Influence of Iron on the Gut Microbiota in Colorectal Cancer
Abstract
1. Introduction
2. Microbiota in Colonic Homeostasis and Disease
2.1. Gut Microbiota
2.2. Bacteria and Cancer
3. Gut Microbiota and Colorectal Cancer
3.1. Pathobionts and Colorectal Cancer
3.2. Symbionts and Colorectal Cancer
4. Dysbiosis and Bacterial Iron Utilization
5. Iron Supplementation, Microbiota, and Colorectal Cancer
6. Molecular Pathological Epidemiology in Colorectal Cancer
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kosumi, K.; Mima, K.; Baba, H.; Ogino, S. Dysbiosis of the Gut Microbiota and Colorectal Cancer: The Key Target of Molecular Pathological Epidemiology. J. Lab. Precis. Med. 2018, 3, 76. [Google Scholar] [CrossRef] [PubMed]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and Colon Cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Seiwert, N.; Heylmann, D.; Hasselwander, S.; Fahrer, J. Mechanism of Colorectal Carcinogenesis Triggered by Heme Iron from Red Meat. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188334. [Google Scholar] [CrossRef]
- Wooldridge, K.G.; Williams, P.H. Iron Uptake Mechanisms of Pathogenic Bacteria. FEMS Microbiol. Rev. 1993, 12, 325–348. [Google Scholar] [CrossRef]
- Mahalhal, A.; Williams, J.M.; Johnson, S.; Ellaby, N.; Duckworth, C.A.; Burkitt, M.D.; Liu, X.; Hold, G.L.; Campbell, B.J.; Pritchard, D.M.; et al. Oral Iron Exacerbates Colitis and Influences the Intestinal Microbiome. PLoS ONE 2018, 13, e0202460. [Google Scholar] [CrossRef]
- Wilson, M.; Dekker, J.W.; Harlaar, J.; Jeekel, H.; Schipperus, M.; Zwaginga, J. The Role of Preoperative Iron Deficiency in Colorectal Cancer Patients: Prevalence and Treatment. Int. J. Colorectal Dis. Clin. Mol. Gastroenterol. Surg. 2017, 32, 1–8. [Google Scholar] [CrossRef]
- Yilmaz, B.; Li, H. Gut Microbiota and Iron: The Crucial Actors in Health and Disease. Pharmaceuticals (Basel) 2018, 11, 98. [Google Scholar] [CrossRef]
- Skaar, E.P. The Battle for Iron between Bacterial Pathogens and their Vertebrate Hosts. PLoS Pathog. 2010, 6. [Google Scholar] [CrossRef]
- Kortman, G.A.M.; Boleij, A.; Swinkels, D.W.; Tjalsma, H. Iron Availability Increases the Pathogenic Potential of Salmonella Typhimurium and Other Enteric Pathogens at the Intestinal Epithelial Interface. PLoS ONE 2012, 7, e29968. [Google Scholar] [CrossRef]
- Jaeggi, T.; Kortman, G.A.M.; Moretti, D.; Chassard, C.; Holding, P.; Dostal, A.; Boekhorst, J.; Timmerman, H.M.; Swinkels, D.W.; Tjalsma, H.; et al. Iron Fortification Adversely Affects the Gut Microbiome, Increases Pathogen Abundance and Induces Intestinal Inflammation in Kenyan Infants. Gut 2015, 64, 731–742. [Google Scholar] [CrossRef]
- Sjödin, K.S.; Domellöf, M.; Lagerqvist, C.; Hernell, O.; Lönnerdal, B.; Szymlek-Gay, E.A.; Sjödin, A.; West, C.E.; Lind, T. Administration of Ferrous Sulfate Drops has Significant Effects on the Gut Microbiota of Iron-Sufficient Infants: A Randomised Controlled Study. Gut 2019, 68, 2095–2097. [Google Scholar]
- Ruan, W.; Engevik, M.A.; Spinler, J.K.; Versalovic, J. Healthy Human Gastrointestinal Microbiome: Composition and Function after a Decade of Exploration. Dig. Dis. Sci. 2020, 65, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut Microbiota: Role in Pathogen Colonization, Immune Responses and Inflammatory Disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Gorbach, S.L. Microbiology of the Gastrointestinal Tract. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of Pathogens and Pathobionts by the Gut Microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Illiano, P.; Brambilla, R.; Parolini, C. The Mutual Interplay of Gut Microbiota, Diet and Human Disease. FEBS J. 2020, 287, 833–855. [Google Scholar] [CrossRef]
- Burns, M.B.; Blekhman, R. Integrating Tumor Genomics into Studies of the Microbiome in Colorectal Cancer. Gut Microbes 2018, 10, 547–552. [Google Scholar] [CrossRef]
- Burns, M.B.; Montassier, E.; Abrahante, J.; Priya, S.; Niccum, D.E.; Khoruts, A.; Starr, T.K.; Knights, D.; Blekhman, R. Colorectal Cancer Mutational Profiles Correlate with Defined Microbial Communities in the Tumor Microenvironment. PLoS Genet. 2018, 14, e1007376. [Google Scholar] [CrossRef]
- Whisner, C.M.; Athena Aktipis, C. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef]
- Parsonnet, J. Bacterial Infection as a Cause of Cancer. Environ. Health Perspect. 1995, 103 (Suppl. 8), 263–268. [Google Scholar]
- Wroblewski, L.E.; Peek, R.M., Jr.; Wilson, K.T. Helicobacter Pylori and Gastric Cancer: Factors that Modulate Disease Risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar]
