the Consumption of Zinc Biofortified Wheat (Triticum aestivum)-Based Diet. J. Agric. Food Chem 2018, 66, 6291–6299. [Google Scholar] [CrossRef]
- Juste Contin Gomes, M.; Stampini Duarte Martino, H.; Tako, E. Effects of Iron and Zinc Biofortified Foods on Gut Microbiota In Vivo (Gallus gallus): A Systematic Review. Nutrients 2021, 13, 189. [Google Scholar] [CrossRef]
- Beasley, J.T.; Johnson, A.A.T.; Kolba, N.; Bonneau, J.P.; Glahn, R.P.; Ozeri, L.; Koren, O.; Tako, E. Nicotianamine-chelated iron positively affects iron status, intestinal morphology and microbial populations in vivo (Gallus gallus). Sci. Rep. 2020, 10, 2297. [Google Scholar] [CrossRef] [Green Version]
- Knez, M.; Tako, E.; Glahn, R.P.; Kolba, N.; de Courcy-Ireland, E.; Stangoulis, J.C.R. Linoleic Acid:Dihomo-gamma-Linolenic Acid Ratio Predicts the Efficacy of Zn-Biofortified Wheat in Chicken (Gallus gallus). J. Agric. Food Chem. 2018, 66, 1394–1400. [Google Scholar] [CrossRef]
- Hou, T.; Tako, E. The In Ovo Feeding Administration (Gallus Gallus)-An Emerging In Vivo Approach to Assess Bioactive Compounds with Potential Nutritional Benefits. Nutrients 2018, 10, 418. [Google Scholar] [CrossRef]
- Dias, D.M.; Costa, N.M.B.; Nutti, M.R.; Tako, E.; Martino, H.S.D. Advantages and limitations of in vitro and in vivo methods of iron and zinc bioavailability evaluation in the assessment of biofortification program effectiveness. Crit. Rev. Food Sci. Nutr. 2018, 58, 2136–2146. [Google Scholar] [CrossRef]
- Mahler, G.J.; Esch, M.B.; Tako, E.; Southard, T.L.; Archer, S.D.; Glahn, R.P.; Shuler, M.L. Oral exposure to polystyrene nanoparticles affects iron absorption. Nat. Nanotechnol. 2012, 7, 264–271. [Google Scholar] [CrossRef]
- Dias, D.M.; Kolba, N.; Hart, J.J.; Ma, M.; Sha, S.T.; Lakshmanan, N.; Nutti, M.R.; Martino, H.S.D.; Glahn, R.P.; Tako, E. Soluble extracts from carioca beans (Phaseolus vulgaris L.) affect the gut microbiota and iron related brush border membrane protein expression in vivo (Gallus gallus). Food Res. Int 2019, 123, 172–180. [Google Scholar] [CrossRef]
- Tako, E.; Glahn, R.P.; Knez, M.; Stangoulis, J.C.R. The effect of wheat prebiotics on the gut bacterial population and iron status of iron deficient broiler chickens. Nutr. J. 2014, 13, 1–10. [Google Scholar] [CrossRef]
- Consortium, I.C.G.S. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–777. [Google Scholar]
- Hou, T.; Kolba, N.; Glahn, R.P.; Tako, E. Intra-Amniotic Administration (Gallus gallus) of Cicer arietinum and Lens culinaris Prebiotics Extracts and Duck Egg White Peptides Affects Calcium Status and Intestinal Functionality. Nutrients 2017, 9, 785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reed, S.; Neuman, H.; Glahn, R.P.; Koren, O.; Tako, E. Characterizing the gut (Gallus gallus) microbiota following the consumption of an iron biofortified Rwandan cream seeded carioca (Phaseolus Vulgaris L.) bean-based diet. PLoS ONE 2017, 12, e0182431. [Google Scholar] [CrossRef] [Green Version]
- Decuypere, E.; Michels, H. Incubation temperature as a management tool: A review. World’s Poult. Sci. Assoc. 1992, 48, 28–38. [Google Scholar] [CrossRef]
- Council, Nutrient Requirement. Nutrient Requirements of Poultry; National Academies Press: Washington, DC, USA, 1994; Volume 42, pp. 57–63. [Google Scholar]
