Effects of Maternal Fiber Intake on Intestinal Morphology, Bacterial Profile and Proteome of Newborns Using Pig as Model
Abstract
1. Introduction
2. Materials and Methods
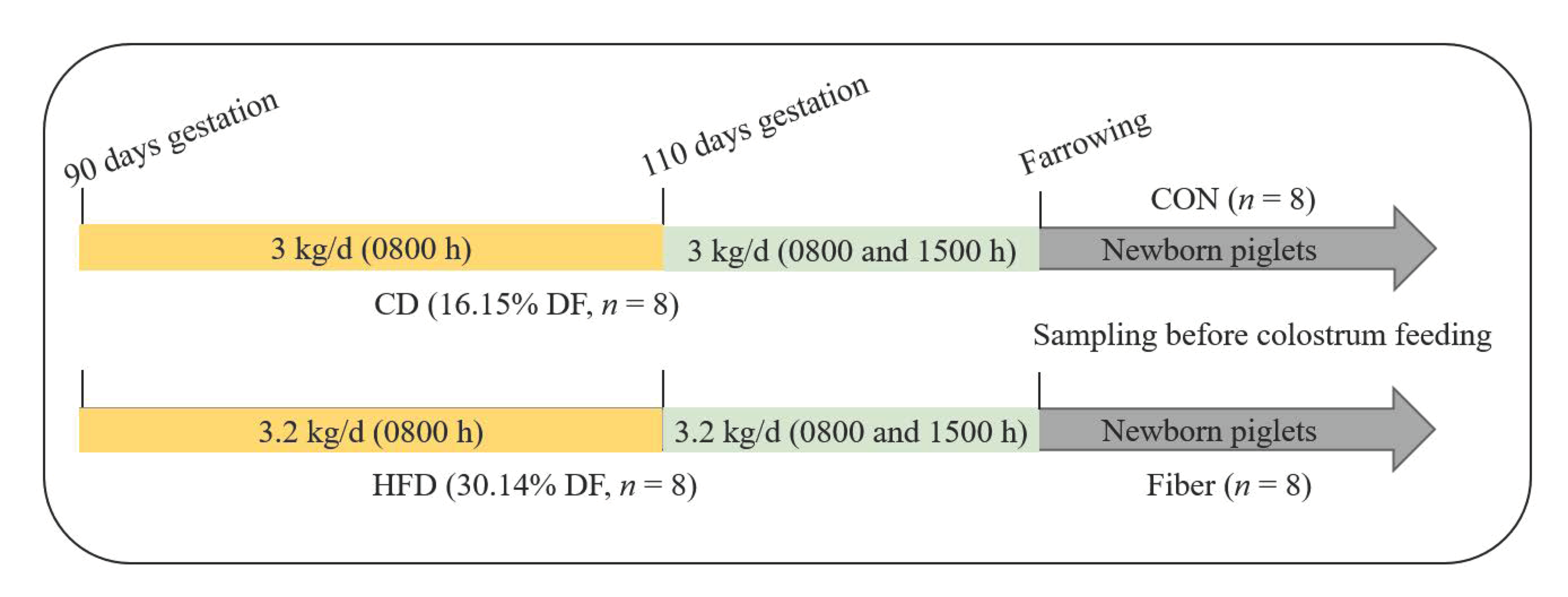
2.1. Experimental Design, Diets and Management
2.2. Sample Collection
2.3. Intestinal Morphology
2.4. Plasma Biochemical Parameters
2.5. The SCFAs Levels in Colonic Digesta
2.6. RNA Extraction and Real-Time Quantitative PCR
2.7. Bacterial Profile Analysis by 16S rRNA Gene Sequencing
2.8. iTRAQ Quantitative Proteomic Analysis
2.8.1. Protein Preparation
2.8.2. Protein Digestion and iTRAQ Labeling
2.8.3. Peptide Fractionation with Strong Cation Exchange (SCX) Chromatography
2.8.4. LC-MS/MS Analysis
2.8.5. Sequence Database Search and Data Analysis
2.8.6. Bioinformatic Analysis
2.9. Correlation Analysis of the Microbiomic and Proteomic Data
2.10. Statistical Analysis
3. Results
3.1. Birth Weights and Organ Index
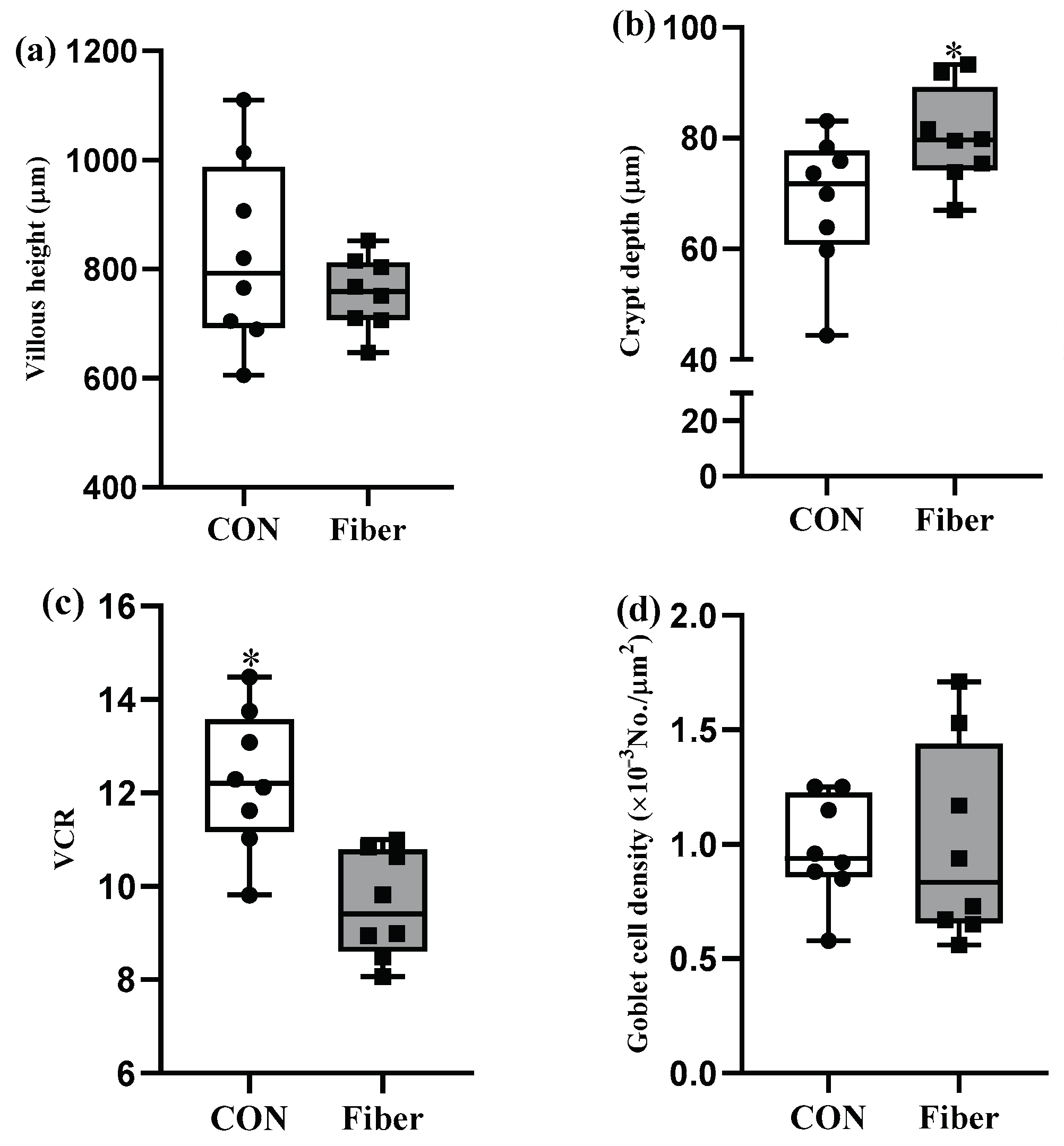
3.2. Intestinal Morphology and Goblet Cell Density
3.3. Plasma Biochemical Parameters
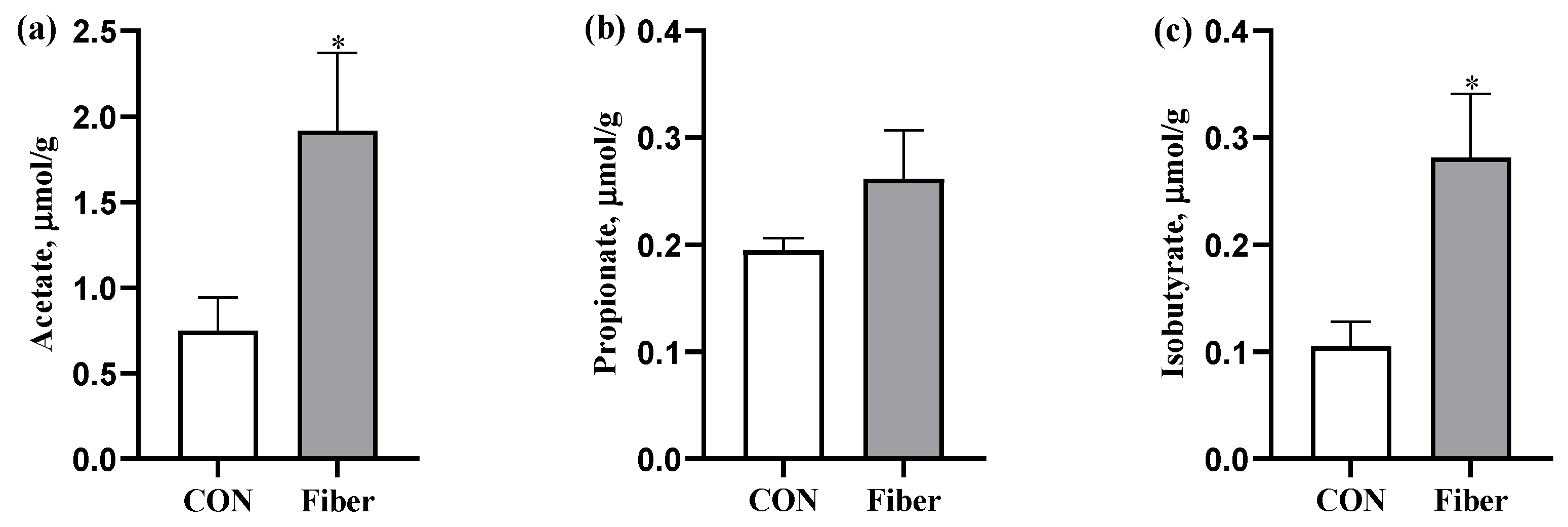
3.4. The SCFAs Levels in Colonic Digesta
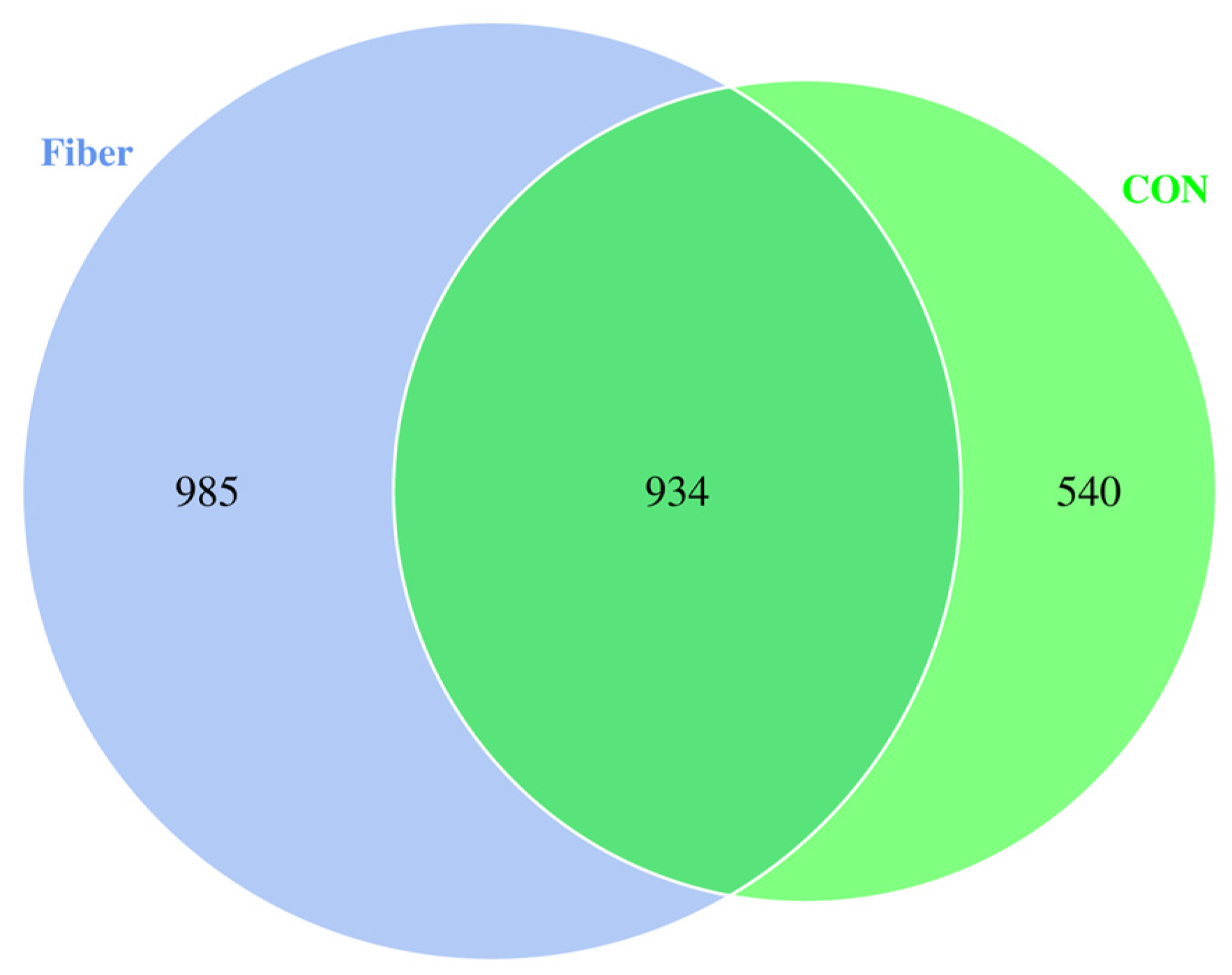
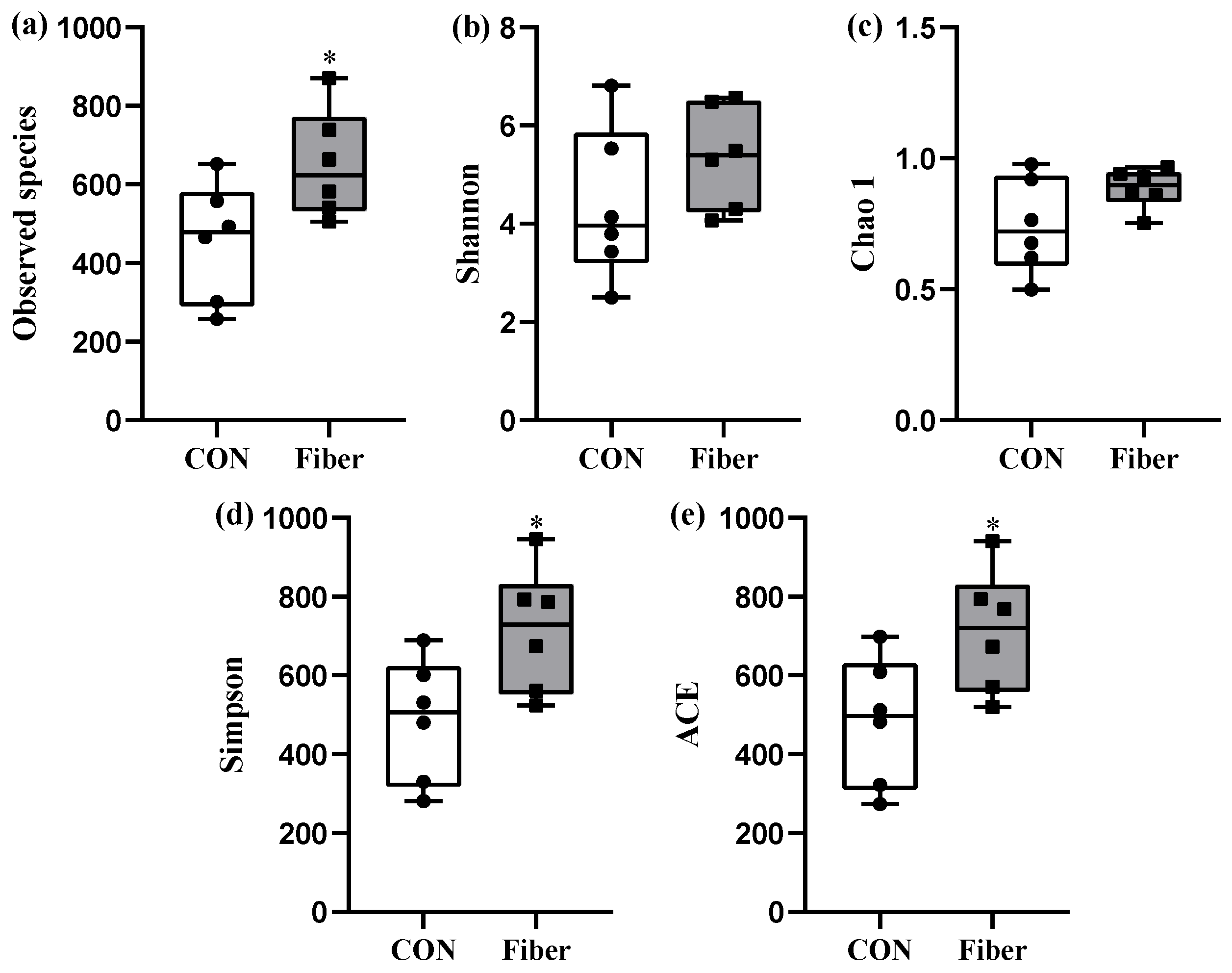
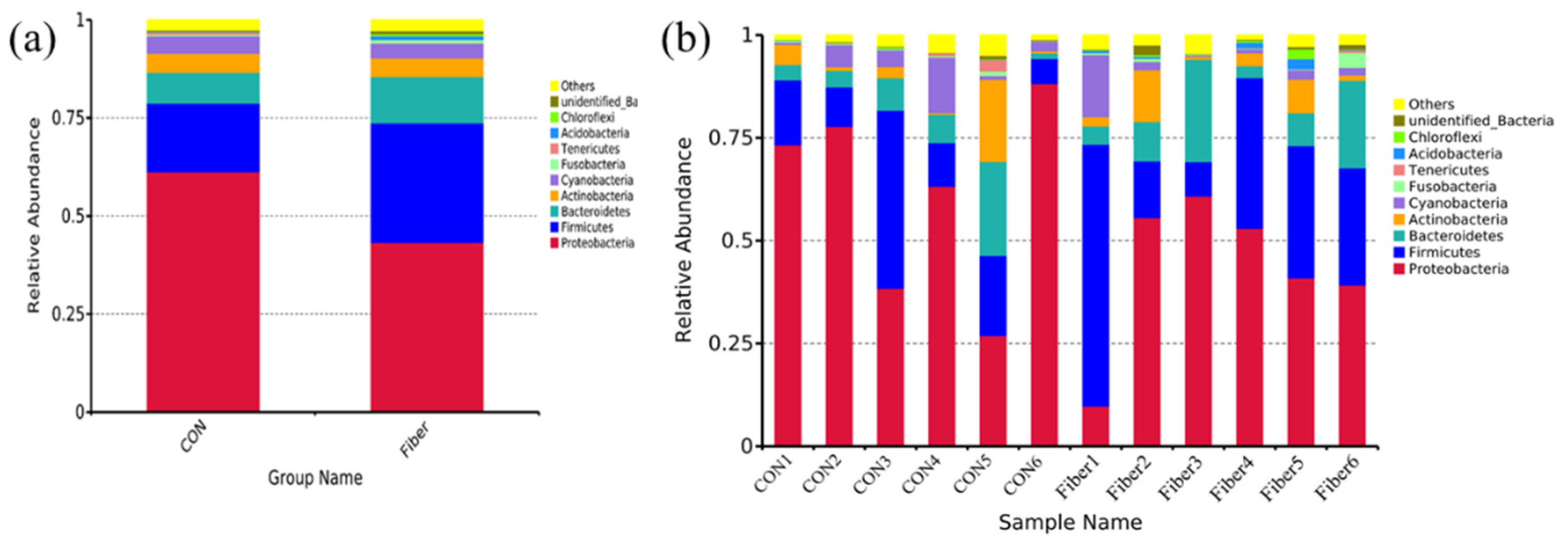
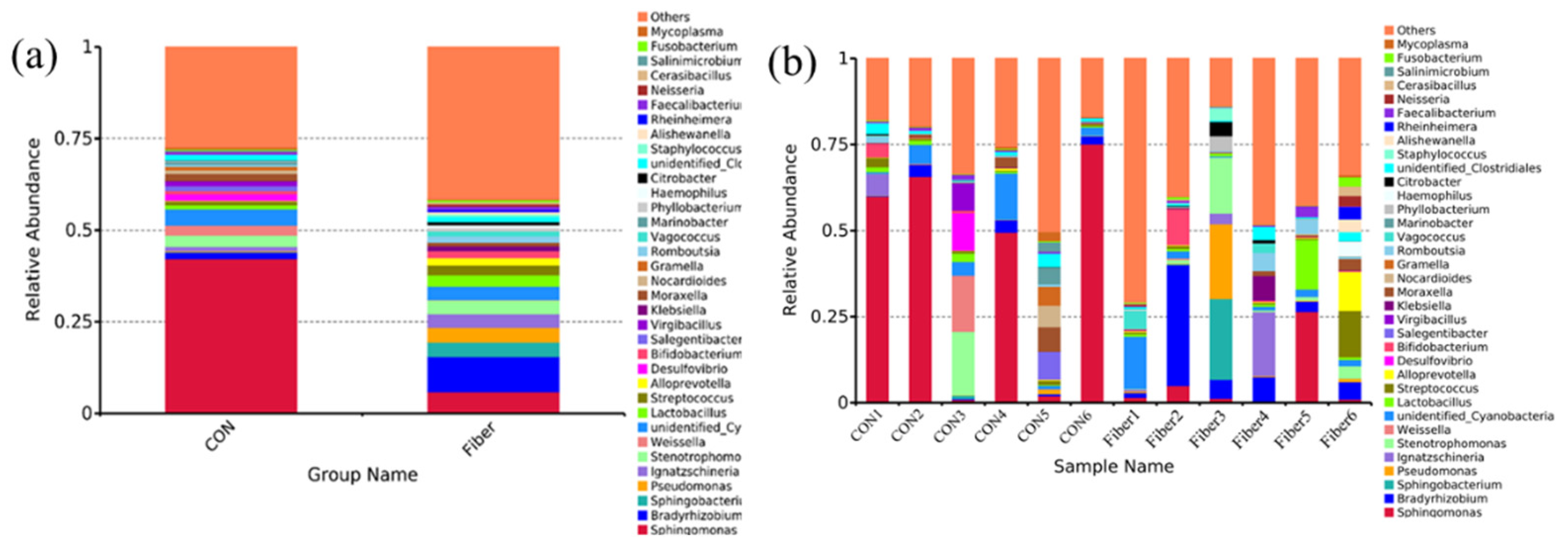
3.5. Bacterial Profile Analysis of Colonic Digesta
3.6. Identification and Quantification of Differentially Abundant Proteins
3.6.1. Protein Profiling
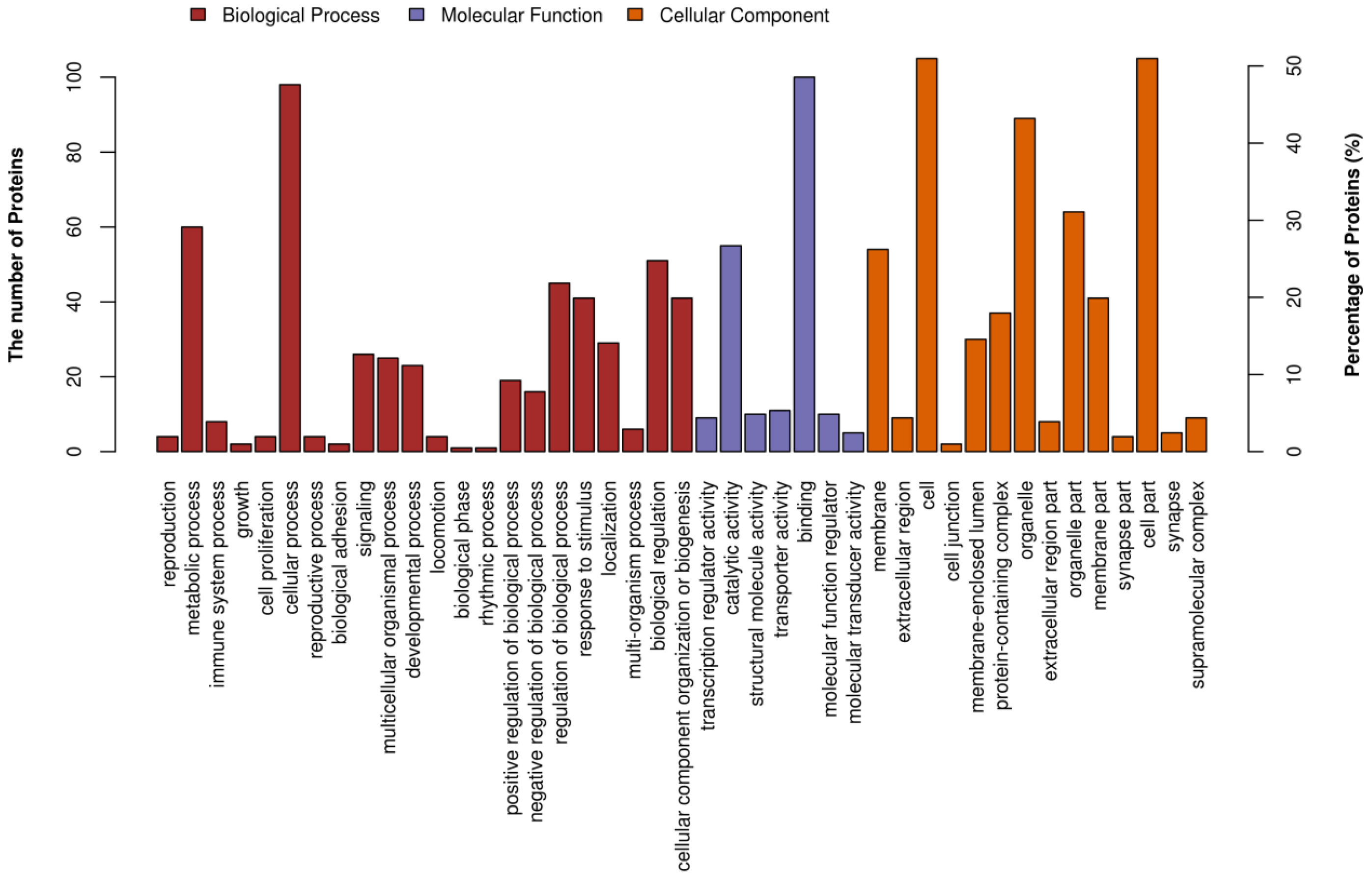
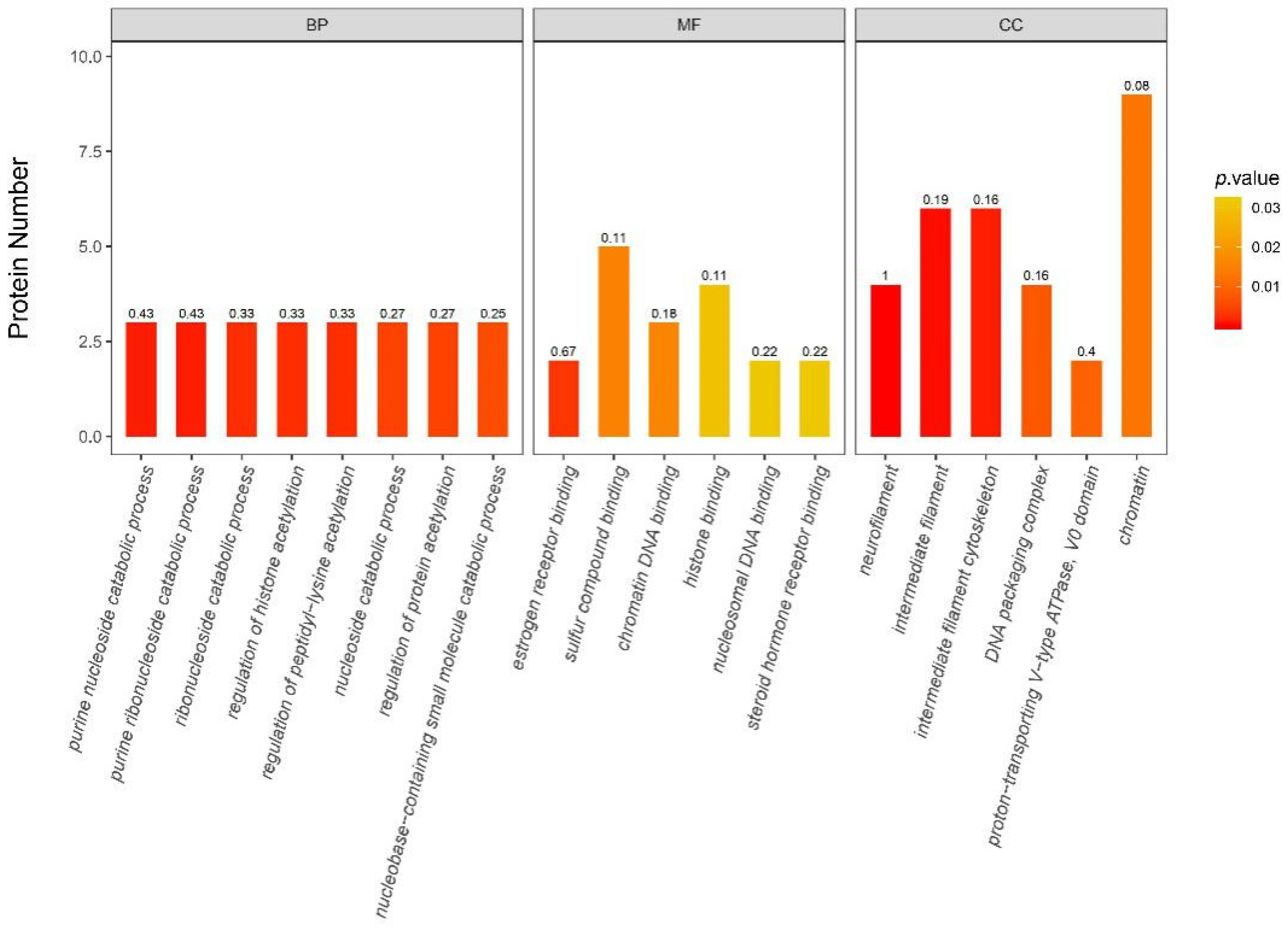
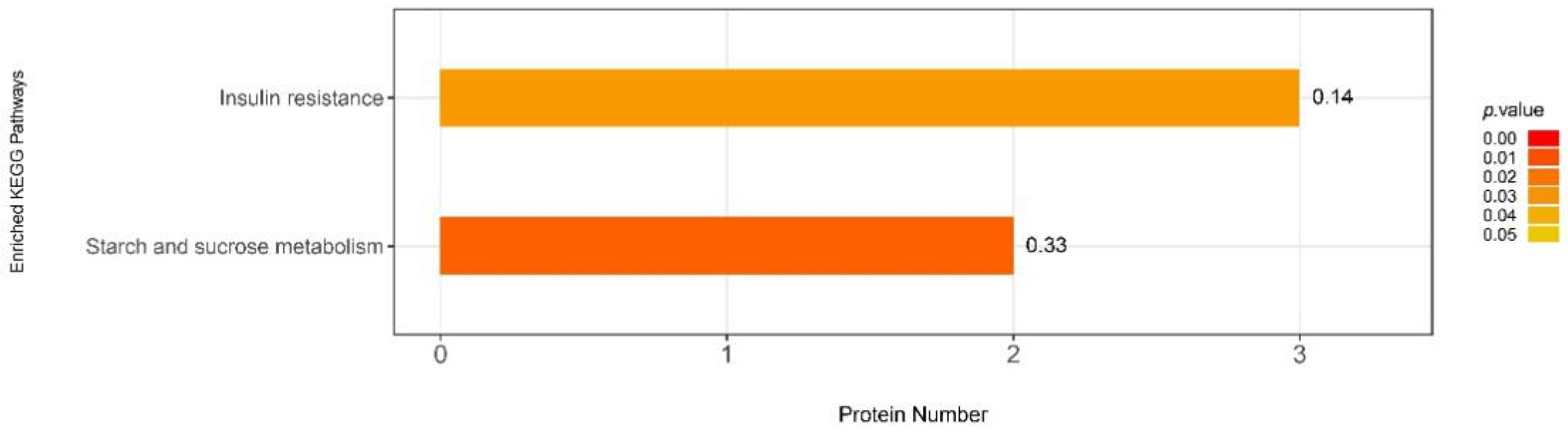
3.6.2. iTRAQ Quantification and Database Enrichment Analyses
3.6.3. Functions of the Selected DAPs
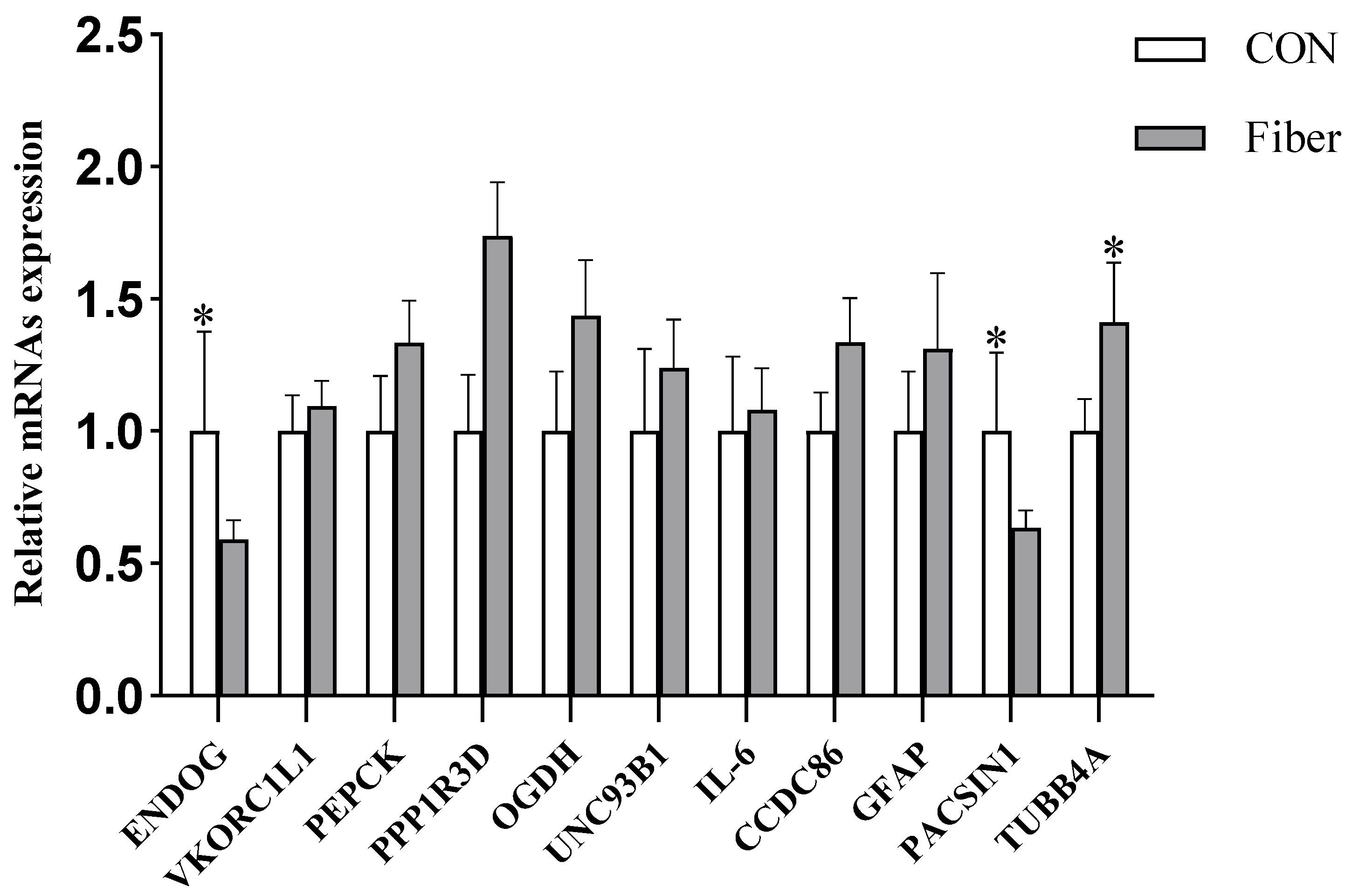
3.6.4. Validation of DAPs by RT-PCR
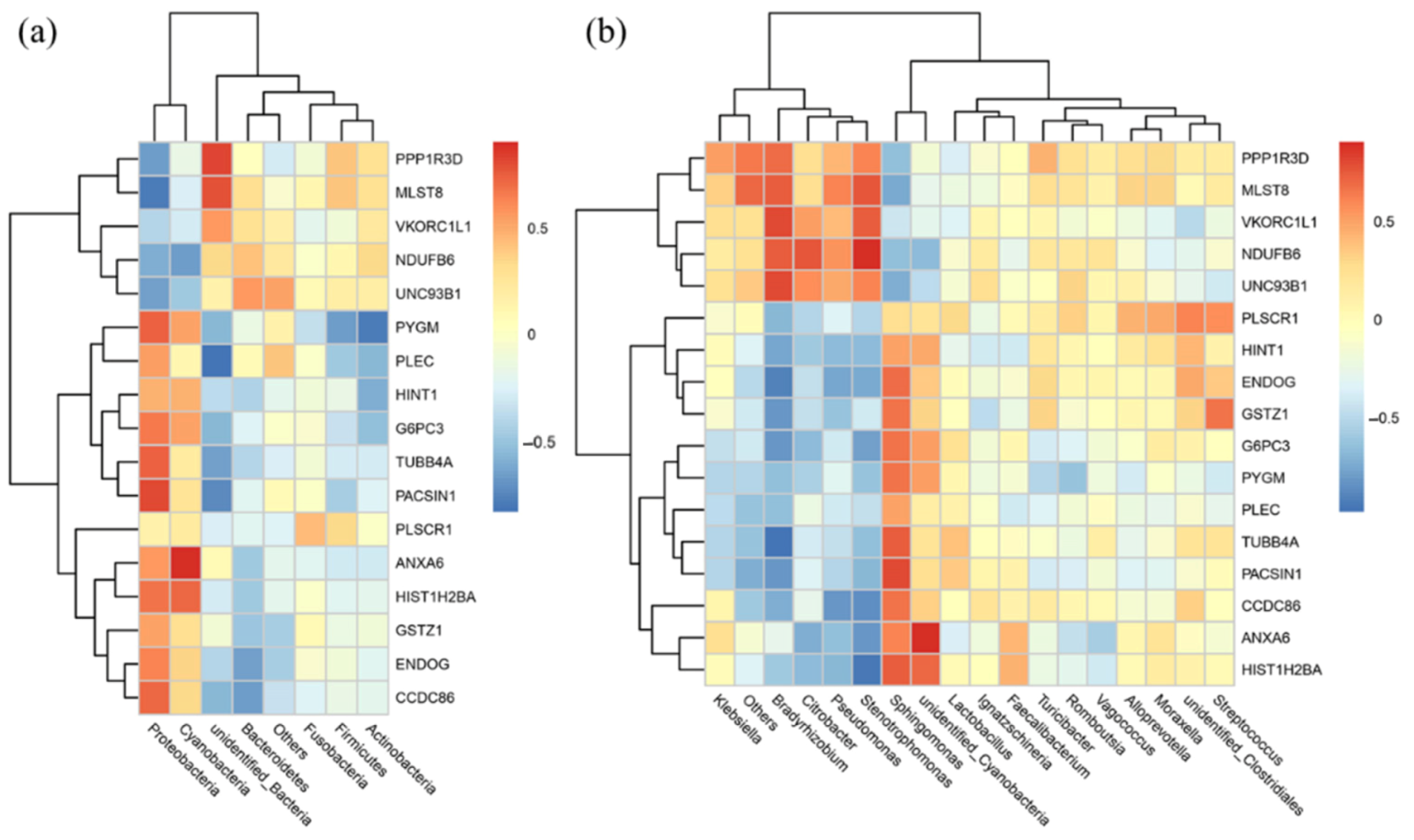
3.7. Correlation Analysis of the Microbiomic and Proteomic Data
4. Discussion
4.1. Cellular Apoptosis
4.2. Oxidative Status
4.3. Energy Metabolism
4.4. Immune and Inflammatory Responses
4.5. Cell Structure and Motility
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Takiishi, T.; Fenero, C.I.M.; Camara, N.O.S. Intestinal barrier and gut microbiota: Shaping our immune responses throughout life. Tissue Barriers 2017, 5, e1373208. [Google Scholar] [CrossRef] [PubMed]
- Guilloteau, P.; Zabielski, R.; Hammon, H.M.; Metges, C.C. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr. Res. Rev. 2010, 23, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yan, C.; Hu, L.; Liu, Y.; Xu, Q.; Wang, R.; Qin, L.; Wu, C.; Fang, Z.; Lin, Y.; et al. Effects of Fat Supplementation during Gestation on Reproductive Performance, Milk Composition of Sows and Intestinal Development of their Offspring. Animals 2019, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Azad, M.A.K.; Bin, P.; Liu, G.; Fang, J.; Li, T.; Yin, Y. Effects of different methionine levels on offspring piglets during late gestation and lactation. Food Funct. 2018, 9, 5843–5854. [Google Scholar] [CrossRef]
- Reyes-Camacho, D.; Vinyeta, E.; Perez, J.F.; Aumiller, T.; Criado, L.; Palade, L.M.; Taranu, I.; Folch, J.M.; Calvo, M.A.; Van der Klis, J.D.; et al. Phytogenic actives supplemented in hyperprolific sows: Effects on maternal transfer of phytogenic compounds, colostrum and milk features, performance and antioxidant status of sows and their offspring, and piglet intestinal gene expression. J. Anim. Sci. 2020, 98. [Google Scholar] [CrossRef]