- Maeda, S.; Mentis, A.F. Pathogenesis of Helicobacter Pylori Infection. Helicobacter 2007, 12, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Khatoon, J.; Rai, R.P.; Prasad, K.N. Role of Helicobacter Pylori in Gastric Cancer: Updates. World J. Gastrointest. Oncol. 2016, 8, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Kusters, J.G.; van Vliet, A.H.; Kuipers, E.J. Pathogenesis of Helicobacter Pylori Infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef]
- Teimoorian, F.; Ranaei, M.; Hajian Tilaki, K.; Shokri Shirvani, J.; Vosough, Z. Association of Helicobacter Pylori Infection with Colon Cancer and Adenomatous Polyps. Iran. J. Pathol. 2018, 13, 325–332. [Google Scholar]
- Bauer, B.; Meyer, T.F. The Human Gastric Pathogen Helicobacter Pylori and its Association with Gastric Cancer and Ulcer Disease. Ulcers 2011. [Google Scholar] [CrossRef]
- Wang, T.C.; Fox, J.G. Helicobacter Pylori and Gastric Cancer: Koch’s Postulates Fulfilled? Gastroenterology 1998, 115, 780–783. [Google Scholar] [CrossRef]
- Akin, H.; Tözün, N. Diet, Microbiota, and Colorectal Cancer. J. Clin. Gastroenterol. 2014, 48 (Suppl. 1), 67. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu. Rev. Microbiol. 2016, 70, 395–411. [Google Scholar] [CrossRef]
- Nelson, H.; Chia, N. Gut Microbiome and Colon Cancer: A Plausible Explanation for Dietary Contributions to Cancer. J. Am. Coll. Surg. 2019, 229, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Kato, I. Gut Microbiota, Inflammation and Colorectal Cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A Bacterial Driver-Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Uronis, J.M.; Mühlbauer, M.; Herfarth, H.H.; Rubinas, T.C.; Jones, G.S.; Jobin, C. Modulation of the Intestinal Microbiota Alters Colitis-Associated Colorectal Cancer Susceptibility. PLoS ONE 2009, 4, e6026. [Google Scholar] [CrossRef]
- Xue, X.; Shah, Y.M. Intestinal Iron Homeostasis and Colon Tumorigenesis. Nutrients 2013, 5, 2333–2351. [Google Scholar] [CrossRef] [PubMed]
- Campisciano, G.; de Manzini, N.; Delbue, S.; Cason, C.; Cosola, D.; Basile, G.; Ferrante, P.; Comar, M.; Palmisano, S. The Obesity-Related Gut Bacterial and Viral Dysbiosis can Impact the Risk of Colon Cancer Development. Microorganisms 2020, 8, 431. [Google Scholar] [CrossRef]
- Park, C.H.; Eun, C.S.; Han, D.S. Intestinal Microbiota, Chronic Inflammation, and Colorectal Cancer. Intest. Res. 2018, 16, 338–345. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J. 2011, 6, 320–329. [Google Scholar] [CrossRef]
- Lin, C.; Cai, X.; Zhang, J.; Wang, W.; Sheng, Q.; Hua, H.; Zhou, X. Role of Gut Microbiota in the Development and Treatment of Colorectal Cancer. Digestion 2019, 100, 72–78. [Google Scholar] [CrossRef]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool Microbiome and Metabolome Differences between Colorectal Cancer Patients and Healthy Adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef]
- Lasry, A.; Zinger, A.; Ben-Neriah, Y. Inflammatory Networks Underlying Colorectal Cancer. Nat. Immunol. 2016, 17, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Klampfer, L. Cytokines, Inflammation and Colon Cancer. Curr. Cancer Drug. Targets 2011, 11, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Kang, D.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Distrutti, E. Bile Acid-Activated Receptors, Intestinal Microbiota, and the Treatment of Metabolic Disorders. Trends Mol. Med. 2015, 21, 702–714. [Google Scholar] [CrossRef]
- Rossocha, M.; Schultz-Heienbrok, R.; von Moeller, H.; Coleman, J.P.; Saenger, W. Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl but Not of its Tauryl Product. Biochemistry 2005, 44, 5739–5748. [Google Scholar] [CrossRef]
- Gérard, P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.M.; Marchesi, J.R. Functional and Comparative Metagenomic Analysis of Bile Salt Hydrolase Activity in the Human Gut Microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef]
- Liu, T.; Song, X.; Khan, S.; Li, Y.; Guo, Z.; Li, C.; Wang, S.; Dong, W.; Liu, W.; Wang, B.; et al. The Gut Microbiota at the Intersection of Bile Acids and Intestinal Carcinogenesis: An Old Story, Yet Mesmerizing. Int. J. Cancer 2020, 146, 1780–1790. [Google Scholar] [CrossRef]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Ung, T.T.; Kim, N.H.; Jung, Y.D. Role of Bile Acids in Colon Carcinogenesis. World J. Clin. Cases 2018, 6, 577–588. [Google Scholar] [CrossRef]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorakova, K.; Garewal, H. Bile Acids as Carcinogens in Human Gastrointestinal Cancers. Mutat. Res. 2005, 589, 47–65. [Google Scholar] [CrossRef] [PubMed]
