Dietary Supplementation of Foxtail Millet Ameliorates Colitis-Associated Colorectal Cancer in Mice via Activation of Gut Receptors and Suppression of the STAT3 Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Animals
2.3. Induction of Colorectal Cancer and Experimental Design
2.4. Analysis of the Principal Nutritional Composition in Cereals
2.5. Evaluation of Colonic Tumors
2.6. Measurement of Cytokines
2.7. Histopathological Evaluation
2.8. Determination of the SCFAs and Tryptophan Metabolites in Feces
2.9. Western Blot Analyses
2.10. Quantitative RT-qPCR
2.11. High-Throughput Sequencing and Bioinformatic Analysis
2.12. Statistical Analysis
3. Results
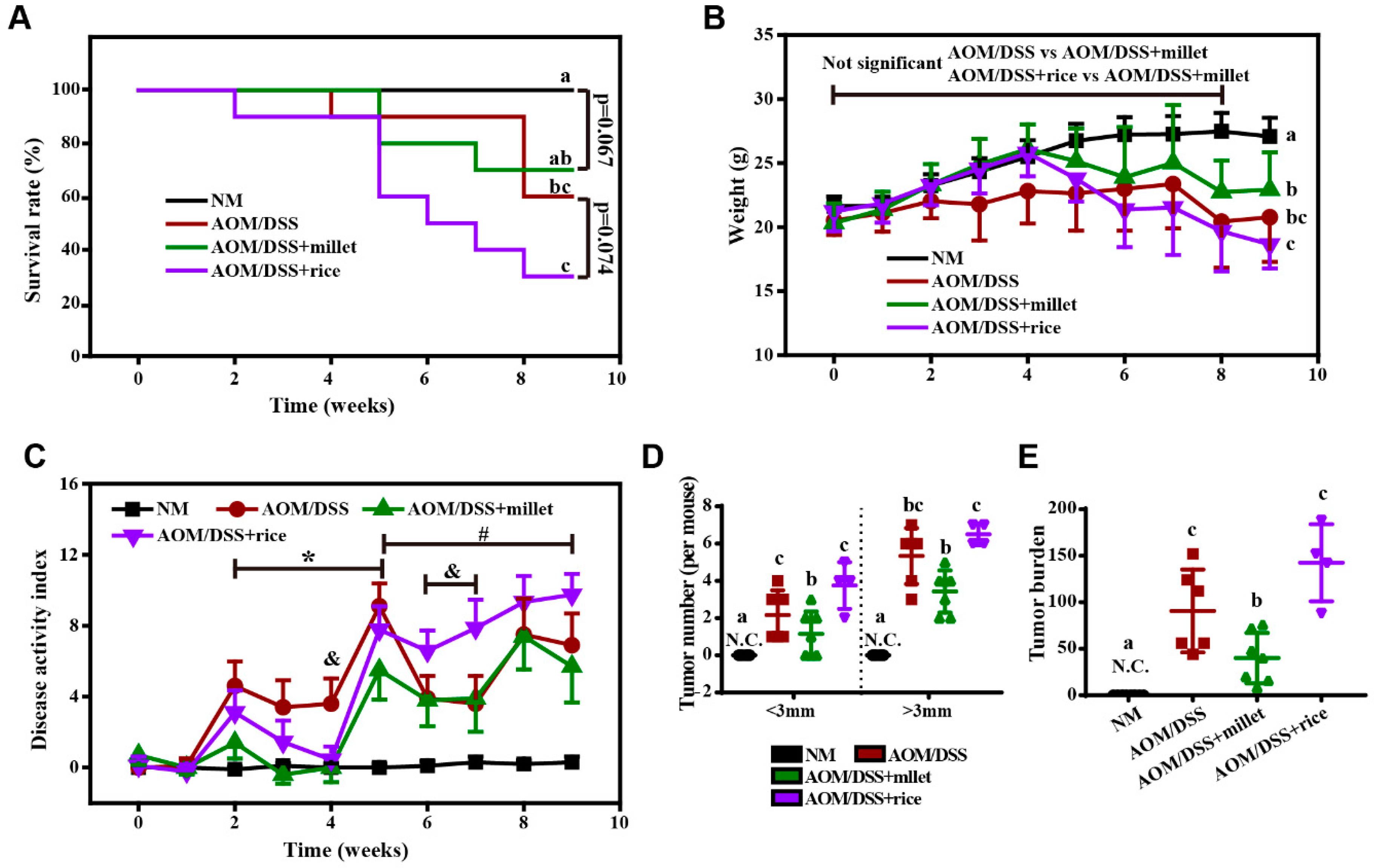
3.1. Foxtail Millet Suppressed the Development of Colorectal Cancer in AOM/DSS-Treated Mice
3.2. Principal Composition of Cereals
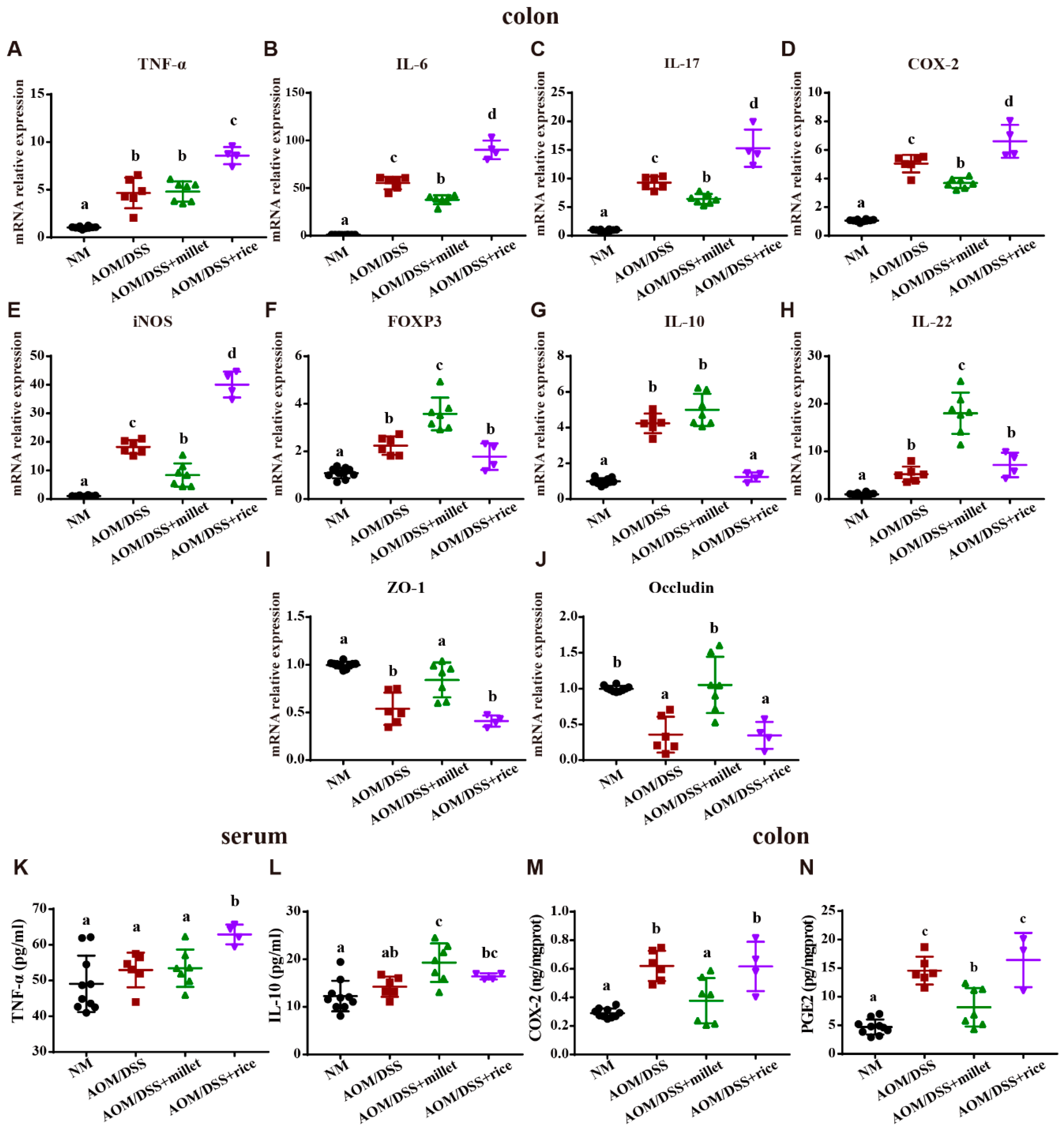
3.3. Foxtail Millet Attenuated Inflammation and Histological Pathology in the AOM/DSS-Treated Mice
3.4. Millet Regulated the Expression of Cytokines and Genes Involved in Colitis-Related Signaling Pathways
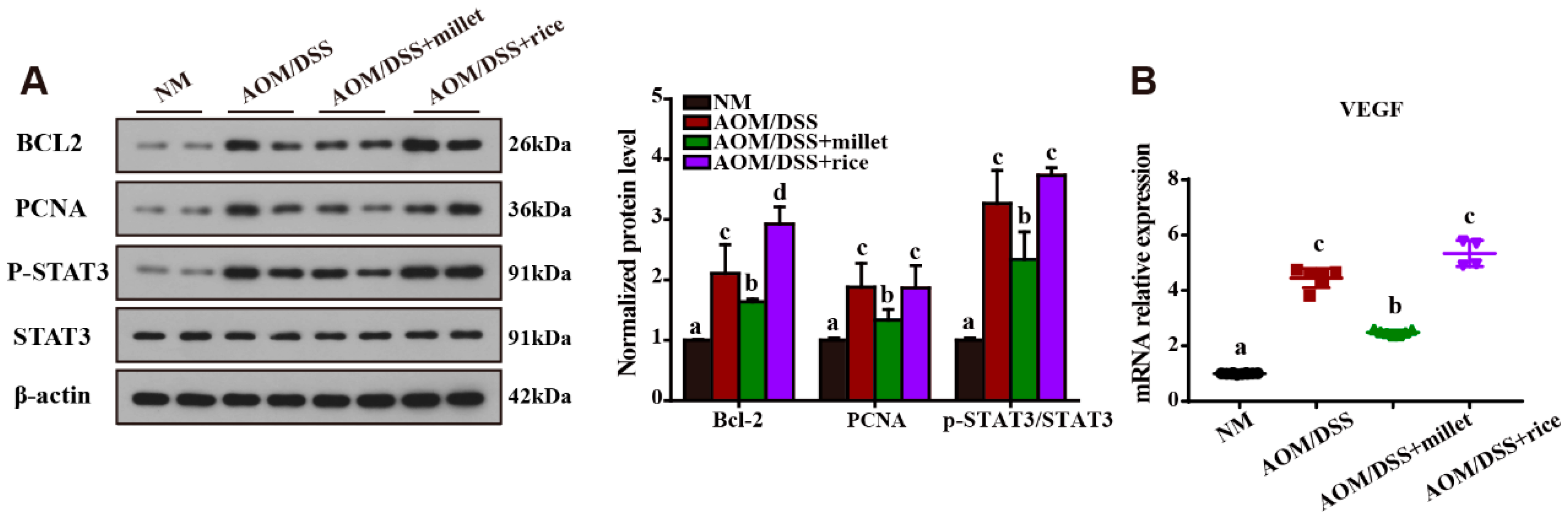
3.5. Inhibition of the Development of Colorectal Cancer by Foxtail Millet
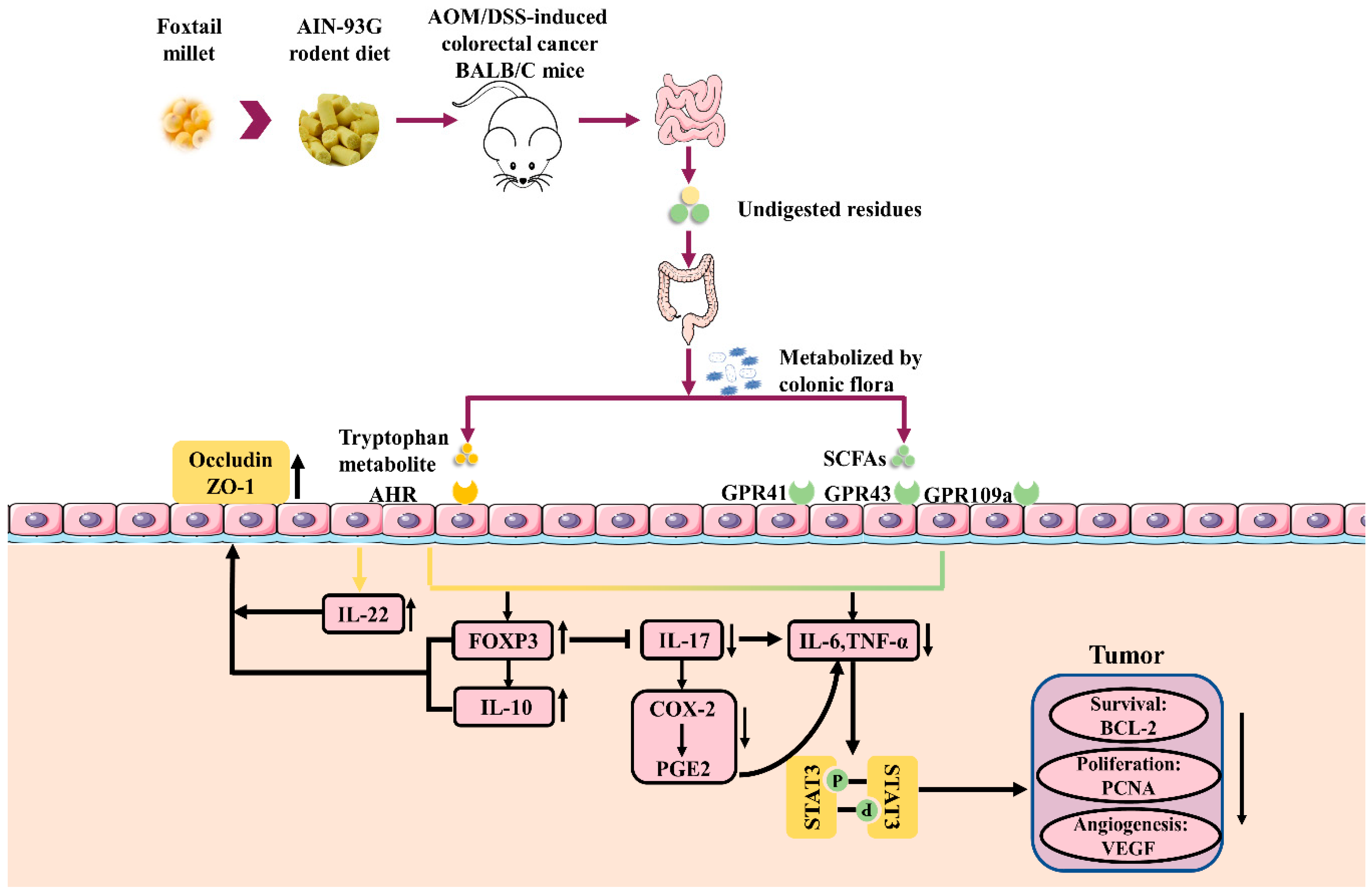
3.6. Microbial Metabolites of Foxtail Millet Activated AHR and GPCRs
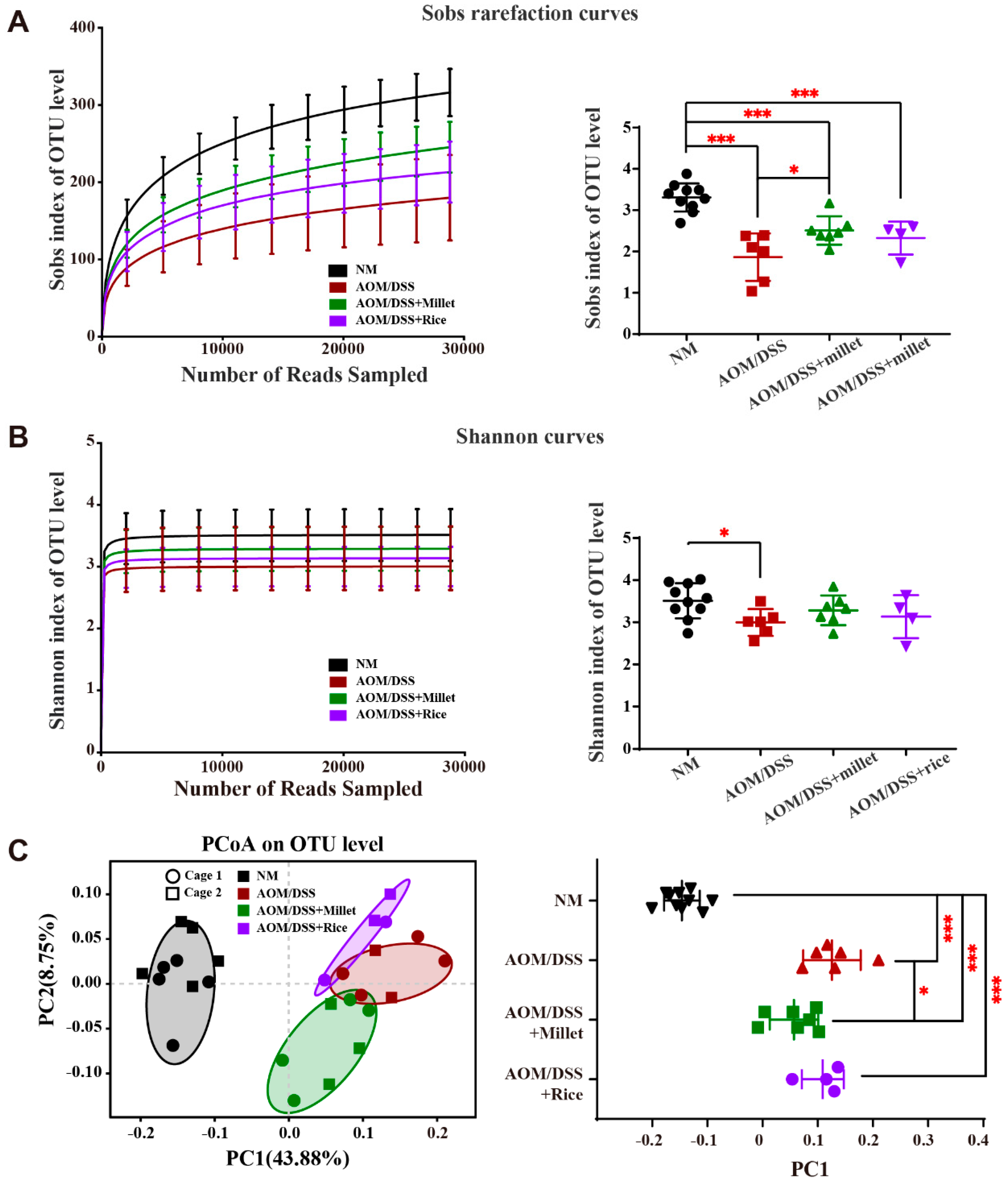
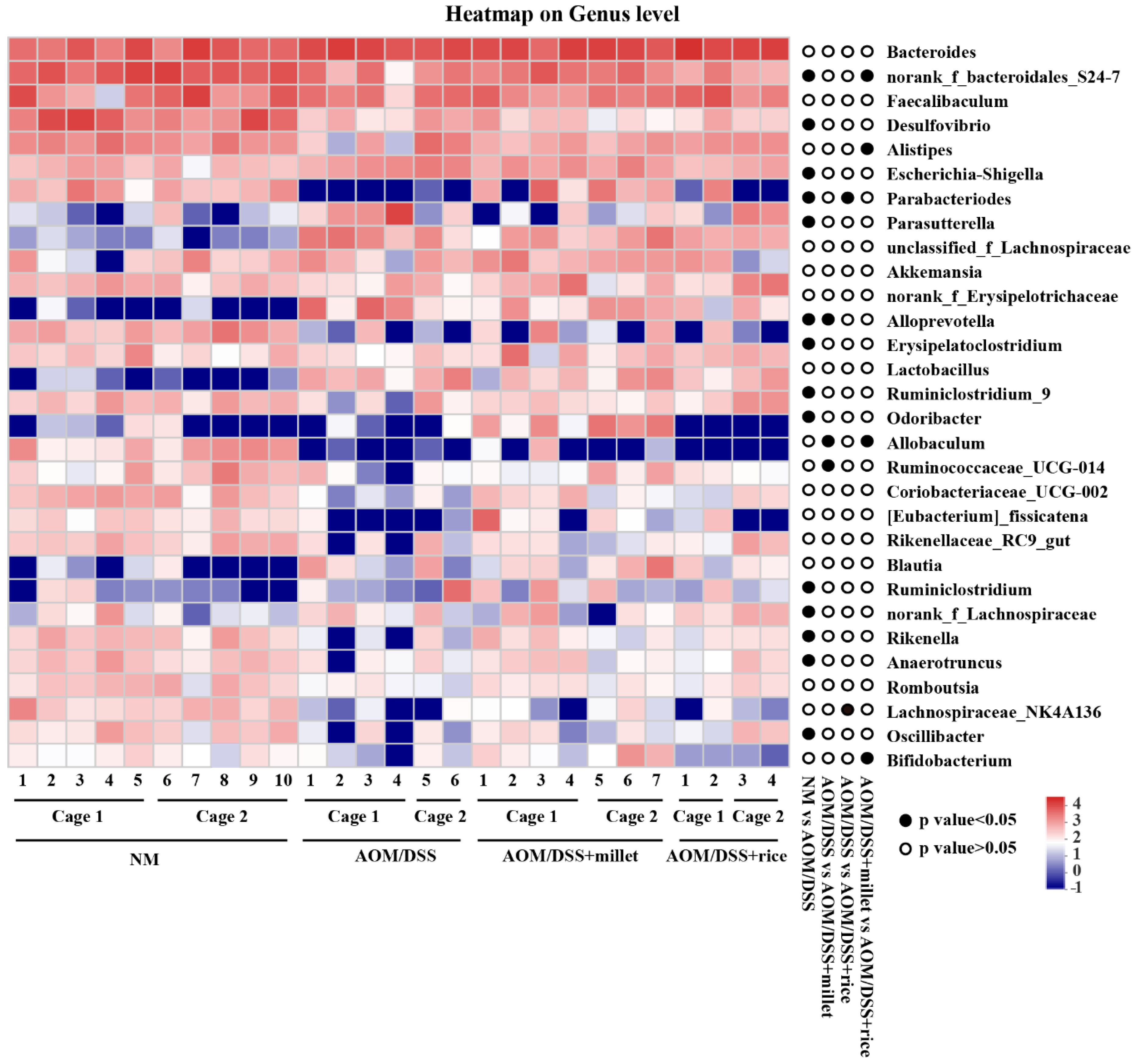
3.7. Effects of Foxtail Millet on the Regulation of Gut Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.S.; Kumar, V.; Al-Abbasi, F.A.; Kamal, M.A.; Anwar, F. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 2020, 64, 51–60. [Google Scholar] [CrossRef]