- Wang, W.; Chen, D.; Yu, B.; Huang, Z.; Mao, X.; Zheng, P.; Luo, Y.; Yu, J.; Luo, J.; Yan, H.; et al. Effects of dietary inulin supplementation on growth performance, intestinal barrier integrity and microbial populations in weaned pigs. Br. J. Nutr. 2020, 124, 296–305. [Google Scholar] [CrossRef]
- Carlson, J.L.; Erickson, J.M.; Lloyd, B.B.; Slavin, J.L. Health Effects and Sources of Prebiotic Dietary Fiber. Curr. Dev. Nutr. 2018, 2, nzy005. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Backhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef]
- Couto, M.R.; Goncalves, P.; Magro, F.; Martel, F. Microbiota-derived butyrate regulates intestinal inflammation: Focus on inflammatory bowel disease. Pharm. Res. 2020, 159, 104947. [Google Scholar] [CrossRef]
- Lin, S.M.; Zhou, X.M.; Zhou, Y.L.; Kuang, W.M.; Chen, Y.J.; Luo, L.; Dai, F.Y. Intestinal morphology, immunity and microbiota response to dietary fibers in largemouth bass, Micropterus salmoide. Fish. Shellfish Immunol. 2020, 103, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, D.; Tian, G.; Zheng, P.; Mao, X.; Yu, J.; He, J.; Huang, Z.; Luo, Y.; Luo, J.; et al. Effects of soluble and insoluble dietary fiber supplementation on growth performance, nutrient digestibility, intestinal microbe and barrier function in weaning piglet. Anim. Feed Sci. Technol. 2020, 260. [Google Scholar] [CrossRef]
- Yu, L.; Zhong, X.; He, Y.; Shi, Y. Butyrate, but not propionate, reverses maternal diet-induced neurocognitive deficits in offspring. Pharm. Res. 2020, 160, 105082. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Kaga, N.; Nakanishi, Y.; Ohno, H.; Miyamoto, J.; Kimura, I.; Hori, S.; Sasaki, T.; Hiramatsu, K.; Okumura, K.; et al. Maternal High Fiber Diet during Pregnancy and Lactation Influences Regulatory T Cell Differentiation in Offspring in Mice. J. Immunol. 2017, 199, 3516–3524. [Google Scholar] [CrossRef]
- Lin, Y.; Li, L.; Li, Y.; Wang, K.; Wei, D.; Xu, S.; Feng, B.; Che, L.; Fang, Z.; Li, J.; et al. Interpretation of Fiber Supplementation on Offspring Testicular Development in a Pregnant Sow Model from a Proteomics Perspective. Int. J. Mol. Sci. 2019, 20, 4549. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, H.; Zhang, L.; Yang, Y.; Lin, Y.; Zhuo, Y.; Fang, Z.; Che, L.; Feng, B.; Xu, S.; et al. Maternal Dietary Fiber Composition during Gestation Induces Changes in Offspring Antioxidative Capacity, Inflammatory Response, and Gut Microbiota in a Sow Model. Int. J. Mol. Sci. 2019, 21, 31. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Hansen, C.F.; Hales, J.; Amdi, C.; Moustsen, V.A. Intrauterine growth-restricted piglets defined by their head shape have impaired survival and growth during the suckling period. Anim. Prod. Sci. 2019, 59. [Google Scholar] [CrossRef]
- Douglas, S.L.; Edwards, S.A.; Kyriazakis, I. Are all piglets born lightweight alike? Morphological measurements as predictors of postnatal performance. J. Anim. Sci. 2016, 94, 3510–3518. [Google Scholar] [CrossRef]
- Ding, X.; Li, H.; Wen, Z.; Hou, Y.; Wang, G.; Fan, J.; Qian, L. Effects of Fermented Tea Residue on Fattening Performance, Meat Quality, Digestive Performance, Serum Antioxidant Capacity, and Intestinal Morphology in Fatteners. Animals 2020, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.M.; Caton, J.S. Role of the Small Intestine in Developmental Programming: Impact of Maternal Nutrition on the Dam and Offspring. Adv. Nutr. 2016, 7, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Voillet, V.; Jegou, M.; SanCristobal, M.; Dou, S.; Rome, V.; Lippi, Y.; Billon, Y.; Pere, M.C.; Boudry, G.; et al. Comparing the intestinal transcriptome of Meishan and Large White piglets during late fetal development reveals genes involved in glucose and lipid metabolism and immunity as valuable clues of intestinal maturity. BMC Genom. 2017, 18, 647. [Google Scholar] [CrossRef] [PubMed]