- de Kok, T.M.; van Faassen, A.; Glinghammar, B.; Pachen, D.M.; Eng, M.; Rafter, J.J.; Baeten, C.G.; Engels, L.G.; Kleinjans, J.C. Bile Acid Concentrations, Cytotoxicity, and pH of Fecal Water from Patients with Colorectal Adenomas. Dig. Dis. Sci. 1999, 44, 2218–2225. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J.D.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, Fibre and Cancer Risk in African Americans and Rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef] [PubMed]
- Molinero, N.; Ruiz, L.; Sánchez, B.; Margolles, A.; Delgado, S. Intestinal Bacteria Interplay with Bile and Cholesterol Metabolism: Implications on Host Physiology. Front. Physiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Nagengast, F.M.; Grubben, M.J.; van Munster, I.P. Role of Bile Acids in Colorectal Carcinogenesis. Eur. J. Cancer 1995, 31A, 1067–1070. [Google Scholar] [CrossRef]
- Dahmus, J.D.; Kotler, D.L.; Kastenberg, D.M.; Kistler, C.A. The Gut Microbiome and Colorectal Cancer: A Review of Bacterial Pathogenesis. J. Gastrointest. Oncol. 2018, 9, 769–777. [Google Scholar] [CrossRef]
- Attene-Ramos, M.S.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. Evidence that Hydrogen Sulfide is a Genotoxic Agent. Mol. Cancer Res. 2006, 4, 9–14. [Google Scholar] [CrossRef]
- Chen, Z.; Ai, L.; Wang, J.; Ren, L.; Yu, Y.; Xu, J.; Chen, H.; Yu, J.; Li, M.; Qin, W.; et al. Probiotics Clostridium Butyricum and Bacillus Subtilis Ameliorate Intestinal Tumorigenesis. Future Microbiol. 2015, 10, 1433–1445. [Google Scholar] [CrossRef]
- Shang, F.; Liu, H. Fusobacterium Nucleatum and Colorectal Cancer: A Review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/Β-Catenin Signaling Via its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Kunzmann, A.T.; Proença, M.A.; Jordao, H.W.; Jiraskova, K.; Schneiderova, M.; Levy, M.; Liska, V.; Buchler, T.; Vodickova, L.; Vymetalkova, V.; et al. Fusobacterium Nucleatum Tumor DNA Levels are Associated with Survival in Colorectal Cancer Patients. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, Y.; Ke, Y.; Li, Y. Prognostic Impact of the Fusobacterium Nucleatum Status in Colorectal Cancers. Medicine (Baltimore) 2019, 98, e17221. [Google Scholar] [CrossRef] [PubMed]
- Cuevas-Ramos, G.; Petit, C.R.; Marcq, I.; Boury, M.; Oswald, E.; Nougayrède, J. Escherichia Coli Induces DNA Damage in Vivo and Triggers Genomic Instability in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11537–11542. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, V.; Dotan, I.; Gophna, U. Carriage of Colibactin-Producing Bacteria and Colorectal Cancer Risk. Trends Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Faïs, T.; Delmas, J.; Barnich, N.; Bonnet, R.; Dalmasso, G. Colibactin: More than a New Bacterial Toxin. Toxins (Basel) 2018, 10, 151. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Boleij, A.; Hechenbleikner, E.M.; Goodwin, A.C.; Badani, R.; Stein, E.M.; Lazarev, M.G.; Ellis, B.; Carroll, K.C.; Albesiano, E.; Wick, E.C.; et al. The Bacteroides Fragilis Toxin Gene is Prevalent in the Colon Mucosa of Colorectal Cancer Patients. Clin. Infect. Dis. 2015, 60, 208–215. [Google Scholar] [CrossRef]
- Haghi, F.; Goli, E.; Mirzaei, B.; Zeighami, H. The Association between Fecal Enterotoxigenic B. Fragilis with Colorectal Cancer. BMC Cancer 2019, 19, 879. [Google Scholar] [CrossRef]
- Purcell, R.V.; Pearson, J.; Aitchison, A.; Dixon, L.; Frizelle, F.A.; Keenan, J.I. Colonization with Enterotoxigenic Bacteroides Fragilis is Associated with Early-Stage Colorectal Neoplasia. PLoS ONE 2017, 12, e0171602. [Google Scholar] [CrossRef]
- Wei, Z.; Cao, S.; Liu, S.; Yao, Z.; Sun, T.; Li, Y.; Li, J.; Zhang, D.; Zhou, Y. Could Gut Microbiota Serve as Prognostic Biomarker Associated with Colorectal Cancer Patients’ Survival? A Pilot Study on Relevant Mechanism. Oncotarget 2016, 7, 46158–46172. [Google Scholar] [CrossRef]