- Liong, E.M.; McDonald, C.M.; Suh, J.; Westcott, J.L.; Wong, C.P.; Signorell, C.; King, J.C. Zinc-Biofortified Wheat Intake and Zinc Status Biomarkers in Men: Randomized Controlled Trial. J. Nutr. 2021, 151, 1817–1823. [Google Scholar] [CrossRef]
- Monteiro, J.P.; Fuzo, C.A.; Ued, F.V.; Kaput, J. Dietary patterns related to zinc and polyunsaturated fatty acids intake are associated with serum linoleic/dihomo-gamma-linolenic ratio in NHANES males and females. Sci. Rep. 2021, 11, 12215. [Google Scholar] [CrossRef]
- Chimhashu, T.; Malan, L.; Baumgartner, J.; van Jaarsveld, P.J.; Galetti, V.; Moretti, D.; Smuts, C.M.; Zimmermann, M.B. Sensitivity of fatty acid desaturation and elongation to plasma zinc concentration: A randomised controlled trial in Beninese children. Br. J. Nutr. 2018, 119, 610–619. [Google Scholar] [CrossRef] [Green Version]
- Knez, M.; Stangoulis, J.C.R.; Zec, M.; Debeljak-Martacic, J.; Pavlovic, Z.; Gurinovic, M.; Glibetic, M. An initial evaluation of newly proposed biomarker of zinc status in humans-linoleic acid: Dihomo-gamma-linolenic acid (LA:DGLA) ratio. Clin. Nutr. ESPEN 2016, 15, 85–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Kolba, N.; Liang, J.; Tako, E. Alterations in gut microflora populations and brush border functionality following intra-amniotic administration (Gallus gallus) of wheat bran prebiotic extracts. Food Funct. 2019, 10, 4834–4843. [Google Scholar] [CrossRef] [PubMed]
- Martino, H.S.D.; Kolba, N.; Tako, E. Yacon (Smallanthus sonchifolius) flour soluble extract improve intestinal bacterial populations, brush border membrane functionality and morphology in vivo (Gallus gallus). Food Res. Int. 2020, 137, 109705. [Google Scholar] [CrossRef] [PubMed]
- Knez, M.; Pantovic, A.; Zekovic, M.; Pavlovic, Z.; Glibetic, M.; Zec, M. Is There a Link between Zinc Intake and Status with Plasma Fatty Acid Profile and Desaturase Activities in Dyslipidemic Subjects? Nutrients 2019, 12, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreini, C.; Banci, L.; Bertini, I.; Rosato, A. Zinc through the Three Domains of Life. J. Proteome Res. 2006, 5, 3173–3178. [Google Scholar] [CrossRef]
- Burrell, A.L.; Dozier, W.A., 3rd; Davis, A.J.; Compton, M.M.; Freeman, M.E.; Vendrell, P.F.; Ward, T.L. Responses of broilers to dietary zinc concentrations and sources in relation to environmental implications. Br. Poult Sci. 2004, 45, 255–263. [Google Scholar] [CrossRef] [PubMed]
- King, J.C. Yet Again, Serum Zinc Concentrations Are Unrelated to Zinc Intakes. J. Nutr. 2018, 148, 1399–1401. [Google Scholar] [CrossRef] [Green Version]
- Harris, W.S. Assessing fatty acid biostatus: Red blood cells or plasma? Lipid Technol. 2013, 25, 179–181. [Google Scholar] [CrossRef]
- Smith, H.E.; Ryan, K.N.; Stephenson, K.B.; Westcott, C.; Thakwalakwa, C.; Maleta, K.; Cheng, J.Y.; Brenna, J.T.; Shulman, R.J.; Trehan, I.; et al. Multiple micronutrient supplementation transiently ameliorates environmental enteropathy in Malawian children aged 12–35 months in a randomized controlled clinical trial. J. Nutr. 2014, 144, 2059–2065. [Google Scholar] [CrossRef] [Green Version]
- Takagishi, T.; Hara, T.; Fukada, T. Recent Advances in the Role of SLC39A/ZIP Zinc Transporters In Vivo. Int J. Mol. Sci. 2017, 18, 2708. [Google Scholar] [CrossRef] [Green Version]