- Eaden, J.A.; Abrams, K.R.; Mayberry, J.F. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut 2001, 48, 526–535. [Google Scholar] [CrossRef]
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef]
- Lamas, B.; Natividad, J.M.; Sokol, H. Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 2018, 11, 1024–1038. [Google Scholar] [CrossRef]
- Rothhammer, V.; Quintana, F.J. The aryl hydrocarbon receptor: An environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 2019, 19, 184–197. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef]
- Sun, M.; Wu, W.; Liu, Z.; Cong, Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J. Gastroenterol. 2017, 52, 1–8. [Google Scholar] [CrossRef]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef] [PubMed]
- De Simone, V.; Franze, E.; Ronchetti, G.; Colantoni, A.; Fantini, M.C.; Di Fusco, D.; Sica, G.S.; Sileri, P.; MacDonald, T.T.; Pallone, F.; et al. Th17-type cytokines, IL-6 and TNF-alpha synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene 2015, 34, 3493–3503. [Google Scholar] [CrossRef] [PubMed]
- Neufert, C.; Becker, C.; Neurath, M.F. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007, 2, 1998–2004. [Google Scholar] [CrossRef] [PubMed]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501–518. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Garrett, W.S.; Chan, A.T. Nutrients, foods, and colorectal cancer prevention. Gastroenterology 2015, 148, 1244–1260.e16. [Google Scholar] [CrossRef]
- Aune, D.; Chan, D.S.; Lau, R.; Vieira, R.; Greenwood, D.C.; Kampman, E.; Norat, T. Dietary fibre, whole grains, and risk of colorectal cancer: Systematic review and dose-response meta-analysis of prospective studies. BMJ 2011, 343, d6617. [Google Scholar] [CrossRef]