- Bahmani, S.; Azarpira, N.; Moazamian, E. Anti-Colon Cancer Activity of Bifidobacterium Metabolites on Colon Cancer Cell Line SW742. Turk. J. Gastroenterol. 2019, 30, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Kim, H.M.; Yang, K.M.; Kim, S.; Kim, S.; An, M.J.; Park, J.J.; Lee, S.K.; Kim, T.I.; Kim, W.H.; et al. Bifidobacterium Lactis Inhibits NF-kappaB in Intestinal Epithelial Cells and Prevents Acute Colitis and Colitis-Associated Colon Cancer in Mice. Inflamm. Bowel Dis. 2010, 16, 1514–1525. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Rivenson, A.; Tomita, M.; Shimamura, S.; Ishibashi, N.; Reddy, B.S. Bifidobacterium Longum, a Lactic Acid-Producing Intestinal Bacterium Inhibits Colon Cancer and Modulates the Intermediate Biomarkers of Colon Carcinogenesis. Carcinogenesis 1997, 18, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Kot, E.; Bezkorovainy, A. Binding of Ferric Iron to the Cell Walls and Membranes of Bifidobacterium Thermophilum: Effect of Free Radicals. J. Agric. Food Chem. 1999, 47, 4606–4610. [Google Scholar] [CrossRef] [PubMed]
- Casanova, M.R.; Azevedo-Silva, J.; Rodrigues, L.R.; Preto, A. Colorectal Cancer Cells Increase the Production of Short Chain Fatty Acids by Propionibacterium Freudenreichii Impacting on Cancer Cells Survival. Front. Nutr. 2018, 5. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Morin, P.J.; Maouyo, D.; Sears, C.L. Bacteroides Fragilis Enterotoxin Induces C-Myc Expression and Cellular Proliferation. Gastroenterology 2003, 124, 392–400. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A Human Colonic Commensal Promotes Colon Tumorigenesis Via Activation of T Helper Type 17 T Cell Responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut Microbiome Development Along the Colorectal Adenoma-Carcinoma Sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Yang, Y.; Jobin, C. Novel Insights into Microbiome in Colitis and Colorectal Cancer. Curr. Opin. Gastroenterol. 2017, 33, 422–427. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Ahmadi Amoli, H.; Ghasemian-Safaei, H. Role of Gut Microbiota in the Pathogenesis of Colorectal Cancer; a Review Article. Gastroenterol. Hepatol. Bed Bench 2018, 11, 101–109. [Google Scholar]
- Ewaschuk, J.B.; Walker, J.W.; Diaz, H.; Madsen, K.L. Bioproduction of Conjugated Linoleic Acid by Probiotic Bacteria Occurs in Vitro and in Vivo in Mice. J. Nutr. 2006, 136, 1483–1487. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Abu Bakar, F. The Association of Streptococcus Bovis/Gallolyticus with Colorectal Tumors: The Nature and the Underlying Mechanisms of its Etiological Role. J. Exp. Clin. Cancer Res. 2011, 30, 11. [Google Scholar] [CrossRef] [PubMed]
- Kannen, V.; Parry, L.; Martin, F.L. Phages Enter the Fight against Colorectal Cancer. Trends Cancer 2019, 5, 577–579. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.; Martel, F. Butyrate and Colorectal Cancer: The Role of Butyrate Transport. Curr. Drug Metab. 2013, 14, 994–1008. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Louis, P.; Flint, H.J. Lactate-Utilizing Bacteria, Isolated from Human Feces, that Produce Butyrate as a Major Fermentation Product. Appl. Environ. Microbiol. 2004, 70, 5810–5817. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus Anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017, 152, 1419–1433.e5. [Google Scholar] [CrossRef]
- Balamurugan, R.; Rajendiran, E.; George, S.; Samuel, G.V.; Ramakrishna, B.S. Real-Time Polymerase Chain Reaction Quantification of Specific Butyrate-Producing Bacteria, Desulfovibrio and Enterococcus Faecalis in the Feces of Patients with Colorectal Cancer. J. Gastroenterol. Hepatol. 2008, 23, 1298–1303. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.; Blugeon, S.; Bridonneau, C.; Furet, J.; Corthier, G.; et al. Faecalibacterium Prausnitzii is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, K.; Ji, G. Faecalibacterium Prausnitzii Prevents Tumorigenesis in a Model of Colitis-Associated Colorectal Cancer. Gastroenterology 2017, 152, S354–S355. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium Nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, G.; Cougnoux, A.; Delmas, J.; Darfeuille-Michaud, A.; Bonnet, R. The Bacterial Genotoxin Colibactin Promotes Colon Tumor Growth by Modifying the Tumor Microenvironment. Gut Microbes 2014, 5, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Huber, A.R.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational Signature in Colorectal Cancer Caused by Genotoxic Pks + E. Coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Bosland, M.; Xia, Y.; Zhang, Y.; Kato, I.; Sun, J. Presence of Salmonella AvrA in Colorectal Tumor and its Precursor Lesions in Mouse Intestine and Human Specimens. Oncotarget 2017, 8, 55104–55115. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Ye, Z.; Liu, X.; Zhao, Y.; Xia, Y.; Steiner, A.; Petrof, E.O.; Claud, E.C.; Sun, J. Salmonella Typhimurium Infection Increases p53 Acetylation in Intestinal Epithelial Cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G784–G794. [Google Scholar] [CrossRef]