- Kimura, T.; Kambe, T. The Functions of Metallothionein and ZIP and ZnT Transporters: An Overview and Perspective. Int. J. Mol. Sci. 2016, 17, 336. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Tepaamorndech, S. The SLC30 family of zinc transporters—A review of current understanding of their biological and pathophysiological roles. Mol. Asp. Med. 2013, 34, 548–560. [Google Scholar] [CrossRef]
- Jeong, J.; Eide, D.J. The SLC39 family of zinc transporters. Mol. Asp. Med. 2013, 34, 612–619. [Google Scholar] [CrossRef] [Green Version]
- Geiser, J.; Venken, K.J.; De Lisle, R.C.; Andrews, G.K. A mouse model of acrodermatitis enteropathica: Loss of intestine zinc transporter ZIP4 (Slc39a4) disrupts the stem cell niche and intestine integrity. PLoS Genet. 2012, 8, e1002766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, A.; Nakagawa, M.; Tsujimura, N.; Miyazaki, S.; Kizu, K.; Goto, T.; Komatsu, Y.; Matsunaga, A.; Shirakawa, H.; Narita, H.; et al. Properties of Zip4 accumulation during zinc deficiency and its usefulness to evaluate zinc status: A study of the effects of zinc deficiency during lactation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R459–R468. [Google Scholar] [CrossRef] [PubMed]
- Koletzko, B.; Abiodun, P.O.; Laryea, M.D.; Bremer, H.J. Fatty acid composition of plasma lipids in Nigerian children with protein-energy malnutrition. Eur. J. Pediatr. 1986, 145, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Ayala, S.; Brenner, R.R. Dietary Zinc Deficiency and Fatty Acid Metabolism in Rats. Acta Physiol. Lat. Am. 1983, 33, 193–204. [Google Scholar] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Coudray, C.; Feillet-Coudray, C.; Gueux, E.; Masur, A.; Rayssiguier, Y. Dietary Inulin Intake and Age Can Affect Intestinal Absorption of Zinc and Copper in Rats. J. Nutr. 2006, 136, 117–122. [Google Scholar] [CrossRef] [Green Version]
- La Reau, A.J.; Suen, G. The Ruminococci: Key symbionts of the gut ecosystem. J. Microbiol. 2018, 56, 199–208. [Google Scholar] [CrossRef]
- Chinda, D.; Nakaji, S.; Fukuda, S.; Sakamoto, J.; Shimoyama, T.; Nakamura, T.; Fujisawa, T.; Terada, A.; Sugawara, K. The Fermentation of Different Dietary Fibers Is Associated with Fecal Clostridia Levels in Men. J. Nutr. 2004, 134, 1881–1886. [Google Scholar] [CrossRef] [Green Version]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The Controversial Role of Human Gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Sorbara, M.T.; Littmann, E.R.; Fontana, E.; Moody, T.U.; Kohout, C.E.; Gjonbalaj, M.; Eaton, V.; Seok, R.; Leiner, I.M.; Pamer, E.G. Functional and Genomic Variation between Human-Derived Isolates of Lachnospiraceae Reveals Inter- and Intra-Species Diversity. Cell Host Microbe 2020, 28, 134–146.e134. [Google Scholar] [CrossRef]
- Sheridan, P.; Martin, J.C.; Lawley, T.D.; Browne, H.; Harris, H.M.B.; Bernalier-Donadille, A.; Duncan, S.; O'Toole, P.W.; Scott, K.P.; Flint, H.J. Polysaccharide utilization loci and nutritional specialization in a dominant group of butyrate-producing human colonic Firmicutes. Microb. Genom. 2016, 2, e000043. [Google Scholar] [CrossRef] [PubMed]
- Edens, F.W.; Parkhurst, C.R.; Casas, I.A.; Dobrogosz, W.J. Principles of ex ovo competitive exclusion and in ovo administration of Lactobacillus reuteri. Poult. Sci. 1997, 76, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Mengheri, E.N.; Vignolini, F.; Pesenti, M.; Brandi, G.; Biavati, B. Bifidobacterium animalis Protects Intestine from Damage Induced by Zinc Deficiency in Rats. J. Nutr. 1997, 129, 2251–2257. [Google Scholar] [CrossRef] [Green Version]