- Goulet, O. Potential role of the intestinal microbiota in programming health and disease. Nutr. Rev. 2015, 73, 32–40. [Google Scholar] [CrossRef]
- Cheng, C.; Wei, H.; Xu, C.; Xie, X.; Jiang, S.; Peng, J. Maternal Soluble Fiber Diet during Pregnancy Changes the Intestinal Microbiota, Improves Growth Performance, and Reduces Intestinal Permeability in Piglets. Appl. Env. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.-J.; Liu, Q.-q.; Liao, S.; Fang, H.-H.; Yin, P.; Xie, S.-W.; Tian, L.-X.; Liu, Y.-J.; Niu, J. Effects of dietary mixed probiotics on growth, non-specific immunity, intestinal morphology and microbiota of juvenile pacific white shrimp, Litopenaeus vannamei. Fish. Shellfish Immunol. 2019, 90, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Konstantinov, S.R.; Favier, C.F.; Zhu, W.Y.; Williams, B.A.; Klüß, J.; Souffrant, W.-B.; de Vos, W.M.; Akkermans, A.D.L.; Smidt, H. Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim. Res. 2004, 53, 317–324. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; Van, V.J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Zheng, X.; Duan, Y.; Dong, H.; Zhang, J. The effect of Lactobacillus plantarum administration on the intestinal microbiota of whiteleg shrimp Penaeus vannamei. Aquaculture 2020, 526. [Google Scholar] [CrossRef]
- Ferrario, C.; Statello, R.; Carnevali, L.; Mancabelli, L.; Milani, C.; Mangifesta, M.; Duranti, S.; Lugli, G.A.; Jimenez, B.; Lodge, S.; et al. How to Feed the Mammalian Gut Microbiota: Bacterial and Metabolic Modulation by Dietary Fibers. Front. Microbiol. 2017, 8, 1749. [Google Scholar] [CrossRef]
- Kong, X.F.; Ji, Y.J.; Li, H.W.; Zhu, Q.; Blachier, F.; Geng, M.M.; Chen, W.; Yin, Y.L. Colonic luminal microbiota and bacterial metabolite composition in pregnant Huanjiang mini-pigs: Effects of food composition at different times of pregnancy. Sci. Rep. 2016, 6, 37224. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, A.; Sasaki, T.; Itoh, K.; Kitahara, T.; Takema, Y.; Hiramatsu, K.; Ishikawa, D.; Shibuya, T.; Kobayashi, O.; Osada, T.; et al. A Soluble Fiber Diet Increases Bacteroides fragilis Group Abundance and Immunoglobulin A Production in the Gut. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mittal, M. Neonatal microbiome—A brief review. J. Matern. Neonatal Med. 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.W.; Clemente, J.C.; Peter, I.; Loos, R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatr. Obes. 2017, 12, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Eviston, D.; Hsu, P.; Marino, E.; Chidgey, A.; Santner-Nanan, B.; Wong, K.; Richards, J.L.; Yap, Y.A.; Collier, F.; et al. Decreased maternal serum acetate and impaired fetal thymic and regulatory T cell development in preeclampsia. Nat. Commun. 2019, 10, 3031. [Google Scholar] [CrossRef]
- Wang, X.; Lin, G.; Liu, C.; Feng, C.; Zhou, H.; Wang, T.; Li, D.; Wu, G.; Wang, J. Temporal proteomic analysis reveals defects in small-intestinal development of porcine fetuses with intrauterine growth restriction. J. Nutr. Biochem. 2014, 25, 785–795. [Google Scholar] [CrossRef]