- Mira-Pascual, L.; Cabrera-Rubio, R.; Ocon, S.; Costales, P.; Parra, A.; Suarez, A.; Moris, F.; Rodrigo, L.; Mira, A.; Collado, M.C. Microbial Mucosal Colonic Shifts Associated with the Development of Colorectal Cancer Reveal the Presence of Different Bacterial and Archaeal Biomarkers. J. Gastroenterol. 2015, 50, 167–179. [Google Scholar] [CrossRef]
- Ng, O. Iron, Microbiota and Colorectal Cancer. Wien. Med. Wochenschr. 2016, 166, 431–436. [Google Scholar] [CrossRef]
- Orrhage, K.M.; Annas, A.; Nord, C.E.; Brittebo, E.B.; Rafter, J.J. Effects of Lactic Acid Bacteria on the Uptake and Distribution of the Food Mutagen Trp-P-2 in Mice. Scand. J. Gastroenterol. 2002, 37, 215–221. [Google Scholar] [CrossRef]
- Wu, X.; Wu, Y.; He, L.; Wu, L.; Wang, X.; Liu, Z. Effects of the Intestinal Microbial Metabolite Butyrate on the Development of Colorectal Cancer. J. Cancer 2018, 9, 2510–2517. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for their Stimulation in the Human Gut. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Bach Knudsen, K.E.; Lærke, H.N.; Hedemann, M.S.; Nielsen, T.S.; Ingerslev, A.K.; Gundelund Nielsen, D.S.; Theil, P.K.; Purup, S.; Hald, S.; Schioldan, A.G.; et al. Impact of Diet-Modulated Butyrate Production on Intestinal Barrier Function and Inflammation. Nutrients 2018, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Bennett, N.; Ahmed, B.; Whelan, J.; Donohoe, D.R. Butyrate Decreases its Own Oxidation in Colorectal Cancer Cells through Inhibition of Histone Deacetylases. Oncotarget 2018, 9, 27280–27292. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vitetta, L. Inflammation-Modulating Effect of Butyrate in the Prevention of Colon Cancer by Dietary Fiber. Clin. Colorectal Cancer 2018, 17, e541–e544. [Google Scholar] [CrossRef] [PubMed]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and Colorectal Cancer: Roles in Carcinogenesis and Clinical Potential. Mol. Aspects Med. 2019, 69, 93–106. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A Gnotobiotic Mouse Model Demonstrates that Dietary Fiber Protects Against Colorectal Tumorigenesis in a Microbiota- and Butyrate-Dependent Manner. Cancer Discov. 2014, 4, 1387–1397. [Google Scholar] [CrossRef]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria Across Populations and Universal Bacterial Markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef]
- Dong, T.S.; Gupta, A. Influence of Early Life, Diet, and the Environment on the Microbiome. Clin. Gastroenterol. Hepatol. 2019, 17, 231–242. [Google Scholar] [CrossRef]
- Song, M.; Chan, A.T. Environmental Factors, Gut Microbiota, and Colorectal Cancer Prevention. Clin. Gastroenterol. Hepatol. 2019, 17, 275–289. [Google Scholar] [CrossRef]
- Huang, C.; Shi, G. Smoking and Microbiome in Oral, Airway, Gut and some Systemic Diseases. J. Transl. Med. 2019, 17, 225. [Google Scholar] [CrossRef]
- Li, R.; Grimm, S.A.; Mav, D.; Gu, H.; Djukovic, D.; Shah, R.; Merrick, B.A.; Raftery, D.; Wade, P.A. Transcriptome and DNA Methylome Analysis in a Mouse Model of Diet-Induced Obesity Predicts Increased Risk of Colorectal Cancer. Cell Rep. 2018, 22, 624–637. [Google Scholar] [CrossRef]
- Weinberg, E.D. Iron Availability and Infection. Biochim. Biophys. Acta 2009, 1790, 600–605. [Google Scholar]
- Sornjai, W.; Nguyen Van Long, F.; Pion, N.; Pasquer, A.; Saurin, J.; Marcel, V.; Diaz, J.J.; Mertani, H.C.; Smith, D.R. Iron and Hepcidin Mediate Human Colorectal Cancer Cell Growth. Chem. Biol. Interact. 2020, 319, 109021. [Google Scholar] [CrossRef] [PubMed]
- Hooda, J.; Shah, A.; Zhang, L. Heme, an Essential Nutrient from Dietary Proteins, Critically Impacts Diverse Physiological and Pathological Processes. Nutrients 2014, 6, 1080–1102. [Google Scholar] [CrossRef] [PubMed]
- Amagloh, F.K.; Atuna, R.A.; McBride, R.; Carey, E.E.; Christides, T. Nutrient and Total Polyphenol Contents of Dark Green Leafy Vegetables, and Estimation of their Iron Bioaccessibility using the in Vitro Digestion/Caco-2 Cell Model. Foods 2017, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Parmanand, B.A.; Kellingray, L.; Le Gall, G.; Basit, A.W.; Fairweather-Tait, S.; Narbad, A. A Decrease in Iron Availability to Human Gut Microbiome Reduces the Growth of Potentially Pathogenic Gut Bacteria; an in Vitro Colonic Fermentation Study. J. Nutr. Biochem. 2019, 67, 20–27. [Google Scholar] [CrossRef]