- Li, S.-C.; Chou, T.-C.; Shih, C.-K. Effects of brown rice, rice bran, and polished rice on colon carcinogenesis in rats. Food Res. Int. 2011, 44, 209–216. [Google Scholar] [CrossRef]
- Lahouar, L.; Pochart, P.; Salem, H.B.; El Felah, M.; Mokni, M.; Magne, F.; Mangin, I.; Suau, A.; Pereira, E.; Hammami, M.; et al. Effect of dietary fibre of barley variety ‘Rihane’ on azoxymethane-induced aberrant crypt foci development and on colonic microbiota diversity in rats. Br. J. Nutr. 2012, 108, 2034–2042. [Google Scholar] [CrossRef]
- Wang, H.-C.; Hung, C.-H.; Hsu, J.-D.; Yang, M.-Y.; Wang, S.-J.; Wang, C.-J. Inhibitory effect of whole oat on aberrant crypt foci formation and colon tumor growth in ICR and BALB/c mice. J. Cereal Sci. 2011, 53, 73–77. [Google Scholar] [CrossRef]
- Sharma, N.; Niranjan, K. Foxtail millet: Properties, processing, health benefits and uses. Food Rev. Int. 2018, 34, 329–363. [Google Scholar] [CrossRef]
- Fujita, S.; Sugimoto, Y.; Yamashita, Y.; Fuwa, H. Physicochemical studies of starch from foxtail millet (Setaria italica Beauv). Food Chem. 1996, 55, 209–213. [Google Scholar] [CrossRef]
- Islam, J.; Sato, S.; Watanabe, K.; Watanabe, T.; Ardiansyah, A.; Hirahara, K.; Aoyama, Y.; Tomita, S.; Aso, H.; Komai, M.; et al. Dietary tryptophan alleviates dextran sodium sulfate-induced colitis through aryl hydrocarbon receptor in mice. J. Nutr. Biochem. 2017, 42, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.S.; Cheon, S.Y.; Roh, S.S.; Lee, M.; An, H.J. Chemopreventive Effect of Aster glehni on Inflammation-Induced Colorectal Carcinogenesis in Mice. Nutrients 2018, 10, 202. [Google Scholar] [CrossRef] [PubMed]