- Suarez-Trujillo, A.; Chen, Y.; Aduwari, C.; Cummings, S.; Kuang, S.; Buhman, K.K.; Hedrick, V.; Sobreira, T.J.P.; Aryal, U.K.; Plaut, K.; et al. Maternal high-fat diet exposure during gestation, lactation, or gestation and lactation differentially affects intestinal morphology and proteome of neonatal mice. Nutr. Res. 2019, 66, 48–60. [Google Scholar] [CrossRef]
- Mao, X.; Hu, H.; Xiao, X.; Chen, D.; Yu, B.; He, J.; Yu, J.; Zheng, P.; Luo, J.; Luo, Y.; et al. Lentinan administration relieves gut barrier dysfunction induced by rotavirus in a weaned piglet model. Food Funct. 2019, 10, 2094–2101. [Google Scholar] [CrossRef]
- Sivagnanam, U.; Palanirajan, S.K.; Gummadi, S.N. The role of human phospholipid scramblases in apoptosis: An overview. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2261–2271. [Google Scholar] [CrossRef]
- Azami, M.; Ranjkesh Adermanabadi, V.; Khanahmad, H.; Mohaghegh, M.A.; Zaherinejad, E.; Aghaei, M.; Jalali, A.; Hejazi, S.H. Immunol.ogy and Genetic of Leishmania infantum: The Role of Endonuclease G in the Apoptosis. J. Res. Med. Sci. 2018, 23, 36. [Google Scholar] [CrossRef]
- Tan, Y.; Wang, Q.; Zhao, B.; She, Y.; Bi, X. GNB2 is a mediator of lidocaine-induced apoptosis in rat pheochromocytoma PC12 cells. Neurotoxicology 2016, 54, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Weiske, J.; Huber, O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J. Biol. Chem. 2006, 281, 27356–27366. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Ni, X.; Wang, Y.; Xie, J.; Zhang, X.; Gu, H.; Zhang, J. Jinhong Tablet Reduces Damage of Int.estinal Mucosal Barrier in Rats with Acute Biliary Infection via Bcl-2/Bax mRNA and Protein Regulation. Evid. Based Complement. Altern. Med. 2017, 2017, 4985926. [Google Scholar] [CrossRef]
- Westhofen, P.; Watzka, M.; Marinova, M.; Hass, M.; Kirfel, G.; Muller, J.; Bevans, C.G.; Muller, C.R.; Oldenburg, J. Human vitamin K 2,3-epoxide reductase complex subunit 1-like 1 (VKORC1L1) mediates vitamin K-dependent intracellular antioxidant function. J. Biol. Chem. 2011, 286, 15085–15094. [Google Scholar] [CrossRef] [PubMed]
- Westhofen, P.; Watzka, M.; Marinova, M.; Hass, M.; Oldenburg, J. New insight into the function of VKORC1L1. Hamostaseologie 2010, 30, S183. [Google Scholar]
- Ma, J.W.; Ji, D.D.; Li, Q.Q.; Zhang, T.; Luo, L. Inhibition of connexin 43 attenuates oxidative stress and apoptosis in human umbilical vein endothelial cells. BMC Pulm. Med. 2020, 20, 19. [Google Scholar] [CrossRef]
- Wang, M.; Huang, H.; Liu, S.; Zhuang, Y.; Yang, H.; Li, Y.; Chen, S.; Wang, L.; Yin, L.; Yao, Y.; et al. Tannic acid modulates intestinal barrier functions associated with intestinal morphology, antioxidative activity, and intestinal tight junction in a diquat-induced mouse model. RSC Adv. 2019, 9, 31988–31998. [Google Scholar] [CrossRef]
- Wen, C.; Guo, Q.; Wang, W.; Duan, Y.; Zhang, L.; Li, J.; He, S.; Chen, W.; Li, F. Taurine Alleviates Int.estinal Injury by Mediating Tight Junction Barriers in Diquat-Challenged Piglet Models. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]
- Van der Beek, C.M.; Canfora, E.E.; Kip, A.M.; Gorissen, S.H.M.; Olde Damink, S.W.M.; van Eijk, H.M.; Holst, J.J.; Blaak, E.E.; Dejong, C.H.C.; Lenaerts, K. The prebiotic inulin improves substrate metabolism and promotes short-chain fatty acid production in overweight to obese men. Metabolism 2018, 87, 25–35. [Google Scholar] [CrossRef]