- Zimmermann, M.B.; Chassard, C.; Rohner, F.; N’goran, E.K.; Nindjin, C.; Dostal, A.; Utzinger, J.; Ghattas, H.; Lacroix, C.; Hurrell, R.F. The Effects of Iron Fortification on the Gut Microbiota in African Children: A Randomized Controlled Trial in Cote D’Ivoire. Am. J. Clin. Nutr. 2010, 92, 1406–1415. [Google Scholar] [CrossRef]
- Rocha, E.R.; Krykunivsky, A.S. Anaerobic Utilization of Fe(III)-xenosiderophores among Bacteroides Species and the Distinct Assimilation of Fe(III)-ferrichrome by Bacteroides Fragilis within the Genus. Microbiologyopen 2017, 6. [Google Scholar] [CrossRef]
- Zafar, H.; Saier, M.H., Jr. Comparative Genomics of Transport Proteins in Seven Bacteroides Species. PLoS ONE 2018, 13, e0208151. [Google Scholar] [CrossRef]
- Huang, Y.; Chassard, C.; Hausmann, M.; von Itzstein, M.; Hennet, T. Sialic Acid Catabolism Drives Intestinal Inflammation and Microbial Dysbiosis in Mice. Nat. Commun. 2015, 6, 8141. [Google Scholar] [CrossRef]
- Vivarelli, S.; Salemi, R.; Candido, S.; Falzone, L.; Santagati, M.; Stefani, S.; Torino, F.; Banna, G.L.; Tonini, G.; Libra, M. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers (Basel) 2019, 11, 38. [Google Scholar] [CrossRef]
- Lee, T.; Clavel, T.; Smirnov, K.; Schmidt, A.; Lagkouvardos, I.; Walker, A.; Lucio, M.; Michalke, B.; Schmitt-Kopplin, P.; Fedorak, R.; et al. Oral Versus Intravenous Iron Replacement Therapy Distinctly Alters the Gut Microbiota and Metabolome in Patients with IBD. Gut 2017, 66, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Zarei, O.; Arabestan, M.R.; Majlesi, A.; Mohammadi, Y.; Alikhani, M.Y. Determination of Virulence Determinants of Escherichia Coli Strains Isolated from Patients with Colorectal Cancer Compared to the Healthy Subjects. Gastroenterol. Hepatol. Bed Bench 2019, 12, 52–59. [Google Scholar] [PubMed]
- Long, S.; Yang, Y.; Shen, C.; Wang, Y.; Deng, A.; Qin, Q.; Qiao, L. Metaproteomics Characterizes Human Gut Microbiome Function in Colorectal Cancer. NPJ Biofilms Microbiomes 2020, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ellermann, M.; Gharaibeh, R.Z.; Maharshak, N.; Peréz-Chanona, E.; Jobin, C.; Carroll, I.M.; Arthur, J.C.; Plevy, S.E.; Fodor, A.A.; Brouwer, C.R.; et al. Dietary Iron Variably Modulates Assembly of the Intestinal Microbiota in Colitis-Resistant and Colitis-Susceptible Mice. Gut Microbes 2020, 11, 32–50. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.J.; Harlaar, J.J.; Jeekel, J.; Schipperus, M.; Zwaginga, J.J. Iron Therapy as Treatment of Anemia: A Potentially Detrimental and Hazardous Strategy in Colorectal Cancer Patients. Med. Hypotheses 2018, 110, 110–113. [Google Scholar] [CrossRef]
- Fowler, A.J.; Ahmad, T.; Phull, M.K.; Allard, S.; Gillies, M.A.; Pearse, R.M. Meta-Analysis of the Association between Preoperative Anaemia and Mortality After Surgery. Br. J. Surg. 2015, 102, 1314–1324. [Google Scholar] [CrossRef]
- Leichtle, S.W.; Mouawad, N.J.; Lampman, R.; Singal, B.; Cleary, R.K. Does Preoperative Anemia Adversely Affect Colon and Rectal Surgery Outcomes? J. Am. Coll. Surg. 2011, 212, 187–194. [Google Scholar] [CrossRef]
- Borstlap, W.A.A.; Buskens, C.J.; Tytgat, K.M.A.J.; Tuynman, J.B.; Consten, E.C.J.; Tolboom, R.C.; Heuff, G.; van Geloven, N.; van Wagensveld, B.A.; Wientjes, C.A.C.A.; et al. Multicentre Randomized Controlled Trial Comparing Ferric(III)Carboxymaltose Infusion with Oral Iron Supplementation in the Treatment of Preoperative Anaemia in Colorectal Cancer Patients. BMC Surg. 2015, 15, 78. [Google Scholar]
- Calleja, J.; Calleja, J.; Delgado, S.; Delgado, S.; del Val, A.; del Val, A.; Hervás, A.; Hervás, A.; Larraona, J.; Larraona, J.; et al. Ferric Carboxymaltose Reduces Transfusions and Hospital Stay in Patients with Colon Cancer and Anemia. Int. J. Colorectal Dis. 2016, 31, 543–551. [Google Scholar] [CrossRef]
- Constante, M.; Fragoso, G.; Calvé, A.; Samba-Mondonga, M.; Santos, M.M. Dietary Heme Induces Gut Dysbiosis, Aggravates Colitis, and Potentiates the Development of Adenomas in Mice. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Hnatyszyn, A.; Hryhorowicz, S.; Kaczmarek-Ryś, M.; Lis, E.; Słomski, R.; Scott, R.J.; Pławski, A. Colorectal Carcinoma in the Course of Inflammatory Bowel Diseases. Hered. Cancer Clin. Pract. 2019, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Tulewicz-Marti, E.; Moniuszko, A.; Rydzewska, G. Management of Anemia in Inflammatory Bowel Disease: A Challenge in Everyday Clinical Practice. Prz. Gastroenterol. 2017, 12, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium Prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the use of F. Prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium Prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef]