- Bibi, S.; de Sousa Moraes, L.F.; Lebow, N.; Zhu, M.J. Dietary Green Pea Protects against DSS-Induced Colitis in Mice Challenged with High-Fat Diet. Nutrients 2017, 9, 509. [Google Scholar] [CrossRef]
- Xiang, N.; Guo, X.; Liu, F.; Li, Q.; Hu, J.; Brennan, C.S. Effect of Light- and Dark-Germination on the Phenolic Biosynthesis, Phytochemical Profiles, and Antioxidant Activities in Sweet Corn (Zea mays L.) Sprouts. Int. J. Mol. Sci. 2017, 18, 1246. [Google Scholar] [CrossRef]
- Meng, S.; Li, Y.; Zang, X.; Jiang, Z.; Ning, H.; Li, J. Effect of TLR2 on the proliferation of inflammation-related colorectal cancer and sporadic colorectal cancer. Cancer Cell Int 2020, 20, 95. [Google Scholar] [CrossRef]
- Benninghoff, A.D.; Hintze, K.J.; Monsanto, S.P.; Rodriguez, D.M.; Hunter, A.H.; Phatak, S.; Pestka, J.J.; Wettere, A.J.V.; Ward, R.E. Consumption of the Total Western Diet Promotes Colitis and Inflammation-Associated Colorectal Cancer in Mice. Nutrients 2020, 12, 544. [Google Scholar] [CrossRef]
- Dieleman, L.A.; Palmen, M.J.; Akol, H.; Bloemena, E.; Pena, A.S.; Meuwissen, S.G.; Van Rees, E.P. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is characterized by Th1 and Th2 cytokines. Clin. Exp. Immunol. 1998, 114, 385–391. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, W.; Yu, N.; Sun, J.; Yu, X.; Li, X.; Xing, Y.; Yan, D.; Ding, Q.; Xiu, Z. Anti-diabetic effect of baicalein is associated with the modulation of gut microbiota in streptozotocin and high-fat-diet induced diabetic rats. J. Funct. Foods 2018, 46, 256–267. [Google Scholar] [CrossRef]