- Rabot, S.; Membrez, M.; Blancher, F.; Berger, B.; Moine, D.; Krause, L.; Bibiloni, R.; Bruneau, A.; Gerard, P.; Siddharth, J.; et al. High fat diet drives obesity regardless the composition of gut microbiota in mice. Sci. Rep. 2016, 6, 32484. [Google Scholar] [CrossRef]
- Most, J.; Goossens, G.H.; Reijnders, D.; Canfora, E.E.; Penders, J.; Blaak, E.E. Gut microbiota composition strongly correlates to peripheral insulin sensitivity in obese men but not in women. Benef. Microbes 2017, 8, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Boby, R.G.; Leelamma, S. Blackgram fiber (Phaseolus mungo): Mechanism of hypoglycemic action. Plant. Foods Hum. Nutr. 2003, 58, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Kim, H.; Jung, M.H.; Hong, S.; Song, J. Consumption of barley beta-glucan ameliorates fatty liver and insulin resistance in mice fed a high-fat diet. Mol. Nutr. Food Res. 2010, 54, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Ishii, M.; Akagawa, M. Propionate suppresses hepatic gluconeogenesis via GPR43/AMPK signaling pathway. Arch. Biochem. Biophys. 2019, 672, 108057. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Villena, C.; Sanz, P.; Garcia-Gimeno, M.A. Structure-Function Analysis of PPP1R3D, a Protein Phosphatase 1 Targeting Subunit, Reveals a Binding Motif for 14-3-3 Proteins which Regulates its Glycogenic Properties. PLoS ONE 2015, 10, e0131476. [Google Scholar] [CrossRef]
- Oligschlaeger, Y.; Miglianico, M.; Dahlmans, V.; Rubio-Villena, C.; Chanda, D.; Garcia-Gimeno, M.A.; Coumans, W.A.; Liu, Y.; Voncken, J.W.; Luiken, J.J.; et al. The interaction between AMPKbeta2 and the PP1-targeting subunit R6 is dynamically regulated by intracellular glycogen content. Biochem. J. 2016, 473, 937–947. [Google Scholar] [CrossRef]
- Mithieux, G.; Andreelli, F.; Magnan, C. Intestinal gluconeogenesis: Key signal of central control of energy and glucose homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 2009, 12, 419–423. [Google Scholar] [CrossRef]
- Loublier, S.; Bayot, A.; Rak, M.; El-Khoury, R.; Benit, P.; Rustin, P. The NDUFB6 subunit of the mitochondrial respiratory chain complex I is required for electron transfer activity: A proof of principle study on stable and controlled RNA interference in human cell lines. Biochem. Biophys. Res. Commun. 2011, 414, 367–372. [Google Scholar] [CrossRef]
- Hoshino, A.; Fujii, H. Redundant promoter elements mediate IL-3-induced expression of a novel cytokine-inducible gene, cyclon. FEBS Lett. 2007, 581, 975–980. [Google Scholar] [CrossRef]
- Chen, R.; Kang, R.; Fan, X.G.; Tang, D. Release and activity of histone in diseases. Cell Death Dis. 2014, 5, e1370. [Google Scholar] [CrossRef]
- Pelka, K.; Bertheloot, D.; Reimer, E.; Phulphagar, K.; Schmidt, S.V.; Christ, A.; Stahl, R.; Watson, N.; Miyake, K.; Hacohen, N.; et al. The Chaperone UNC93B1 Regulates Toll-like Receptor Stability Independently of Endosomal TLR Transport. Immunity 2018, 48, 911–922. [Google Scholar] [CrossRef]
- Casrouge, A.; Zhang, S.Y.; Eidenschenk, C.; Jouanguy, E.; Puel, A.; Yang, K.; Alcais, A.; Picard, C.; Mahfoufi, N.; Nicolas, N.; et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 2006, 314, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Vissers, J.P.C.; Nanda, J.; Stewart, G.D.; Husi, H.; Habib, F.K.; Hammond, D.E.; Gethings, L.A. The influence of hypoxia on the prostate cancer proteome. Clin. Chem. Lab. Med. 2020, 58, 980–993. [Google Scholar] [CrossRef] [PubMed]