- Ze, X.; Duncan, S.H.; Louis, P.; Flint, H.J. Ruminococcus Bromii is a Keystone Species for the Degradation of Resistant Starch in the Human Colon. ISME J. 2012, 6, 1535–1543. [Google Scholar] [CrossRef]
- Golonka, R.M.; Xiao, X.; Abokor, A.A.; Joe, B.; Vijay-Kumar, M. Altered Nutrient Status Reprograms Host Inflammation and Metabolic Health Via Gut Microbiota. J. Nutr. Biochem. 2020, 80, 108360. [Google Scholar] [CrossRef]
- Danese, S.; Mantovani, A. Inflammatory Bowel Disease and Intestinal Cancer: A Paradigm of the Yin-Yang Interplay between Inflammation and Cancer. Oncogene 2010, 29, 3313–3323. [Google Scholar] [CrossRef]
- Ogino, S.; Nowak, J.A.; Hamada, T.; Milner, D.A.; Nishihara, R. Insights into Pathogenic Interactions among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu. Rev. Pathol. 2019, 14, 83–103. [Google Scholar] [CrossRef]
- Hamada, T.; Nowak, J.A.; Milner, D.A.; Song, M.; Ogino, S. Integration of Microbiology, Molecular Pathology, and Epidemiology: A New Paradigm to Explore the Pathogenesis of Microbiome-Driven Neoplasms. J. Pathol. 2019, 247, 615–628. [Google Scholar] [CrossRef]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y.; et al. Dietary Patterns and Risk of Colorectal Cancer Subtypes Classified by Fusobacterium Nucleatum in Tumor Tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef]
- Liu, L.; Tabung, F.K.; Zhang, X.; Nowak, J.A.; Qian, Z.R.; Hamada, T.; Nevo, D.; Bullman, S.; Mima, K.; Kosumi, K.; et al. Diets that Promote Colon Inflammation Associate with Risk of Colorectal Carcinomas that Contain Fusobacterium Nucleatum. Clin. Gastroenterol. Hepatol. 2018, 16, 1622–1631.e3. [Google Scholar]
- Huang, P.; Liu, Y. A Reasonable Diet Promotes Balance of Intestinal Microbiota: Prevention of Precolorectal Cancer. BioMed Res. Int. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kellingray, L.; Tapp, H.S.; Saha, S.; Doleman, J.F.; Narbad, A.; Mithen, R.F. Consumption of a Diet Rich in Brassica Vegetables is Associated with a Reduced Abundance of Sulphate-Reducing Bacteria: A Randomised Crossover Study. Mol. Nutr. Food Res. 2017, 61, 1600992. [Google Scholar] [CrossRef] [PubMed]
- Eslami, M.; Yousefi, B.; Kokhaei, P.; Hemati, M.; Nejad, Z.R.; Arabkari, V.; Namdar, A. Importance of Probiotics in the Prevention and Treatment of Colorectal Cancer. J. Cell. Physiol. 2019, 234, 17127–17143. [Google Scholar] [CrossRef] [PubMed]
- Zaharuddin, L.; Mokhtar, N.; Muhammad Nawawi, K.N.; Raja Ali, R.A. A Randomized Double-Blind Placebo-Controlled Trial of Probiotics in Post-Surgical Colorectal Cancer. BMC Gastroenterol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
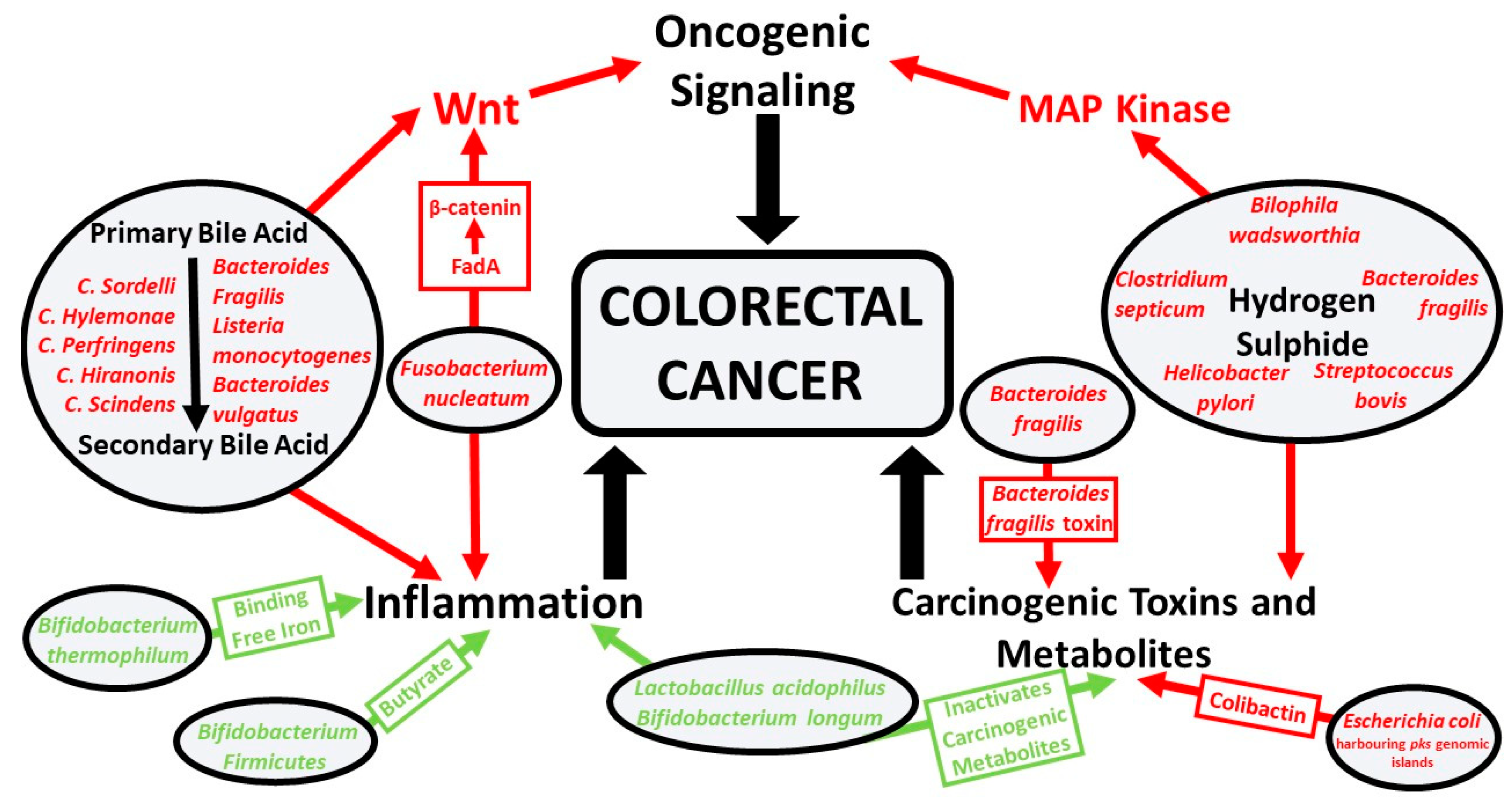
| Phylum | Class | Order | Family | Genus | Species | Symbiont or Pathobiont | Potential Mechanism |
|---|---|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | Bifidobacterium bifidum | Symbiont | Produces metabolites that can inhibit colorectal cancer cell growth [72]. |
| Bifidobacterium lactis | Symbiont | Inhibits NF-κB signaling, limiting colitis-associated colorectal cancer [73]. | |||||
| Bifidobacterium longum | Symbiont | Lactic acid-producing bacteria that inhibits colorectal tumor cell proliferation through modulation of MAP kinase oncogenic pathway [74]. | |||||
| Bifidobacterium thermophilum | Symbiont | Binds free iron, reducing iron for pathogenic bacteria and reactive oxygen species production [75]. | |||||
| Propionibacteriales | Propionibacteriaceae | Propionibacterium | Propionibacterium freudenreichii | Symbiont | Exerts a protective effect against colorectal cancer through the production of short-chain fatty acids, acetate, and propionate [76]. | ||
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Enterotoxigenic Bacteroides fragilis | Pathobiont | Degrades E-cadherin and activates beta-catenin signaling, upregulating c-Myc expression and contributing to colonic cellular proliferation [77]. Secretes B. fragilis toxin that can induce STAT3 signaling [78]. |
| Bacteroides vulgatus | Pathobiont | Involved in the hydrolysis of primary bile acids to secondary bile acids, which is associated with colitis and oncogenic signaling [49,79]. Correlated with systemic inflammation and colorectal cancer tumor stage [79]. | |||||