- Roselli, M.F.A.; Garaguso, I.; Britti, M.S.; Mengheri, E. Zinc Oxide Protects Cultured Enterocytes from the Damage Induced by Escherichia coli. J. Nutr. 2003, 133, 4077–4082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, J.C. Zinc: An essential but elusive nutrient. Am. J. Clin. Nutr. 2011, 94, 679S–684S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, R.J. Assessment of Marginal Zinc Status in Humans. J. Nutr. 2000, 130, 1350S–1354S. [Google Scholar] [CrossRef]
- Knez, M.; Stangoulis, J.C.R.; Glibetic, M.; Tako, E. The Linoleic Acid: Dihomo-gamma-Linolenic Acid Ratio (LA:DGLA)—An Emerging Biomarker of Zn Status. Nutrients 2017, 9, 825. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.; Stewart, L.; Blanchard, J.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
| Analyte | Organ | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Base Pair | GI Identifier |
|---|---|---|---|---|---|
| ZnT1 | duodenum | GGTAACAGAGCTGCCTTAACT | GGTAACAGAGCTGCCTTAACT | 105 | 54109718 |
| ZnT5 | duodenum | TGGTTGGTATCTGTGCCTTTAG | GGCTGTGTCCATGGTAAGATT | 99 | 56555150 |
| ZnT7 | duodenum | GGAAGATGTCAGGATGGTTCA | CGAAGGACAAATTGAGGCAAAG | 87 | 56555152 |
| ZIP1 | duodenum | TGCCTCAGTTTCCCTCAC | GGCTCTTAAGGGCACTTCT | 144 | XM_015298606.1 |
| ZIP4 | duodenum | TCTCCTTAGCAGACAATTGAG | GTGACAAACAAGTAGGCGAAAC | 95 | 107050877 |
| ZIP6 | duodenum | GCTACTGGGTAATGGTGAAGAA | GCTGTGCCAGAACTGTAGAA | 99 | 66735072 |
| ZIP9 | duodenum | CTAAGCAAGAGCAGCAAAGAAG | CATGAACTGTGGCAACGTAAAG | 100 | 237874618 |
| Δ6-desaturase | liver | GGCGAAAGTCAGCCTATTGA | AGGTGGGAAGATGAGGAAGA | 93 | 261865208 |
| 18S | duodenum, liver | GCAAGACGAACTAAAGCGAAAG | TCGGAACTACGACGGTATCT | 100 | 7262899 |
| Reference | Animal Model | Number of Subjects | Treatment or Intervention | Duration (Weeks) | Zn Status Measures | |||
|---|---|---|---|---|---|---|---|---|
| LA:DGLA (Erythrocyte) | Zn-Related Gene Expression | Gut Microbiota Evaluation Method | Other (Not Included in ZSI) | |||||
| Beasley et al., 2020 [20] | Cornish Cross broiler (Gallus gallus) | 30 (n = 15 per group) | Nicotianamine-enhanced Zn- and Fe-biofortified wheat (Triticum aestivum L.) | 6 | Yes | ZnT1 ZnT5 ZnT7 ZIP1 ZIP4 ZIP6 ZIP9 Δ6-desaturase | 16s rRNA gene sequencing | Serum Liver Nail Feathers |
| Knez et al., 2018 [21] | Cornish Cross broiler (Gallus gallus) | 30 (n = 15 per group) | Zn-biofortified wheat (Triticum aestivum) | 6 | Yes | ZnT1 ZnT5 ZnT7 ZIP4 ZIP6 ZIP9 Δ6-desaturase | N/A | Serum Nail Feathers |
| Reed et al., 2018 [18] | Cornish Cross broiler (Gallus gallus) | 30 (n = 15 per group) | Zn-biofortified wheat (Triticum aestivum) | 6 | Yes | ZnT1 ZnT5 ZnT7 ZIP4 ZIP6 ZIP9 Δ6-desaturase | 16s rRNA gene sequencing | Serum Nail Feathers |
| Reed et al., 2015 [17] | Cornish Cross broiler (Gallus gallus) | 24 (n = 12 per group) | Zn-adequate control diet versus Zn-deficient diet (Zn carbonate as Zn source) | 4 | Yes | ZnT1 ZnT5 ZnT7 ZIP6 ZIP9 Δ6-desaturase | 16s rRNA gene sequencing | Serum Nail Feathers |
| Reed et al., 2014 [13] | Cornish Cross broiler (Gallus gallus) | 24 (n = 12 per group) | Zn-adequate control diet versus Zn-deficient diet (Zn carbonate as Zn source) | 4 | Yes | ZnT1 ZnT5 ZnT7 ZIP6 ZIP9 Δ6-desaturase | N/A | Serum Nail Feathers |