- Lee, C.W.; Chen, H.J.; Chien, Y.H.; Hsia, S.M.; Chen, J.H.; Shih, C.K. Synbiotic Combination of Djulis (Chenopodium formosanum) and Lactobacillus acidophilus Inhibits Colon Carcinogenesis in Rats. Nutrients 2019, 12, 103. [Google Scholar] [CrossRef]
- Ma, H.; Zhang, B.; Hu, Y.; Wang, J.; Liu, J.; Qin, R.; Lv, S.; Wang, S. Correlation analysis of intestinal redox state with the gut microbiota reveals the positive intervention of tea polyphenols on hyperlipidemia in high fat diet fed mice. J. Agric. Food Chem. 2019, 67, 7325–7335. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Ou, D.; Li, H.; Li, W.; Wu, X.; Wang, Y.Q.; Liu, Y.D. Salt-tolerance aerobic granular sludge: Formation and microbial community characteristics. Bioresour. Technol. 2018, 249, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Yuan, B.; Wu, X.; Du, H.; Gu, M.; Han, Y.; Yang, W.; Song, M.; Xiao, H. Dietary Intake of Pleurotus eryngii Ameliorated Dextran-Sodium-Sulfate-Induced Colitis in Mice. Mol. Nutr. Food Res. 2019, 63, e1801265. [Google Scholar] [CrossRef] [PubMed]
- Deuring, J.J.; de Haar, C.; Kuipers, E.J.; Peppelenbosch, M.P.; van der Woude, C.J. The cell biology of the intestinal epithelium and its relation to inflammatory bowel disease. Int. J. Biochem. Cell Biol. 2013, 45, 798–806. [Google Scholar] [CrossRef] [PubMed]
- Vidal, A.C.; Lund, P.K.; Hoyo, C.; Galanko, J.; Burcal, L.; Holston, R.; Massa, B.; Omofoye, O.; Sandler, R.S.; Keku, T.O. Elevated C-peptide and insulin predict increased risk of colorectal adenomas in normal mucosa. BMC Cancer 2012, 12, 389. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, Y.; Wan, P.; Chen, D.; Ding, Y.; Ran, L.; Mi, J.; Lu, L.; Zhang, Z.; Li, X.; et al. Gut microbiota modulation and anti-inflammatory properties of anthocyanins from the fruits of Lycium ruthenicum Murray in dextran sodium sulfate-induced colitis in mice. Free Radic. Biol. Med. 2019, 136, 96–108. [Google Scholar] [CrossRef]
- Kanwal, S.; Joseph, T.P.; Aliya, S.; Song, S.; Saleem, M.Z.; Nisar, M.A.; Wang, Y.; Meyiah, A.; Ma, Y.; Xin, Y. Attenuation of DSS induced colitis by Dictyophora indusiata polysaccharide (DIP) via modulation of gut microbiota and inflammatory related signaling pathways. J. Funct. Foods 2020, 64, 103641. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Wang, D.; Dubois, R.N. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene 2010, 29, 781–788. [Google Scholar] [CrossRef]
- Hyun, Y.S.; Han, D.S.; Lee, A.R.; Eun, C.S.; Youn, J.; Kim, H.Y. Role of IL-17A in the development of colitis-associated cancer. Carcinogenesis 2012, 33, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, L.; Zhang, Q.; Liu, S.; Ge, D.; You, Z. Interleukin-17 Indirectly Promotes M2 Macrophage Differentiation through Stimulation of COX-2/PGE2 Pathway in the Cancer Cells. Cancer Res. Treat. 2014, 46, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.P.; Singh, U.P.; Singh, B.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S. Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS ONE 2011, 6, e23522. [Google Scholar] [CrossRef] [PubMed]
- Pandiyan, P.; Zhu, J. Origin and functions of pro-inflammatory cytokine producing Foxp3+ regulatory T cells. Cytokine 2015, 76, 13–24. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol Hepatol 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- Garrett, W.S. The gut microbiota and colon cancer. Science 2019, 364, 1133–1135. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Q.; Cheng, L.; Buch, H.; Zhang, F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019, 12, 1109–1125. [Google Scholar] [CrossRef]