| Rikenellaceae | Alistipes | Alistipes finegoldii | Pathobiont | Linked to colitis-associated colorectal cancer; in vivo studies have shown to be through activation of IL-6/STAT3 signaling [80]. | |||
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | Enterococcus faecalis | Pathobiont | Can produce reactive oxygen and nitrogen species that can contribute to DNA damage and colonic inflammation [81]. |
| Lactobacillaceae | Lactobacillus | Lactobacillus acidophilus | Symbiont | Can produce conjugated linoleic acids from linoleic acid. Fatty acids produced by these species act on colonocytes, possessing antiproliferative and proapoptotic mechanisms [82]. | |||
| Lactobacillus casei | |||||||
| Lactobacillus delbrueckii | |||||||
| Lactobacillus plantarum | |||||||
| Streptococcaceae | Streptococcus | Streptococcus bovis/gallolyticus | Pathobiont | Can induce production of cytokines that lead to free radical production, colonic inflammation, and increased angiogenesis. Contributes to pro-proliferative signaling via MAP kinase and COX-2/prostaglandin induced cellular proliferation and inhibited apoptosis [83]. | |||
| Bacillales | Bacillaceae | Bacillus | Bacillus subtilis | Symbiont | Probiotic bacteria that can inhibit the proliferation of colorectal cancer cells, induces cell cycle arrest, and promotes apoptosis. Shown to reduce inflammation and aids in immune homeostasis [59]. | ||
| Clostridia | Clostridiales | Clostridiaceae | Clostridium | Clostridium butyricum | Symbiont | Produces butyrate, which possesses anticancer properties, inducing cell differentiation and apoptosis, as well as inhibiting cellular proliferation [84,85]. | |
| Lachnospiraceae | Eubacterium | Eubacterium rectale | Symbiont | Anti-inflammatory butyrate-producing bacteria [86]. | |||
| Roseburia | Roseburia intestinalis | ||||||
| Peptostreptococcaceae | Peptostreptococcus | Peptostreptococcus anaerobius | Pathobiont | Has been shown to interact with Toll-like receptors 2 and 4 on colonic cells. Elevating levels of reactive oxygen species, promoting cell proliferation and increasing colonic dysplasia [87]. | |||
| Ruminococcaceae | Faecalibacterium | Faecalibacterium prausnitzii | Symbiont | Butyrate-producing bacteria, shown to be anti-inflammatory and able to inhibit colorectal tumorigenesis [88,89,90]. | |||
| Fusobacteria | Fusobacteriia | Fusobacteriales | Fusobacteriaceae | Fusobacterium | Fusobacterium nucleatum | Pathobiont | FadA medicated activation of beta-catenin can contribute to inflammatory and oncogenic signaling [61]. Fap2 protein present on F. nucleatum binds to TIGIT on the antitumor immune cells, natural killer cells, and T-cells, which can limit tumor immunosurveillance [91]. |
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Bilophila | Bilophila wadsworthia | Pathobiont | Proinflammatory sulfate-reducing bacteria capable of producing genotoxic hydrogen sulfide [92]. |
| Gammaproteobacteria | Enterobacterales | Enterobacteriaceae | Escherichia | Escherichia coli (harboring pks pathogenicity islands) | Pathobiont | Produces the bacterial genotoxin colibactin that promotes the growth of colonic tumor cells. Colibactin induces DNA interstrand crosslinks and double-strand breaks [93,94]. | |
| Salmonella | Salmonella typhimurium | Pathobiont | Produces the bacterial protein AvrA, which is associated with inflammation and colorectal cancer, through modulation of the p53 pathway [95,96]. | ||||
| Verrucomicrobia | Verrucomicrobiae | Verrucomicrobiales | Akkermansiaceae | Akkermansia | Akkermansia muciniphila | Pathobiont | A mucin-degrading bacterium that contributes to colonic inflammation [97]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phipps, O.; Al-Hassi, H.O.; Quraishi, M.N.; Kumar, A.; Brookes, M.J. Influence of Iron on the Gut Microbiota in Colorectal Cancer. Nutrients 2020, 12, 2512. https://doi.org/10.3390/nu12092512
Phipps O, Al-Hassi HO, Quraishi MN, Kumar A, Brookes MJ. Influence of Iron on the Gut Microbiota in Colorectal Cancer. Nutrients. 2020; 12(9):2512. https://doi.org/10.3390/nu12092512
Chicago/Turabian StylePhipps, Oliver, Hafid O. Al-Hassi, Mohammed N. Quraishi, Aditi Kumar, and Matthew J. Brookes. 2020. "Influence of Iron on the Gut Microbiota in Colorectal Cancer" Nutrients 12, no. 9: 2512. https://doi.org/10.3390/nu12092512
APA StylePhipps, O., Al-Hassi, H. O., Quraishi, M. N., Kumar, A., & Brookes, M. J. (2020). Influence of Iron on the Gut Microbiota in Colorectal Cancer. Nutrients, 12(9), 2512. https://doi.org/10.3390/nu12092512