- Bouameur, J.E.; Favre, B.; Borradori, L. Plakins, a versatile family of cytolinkers: Roles in skin integrity and in human diseases. J. Investig. Derm. 2014, 134, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Karashima, T.; Tsuruta, D.; Hamada, T.; Ishii, N.; Ono, F.; Hashikawa, K.; Ohyama, B.; Natsuaki, Y.; Fukuda, S.; Koga, H.; et al. Int.eraction of plectin and intermediate filaments. J. Derm. Sci. 2012, 66, 44–50. [Google Scholar] [CrossRef]
- Bai, X.; Zheng, X. Tip-to-tip interaction in the crystal packing of PACSIN 2 is important in regulating tubulation activity. Protein Cell 2013, 4, 695–701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, D.; Liu, X.; Liu, T.; Liu, H.; Tong, L.; Jia, S.; Wang, Y.F. Neurochemical regulation of the expression and function of glial fibrillary acidic protein in astrocytes. Glia 2020, 68, 878–897. [Google Scholar] [CrossRef]
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Ruegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004, 6, 1122–1128. [Google Scholar] [CrossRef]
- Pan, F.; Xu, X.; Zhang, L.L.; Luo, H.J.; Chen, Y.; Long, L.; Wang, X.; Zhuang, P.T.; Li, E.M.; Xu, L.Y. Dietary riboflavin deficiency induces genomic instability of esophageal squamous cells that is associated with gut microbiota dysbiosis in rats. Food Funct. 2020, 11, 10070–10083. [Google Scholar] [CrossRef]
- Tarnopolsky, M.A. Myopathies Related to Glycogen Metabolism Disorders. Neurotherapeutics 2018, 15, 915–927. [Google Scholar] [CrossRef]
| CD | HFD | |
|---|---|---|
| Corn (crude protein 8.2%) | 73.50 | 49.83 |
| Soybean meal (crude protein 46%) | 16.80 | 11.30 |
| Wheat bran | 2.90 | 12.00 |
| Soybean hull | 2.90 | 12.00 |
| Sugar beet pulp | 0.00 | 11.40 |
| L-Lysine-HCl (70%) | 0.02 | 0.00 |
| DL-Methionine (98.5%) | 0.01 | 0.04 |
| L-Threonine (98.5%) | 0.00 | 0.04 |
| CaCO3 | 1.08 | 0.67 |
| CaHPO4 | 1.70 | 1.63 |
| Sodium chloride | 0.40 | 0.40 |
| Choline chloride (50%) | 0.14 | 0.14 |
| Vitamin and mineral premix 1 | 0.55 | 0.55 |
| Total | 100.00 | 100.00 |
| Nutrient levels | ||
| Digestible energy, MJ/kg | 13.31 | 12.47 |
| Crude protein, % | 14.23 | 13.29 |
| Ether extract, % | 3.07 | 2.82 |
| Crude fiber, % | 2.87 | 8.05 |
| Neutral detergent fiber, % | 10.11 | 21.93 |
| Dietary fiber, % | 16.15 | 30.14 |
| Available P, % | 0.46 | 0.43 |
| Lysine, % | 0.57 | 0.53 |
| Methionine, % | 0.22 | 0.21 |
| Threonine, % | 0.47 | 0.44 |
| Tryptophan, % | 0.15 | 0.18 |
| CON | Fiber | p-Value | |
|---|---|---|---|
| Birth weight, kg | 1.29 ± 0.03 | 1.28 ± 0.04 | 0.834 |
| Body length, cm | 35.9 ± 0.7 | 37.9 ± 0.6 | 0.047 |
| Head length, cm | 10.9 ± 0.2 | 12.2 ± 0.3 | 0.002 |
| Heart weight, g | 8.29 ± 0.52 | 8.92 ± 0.28 | 0.302 |
| Liver weight, g | 31.79 ± 2.15 | 28.51 ± 1.73 | 0.255 |
| Spleen weight, g | 0.98 ± 0.05 | 1.10 ± 0.05 | 0.095 |
| Kidney weight, g | 8.74 ± 0.55 | 8.66 ± 0.63 | 0.924 |
| Stomach weight, g | 5.63 ± 0.35 | 6.60 ± 0.65 | 0.207 |
| Pancreas weight, g | 1.41 ± 0.11 | 1.50 ± 0.08 | 0.569 |
| Large intestinal weight, g | 12.45 ± 1.66 | 14.20 ± 1.26 | 0.414 |
| Small intestinal weight, g | 33.07 ± 2.75 | 36.85 ± 2.69 | 0.343 |
| Small intestinal length, cm | 286.8 ± 11.7 | 291.8 ± 3.3 | 0.725 |
| CON | Fiber | p-Value | |
|---|---|---|---|
| ALT, U/L | 14.38 ± 2.77 | 14.00 ± 1.95 | 0.913 |
| AST, U/L | 36.87 ± 9.05 | 28.50 ± 3.19 | 0.398 |
| GGT, U/L | 42.00 ± 7.73 | 51.50 ± 6.30 | 0.357 |