- Osaka, T.; Moriyama, E.; Arai, S.; Date, Y.; Yagi, J.; Kikuchi, J.; Tsuneda, S. Meta-Analysis of Fecal Microbiota and Metabolites in Experimental Colitic Mice during the Inflammatory and Healing Phases. Nutrients 2017, 9, 1329. [Google Scholar] [CrossRef] [PubMed]



| NM/AOM-DSS | AOM/DSS + Millet | AOM/DSS + Rice | |
|---|---|---|---|
| Ingredient (g/kg Diet) | |||
| Millet flour | - | 397.0 | - |
| Rice flour | - | - | 397.0 |
| Corn starch | 397.0 | - | - |
| Casein | 200.0 | 200.0 | 200.0 |
| L-Cystine | 3.0 | 3.0 | 3.0 |
| Maltodextrin | 132.0 | 132.0 | 132.0 |
| Sucrose | 100.0 | 100.0 | 100.0 |
| Soybean oil | 70.0 | 70.0 | 70.0 |
| Cellulose | 60.0 | 30.0 | 30.0 |
| Mineral mix (AIN-93G-MX) | 35.0 | 35.0 | 35.0 |
| Vitamin mix (AIN-93G-VX) | 10.0 | 10.0 | 10.0 |
| Choline Bitartrate | 2.5 | 2.5 | 2.5 |
| Butylhydroquinone | 0.014 | 0.014 | 0.014 |
| Analyzed nutritional composition (g/kg diet) | |||
| Protein | 200.0 | 236.0 | 223.0 |
| Fat | 70.0 | 85.0 | 73.0 |
| Total starch | 329.1 | 214.1 | 288.3 |
| Total dietary fiber | 60.0 | 60.0 | 48.0 |
| Tryptophan | 1.25 | 1.85 | 1.35 |
| Total phenolics (mg GAE/100g) | 0.0019 | 0.0282 | 0.0084 |
| Composition | Content (g/100 g) | ||
|---|---|---|---|
| Foxtail Millet | Rice | Corn Starch | |
| Moisture | 9.58 | 13.00 | 13.54 |
| Ash | 1.16 | 0.40 | 0.17 |
| Protein | 10.51 | 7.20 | 1.32 |
| Fat | 4.32 | 1.13 | 0.42 |
| Total starch | 53.93 | 72.64 | 82.90 |