| Reference | Zn Content (μg Zn/g) | Zn Status Measures | |||
|---|---|---|---|---|---|
| LA:DGLA (Erythrocyte) | Zn-Related Gene Expression | Gut Microbiota Modulation | Other (Not Included in ZSI) | ||
| Beasley et al., 2020 [20] | Control: 16.6 ± 0.06 (standard wheat) Biofortified: 19.2 ± 0.03 (nicotianamine-enhanced Zn- and Fe-biofortified wheat) | In the biofortified relative to the control group: ↓ LA:DGLA at 2 weeks ↓ LA:DGLA at 4 weeks onwards (trend, not significant) | In the biofortified relative to the control group: ↔ ZnT1 ↔ ZnT5 ↔ ZnT7 ↔ ZIP1 ↔ ZIP4 ↔ ZIP6 ↔ ZIP9 ↔ Δ6-desaturase | In the biofortified relative to the control group: ↓ α-diversity Change in β-diversity At the phyla level: ↑ 1.9-fold the proportion of Actinobacteria ↓ 1.2- and 2.0-fold, respectively, the proportion of Firmicutes and Proteobacteria At the family level: ↑ abundance of Enterococcaceae ↓ 1.7-fold the proportion of Lachnospiraceae At the genera level: ↑ Enterococcus abundance ↓ Dorea abundance ↑ 1.9- and 1.5-fold, respectively, proportion of Bifidobacterium and Lactobacillus ↓ proportion of Streptococcus (1.7-fold), Coprococcus (1.4-fold), Ruminococcus (1.2-fold) Faecalibacterium (2-fold), and Escherichia (2-fold) | In the biofortified relative to the control group: ↔ Serum ↔ Liver ↔ Nail ↔ Feathers |
| Knez et al., 2018 [21] | Low Zn: 32.8 ± 0.17 (standard wheat) High Zn: 46.5 ± 0.99 (Zn-biofortified wheat) | In the high-Zn relative to the low-Zn group: ↓ LA:DGLA (2 weeks onwards) | In the high-Zn relative to the low-Zn group: ↓ ZnT1 ↓ ZnT5 ↓ ZnT7 ↓ ZIP4 ↓ ZIP6 ↓ ZIP9 ↓ Δ6-desaturase | See Reed et al., 2018 [18] | In the high-Zn relative to the low-Zn group: ↑ Serum Zn (2 weeks onwards) ↑ Feather Zn ↑ Nail Zn |
| Reed et al., 2018 [18] | Low Zn: 32.8 ± 0.17 (standard wheat) High Zn: 46.5 ± 0.99 (Zn-biofortified wheat) | See Knez et al., 2018 [21] | See Knez et al., 2018 [21] | In the high-Zn relative to the low-Zn group: ↔ α-diversity Change in β-diversity At the phyla level: ↔ Firmicutes, Actinobacteria, and Proteobacteria At the genera level: ↑ Dorea, Clostridiales, unclassified Clostridiales, Ruminococcus, Lachnospiraceae, and unclassified Lachnospiraceae ↓ Lactococcus, Verrucomicrobium, Bacteroides, Bacteroidales, and unclassified Bacteroidales At the species level: ↑ Lactobacillus reuteri ↓ Akkermansia muciniphila | See Knez et al., 2018 [21] |
| Reed et al., 2015 [17] | Zn deficient: 2.5 ± 0.02 Zn adequate (control): 42 ± 0.25 | See Reed et al., 2014 [13] | See Reed et al., 2014 [13] | In the Zn-adequate relative to the Zn-deficient group: ↑ α-diversity (species richness and diversity) Changes (expansion) in β-diversity At the phyla level: ↑ Firmicutes ↓ Proteobacteria At the family level: ↑ Peptostreptococcaceae and unclassified Clostridiales ↓ Enterococcaceae and Enterobacteriaceae At the genera level: ↑ unclassified Clostridiales and unclassified Peptostreptococcaceae ↓ Enterococcus, unclassified Enterococcus, unclassified Enterobacteriaceae, and unclassified Ruminococcaceae At the species level: ↑ Ruminococcus lactaris, Enterococcus sp., Clostridium lactatifermentans, and Clostridium clostridioforme ↓ Clostridium indolis and an unclassified member of the Bacteroidales (Unclassified S24–7) | See Reed et al., 2014 [13] |