| TBA, µmol/L | 4.95 ± 0.78 | 14.83 ± 6.98 | 0.182 |
| GLU, mmol/L | 2.78 ± 0.16 | 2.79 ± 0.21 | 0.961 |
| Urea, mmol/L | 2.97 ± 0.23 | 3.01 ± 0.12 | 0.884 |
| CREA, mmol/L | 72.45 ± 4.25 | 89.60 ± 9.95 | 0.003 |
| TG, mmol/L | 0.15 ± 0.02 | 0.12 ± 0.01 | 0.156 |
| CON | Fiber | p-Value | |
|---|---|---|---|
| Proteobacteria | 63.91 ± 8.13 | 43.25 ± 7.53 | 0.132 |
| Firmicutes | 23.74 ± 8.65 | 30.52 ± 7.99 | 0.589 |
| Bacteroidetes | 4.04 ± 1.22 | 11.85 ± 3.71 | 0.065 |
| Actinobacteria | 1.56 ± 0.77 | 4.69 ± 1.91 | 0.132 |
| Cyanobacteria | 4.30 ± 2.02 | 3.78 ± 2.29 | 0.818 |
| Fusobacteria | 0.21 ± 0.06 | 0.87 ± 0.56 | 0.485 |
| Tenericutes | 0.17 ± 0.10 | 0.14 ± 0.10 | 0.699 |
| Acidobacteria | 0.04 ± 0.02 | 0.85 ± 0.37 | 0.015 |
| Chloroflexi | 0.12 ± 0.07 | 0.60 ± 0.37 | 0.180 |
| unidentified_Bacteria | 0.12 ± 0.03 | 0.74 ± 0.35 | 0.065 |
| Others | 1.78 ± 0.60 | 2.71 ± 0.48 | 0.310 |
| Accession | Description | Gene Name | Protein Name | Fold Change | p-Value |
|---|---|---|---|---|---|
| Cellular apoptosis | |||||
| F4ZS19 | Phospholipid scramblase OS = Sus scrofa OX = 9823 GN = PLSCR1 PE = 2 SV = 1 − [F4ZS19_PIG] | PLSCR1 | Phospholipid scramblase | −0.81 | 0.0393 |
| F1RR64 | Endonuclease G OS = Sus scrofa OX = 9823 GN = ENDOG PE = 1 SV=1 − [F1RR64_PIG] | ENDOG | Endonuclease G | −0.63 | 0.0102 |
| F1S0V3 | Annexin OS = Sus scrofa OX = 9823 GN = ANXA6 PE = 1 SV=3 − [F1S0V3_PIG] | ANXA6 | Annexin | −0.54 | 0.0472 |
| A0A0B8RTB1 | Apoptosis antagonizing transcription factor OS = Sus scrofa domesticus OX = 9825 GN = AATF PE = 4 SV = 1 − [A0A0B8RTB1_PIG] | AATF | Apoptosis antagonizing transcription factor | −0.82 | 0.0436 |
| F1RKI3 | Histidine triad nucleotide binding protein 1 OS = Sus scrofa OX = 9823 GN = HINT1 PE = 1 SV = 1 − [F1RKI3_PIG] | HINT1 | Histidine triad nucleotide binding protein 1 | −0.71 | 0.0291 |
| Oxidative status | |||||
| I3LQ80 | Vitamin K epoxide reductase complex, subunit 1-like 1 OS = Sus scrofa OX = 9823 GN = VKORC1L1 PE = 1 SV = 2 − [I3LQ80_PIG] | VKORC1L1 | Vitamin K epoxide reductase complex, subunit 1-like 1 | +1.26 | 0.0230 |
| F1S2N0 | Glutathione S-transferase zeta 1 OS = Sus scrofa OX = 9823 GN = GSTZ1 PE = 1 SV = 2 − [F1S2N0_PIG] | GSTZ1 | Glutathione S-transferase zeta 1 | −0.82 | 0.0355 |
| Energy metabolism | |||||
| Q29265 | Phosphoenolpyruvate carboxykinase, cytosolic (Fragment) OS = Sus scrofa OX = 9823 PE = 2 SV = 1 − [Q29265_PIG] | PEPCK | Phosphoenolpyruvate carboxykinase, cytosolic (Fragment) | +1.35 | 0.0061 |
| K9IVI1 | 2-oxoglutarate dehydrogenase, mitochondrial OS = Sus scrofa OX = 9823 GN = OGDH PE = 1 SV = 1 − [K9IVI1_PIG] | OGDH | 2-oxoglutarate dehydrogenase, mitochondrial | +1.57 | 0.0346 |
| F1RJ12 | Protein phosphatase 1 regulatory subunit OS = Sus scrofa OX = 9823 GN = PPP1R3D PE = 4 SV = 1 − [F1RJ12_PIG] | PPP1R3D | Protein phosphatase 1 regulatory subunit | +1.21 | 0.0201 |
| Q2EN80 | NADH dehydrogenase 1 beta subcomplex 6 OS = Sus scrofa OX = 9823 GN = NDUFB6 PE = 1 SV = 1 − [Q2EN80_PIG] | NDUFB6 | NADH dehydrogenase 1 beta subcomplex 6 | +1.21 | 0.0034 |
| F1S1J5 | Glucose-6-phosphatase OS = Sus scrofa OX = 9823 GN = G6PC3 PE = 3 SV = 1 − [F1S1J5_PIG] | G6PC3 | Glucose-6-phosphatase | −0.80 | 0.0109 |
| F1RQQ8 | Alpha-1,4 glucan phosphorylase OS = Sus scrofa OX = 9823 GN = PYGM PE = 1 SV = 2 − [F1RQQ8_PIG] | PYGM | Alpha-1,4 glucan phosphorylase | −0.77 | 0.0380 |