| Total dietary fiber | 8.15 | 5.14 | 0.58 |
| Soluble dietary fiber | 0.81 | 0.86 | 0.36 |
| Insoluble dietary fiber | 7.34 | 4.28 | 0.22 |
| Tryptophan | 0.18 | 0.06 | 0.03 |
| Total phenolics (mg GAE/100 g) | 39.05 | 11.17 | 4.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Xu, Y.; Liu, S.; Lv, H.; Hu, Y.; Wang, Y.; Li, Z.; Wang, J.; Ji, X.; Ma, H.; et al. Dietary Supplementation of Foxtail Millet Ameliorates Colitis-Associated Colorectal Cancer in Mice via Activation of Gut Receptors and Suppression of the STAT3 Pathway. Nutrients 2020, 12, 2367. https://doi.org/10.3390/nu12082367
Zhang B, Xu Y, Liu S, Lv H, Hu Y, Wang Y, Li Z, Wang J, Ji X, Ma H, et al. Dietary Supplementation of Foxtail Millet Ameliorates Colitis-Associated Colorectal Cancer in Mice via Activation of Gut Receptors and Suppression of the STAT3 Pathway. Nutrients. 2020; 12(8):2367. https://doi.org/10.3390/nu12082367
Chicago/Turabian StyleZhang, Bowei, Yingchuan Xu, Shuang Liu, Huan Lv, Yaozhong Hu, Yaya Wang, Zhi Li, Jin Wang, Xuemeng Ji, Hui Ma, and et al. 2020. "Dietary Supplementation of Foxtail Millet Ameliorates Colitis-Associated Colorectal Cancer in Mice via Activation of Gut Receptors and Suppression of the STAT3 Pathway" Nutrients 12, no. 8: 2367. https://doi.org/10.3390/nu12082367
APA StyleZhang, B., Xu, Y., Liu, S., Lv, H., Hu, Y., Wang, Y., Li, Z., Wang, J., Ji, X., Ma, H., Wang, X., & Wang, S. (2020). Dietary Supplementation of Foxtail Millet Ameliorates Colitis-Associated Colorectal Cancer in Mice via Activation of Gut Receptors and Suppression of the STAT3 Pathway. Nutrients, 12(8), 2367. https://doi.org/10.3390/nu12082367