| Reed et al., 2014 [13] | Zn deficient: 2.5 ± 0.02 Zn adequate (control): 42 ± 0.25 | In the Zn-adequate relative to the Zn-deficient group: ↓ LA:DGLA (1 week onwards) | In the Zn-adequate relative to the Zn-deficient group: ↔ ZnT1 ↔ ZnT5 ↔ ZnT7 ↔ ZIP6 ↔ ZIP9 ↑ Δ6-desaturase | See Reed et al., 2015 [17] | In the Zn-adequate relative to the Zn-deficient group: ↑ Serum Zn ↑ Feather Zn ↑ Nail Zn |
| LA:DGLA | Zn-Related Gene Expression | Gut Bacteria Genera |
|---|---|---|
| LA:DGLA ratio in erythrocyte |
|
|
| Predicted Probability of Zn Adequacy (p) | Estimated Zn Status |
|---|---|
| 0 ≤ p ≤ 0.2 | Severely Zn deficient |
| 0.2 < p ≤ 0.4 | Moderately Zn deficient |
| 0.4 < p ≤ 0.6 | Mildly Zn deficient |
| 0.6 < p ≤ 0.8 | Minimally Zn adequate |
| 0.8 < p ≤ 1 | Zn adequate |
| Hypothetical Subject | LA:DGLA (x1) | Δ6-Desaturase (x2) | Blautia (x3) | Predicted Probability of Zn Adequacy (p) | Estimated Zn Status | |||
|---|---|---|---|---|---|---|---|---|
| Percentile | Value (AU) | Percentile | Value (AU) | Percentile | Value (AU) | |||
| Subject 1A | 50 | 50 | 50 | 192 | 50 | 0.021 | 0.59 | Mildly Zn deficient |
| Subject 1B | 20 | 38 | 50 | 192 | 50 | 0.021 | 0.64 | Minimally Zn adequate |
| Subject 1C | 50 | 50 | 80 | 249 | 50 | 0.021 | 0.25 | Moderately Zn deficient |
| Subject 1D | 50 | 50 | 50 | 192 | 80 | 0.035 | 0.73 | Minimally Zn adequate |
| Hypothetical Subject | LA:DGLA (x1) | Δ6-Desaturase (x2) | Unclassified Lachnospiraceae (x3) | Predicted Probability of Zn Adequacy (p) | Estimated Zn Status | |||
|---|---|---|---|---|---|---|---|---|
| Percentile | Value (AU) | Percentile | Value (AU) | Percentile | Value (AU) | |||
| Subject 2A | 50 | 50 | 50 | 192 | 20 | 0.013 | 0.33 | Moderately Zn deficient |
| Subject 2B | 20 | 38 | 50 | 192 | 20 | 0.013 | 0.37 | Moderately Zn deficient |
| Subject 2C | 50 | 50 | 20 | 153 | 20 | 0.013 | 0.62 | Minimally Zn adequate |
| Subject 2D | 50 | 50 | 50 | 192 | 80 | 0.034 | 0.90 | Zn adequate |
| Hypothetical Subject | LA:DGLA (x1) | Δ6-Desaturase (x2) | ZIP9 (x3) | Predicted Probability of Zn Adequacy (p) | Estimated Zn Status | |||
|---|---|---|---|---|---|---|---|---|
| Percentile | Value (AU) | Percentile | Value (AU) | Percentile | Value (AU) | |||
| Subject 3A | 80 | 70 | 50 | 197 | 50 | 31 | 0.28 | Moderately Zn deficient |
| Subject 3B | 20 | 38 | 50 | 197 | 50 | 31 | 0.67 | Minimally Zn adequate |
| Subject 3C | 80 | 70 | 20 | 153 | 50 | 31 | 0.60 | Mildly Zn deficient |
| Subject 3D | 80 | 70 | 50 | 197 | 90 | 45 | 0.016 | Severely Zn deficient |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.; Bar, H.; Tako, E. Zinc Status Index (ZSI) for Quantification of Zinc Physiological Status. Nutrients 2021, 13, 3399. https://doi.org/10.3390/nu13103399
Cheng J, Bar H, Tako E. Zinc Status Index (ZSI) for Quantification of Zinc Physiological Status. Nutrients. 2021; 13(10):3399. https://doi.org/10.3390/nu13103399
Chicago/Turabian StyleCheng, Jacquelyn, Haim Bar, and Elad Tako. 2021. "Zinc Status Index (ZSI) for Quantification of Zinc Physiological Status" Nutrients 13, no. 10: 3399. https://doi.org/10.3390/nu13103399
APA StyleCheng, J., Bar, H., & Tako, E. (2021). Zinc Status Index (ZSI) for Quantification of Zinc Physiological Status. Nutrients, 13(10), 3399. https://doi.org/10.3390/nu13103399