| G3CKJ2 | Glyceraldehyde-3-phosphate dehydrogenase (Fragment) OS = Sus scrofa OX = 9823 GN = GAPDH PE = 2 SV = 1 − [G3CKJ2_PIG] | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase (Fragment) | −0.76 | 0.0030 |
| Immune and inflammatory responses | |||||
| I3LEA8 | Unc-93 homolog B1, TLR signaling regulator OS = Sus scrofa OX = 9823 GN = UNC93B1 PE = 4 SV = 2 − [I3LEA8_PIG] | UNC93B1 | Unc-93 homolog B1, TLR signaling regulator | +1.83 | 0.0264 |
| A0A287AP27 | Uncharacterized protein OS = Sus scrofa OX = 9823 GN = CDK9 PE = 4 SV = 1 − [A0A287AP27_PIG] | CDK9 | none | +1.40 | 0.0079 |
| Q2TJZ7 | Microsomal prostaglandin E synthase-1 OS = Sus scrofa OX = 9823 GN = PTGES PE = 1 SV = 1 − [Q2TJZ7_PIG] | PTGES | Microsomal prostaglandin E synthase-1 | +1.39 | 0.0259 |
| Q5W7K7 | Interleukin 6 (Fragment) OS = Sus scrofa OX = 9823 GN = IL-6 PE = 2 SV = 1 − [Q5W7K7_PIG] | IL-6 | Interleukin 6 (Fragment) | +1.26 | 0.0128 |
| F1S9T2 | Uncharacterized protein OS = Sus scrofa OX = 9823 GN = IFI44 PE = 1 SV = 2 − [F1S9T2_PIG] | IFI44 | none | +1.24 | 0.0122 |
| F1RIC1 | Coiled-coil domain containing 86 OS = Sus scrofa OX = 9823 GN = CCDC86 PE = 4 SV = 1 − [F1RIC1_PIG] | CCDC86 | Coiled-coil domain containing 86 | −0.80 | 0.0107 |
| F1RTQ6 | Histone H2B OS = Sus scrofa OX = 9823 GN = HIST1H2BA PE = 1 SV = 1 − [F1RTQ6_PIG] | HIST1H2BA | Histone H2B | −0.80 | 0.0162 |
| Cell structure and motility | |||||
| F1RFA8 | MTOR associated protein, LST8 homolog OS = Sus scrofa OX = 9823 GN = MLST8 PE = 1 SV = 3− [F1RFA8_PIG] | MLST8 | MTOR associated protein, LST8 homolog | +1.51 | 0.0162 |
| F2Z5K5 | Tubulin beta chain OS = Sus scrofa OX = 9823 GN = TUBB4A PE = 1 SV = 2 − [F2Z5K5_PIG] | TUBB4A | Tubulin beta chain | −0.80 | 0.0264 |
| K9IVQ6 | Plectin (Fragment) OS = Sus scrofa OX = 9823 GN = PLEC PE = 2 SV = 1 − [K9IVQ6_PIG] | PLEC | Plectin (Fragment) | −0.73 | 0.0451 |
| A0A287AHG5 | Protein kinase C and casein kinase substrate in neurons 1 OS = Sus scrofa OX = 9823 GN = PACSIN1 PE = 1 SV = 1 − [A0A287AHG5_PIG] | PACSIN1 | Protein kinase C and casein kinase substrate in neurons 1 | −0.68 | 0.0083 |
| A0A287AYM2 | Glial fibrillary acidic protein OS = Sus scrofa OX = 9823 GN = GFAP PE = 1 SV = 1 − [A0A287AYM2_PIG] | GFAP | Glial fibrillary acidic protein | −0.56 | 0.0146 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Peng, X.; Liu, Y.; Wu, Q.; Zhou, Q.; Hu, L.; Fang, Z.; Lin, Y.; Xu, S.; Feng, B.; et al. Effects of Maternal Fiber Intake on Intestinal Morphology, Bacterial Profile and Proteome of Newborns Using Pig as Model. Nutrients 2021, 13, 42. https://doi.org/10.3390/nu13010042
He Y, Peng X, Liu Y, Wu Q, Zhou Q, Hu L, Fang Z, Lin Y, Xu S, Feng B, et al. Effects of Maternal Fiber Intake on Intestinal Morphology, Bacterial Profile and Proteome of Newborns Using Pig as Model. Nutrients. 2021; 13(1):42. https://doi.org/10.3390/nu13010042
Chicago/Turabian StyleHe, Ying, Xie Peng, Yang Liu, Qing Wu, Qiang Zhou, Liang Hu, Zhengfeng Fang, Yan Lin, Shengyu Xu, Bin Feng, and et al. 2021. "Effects of Maternal Fiber Intake on Intestinal Morphology, Bacterial Profile and Proteome of Newborns Using Pig as Model" Nutrients 13, no. 1: 42. https://doi.org/10.3390/nu13010042
APA StyleHe, Y., Peng, X., Liu, Y., Wu, Q., Zhou, Q., Hu, L., Fang, Z., Lin, Y., Xu, S., Feng, B., Li, J., Zhuo, Y., Wu, D., & Che, L. (2021). Effects of Maternal Fiber Intake on Intestinal Morphology, Bacterial Profile and Proteome of Newborns Using Pig as Model. Nutrients, 13(1), 42. https://doi.org/10.3390/nu13010042







