Effect of a Flaxseed Lignan Intervention on Circulating Bile Acids in a Placebo-Controlled Randomized, Crossover Trial
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Design
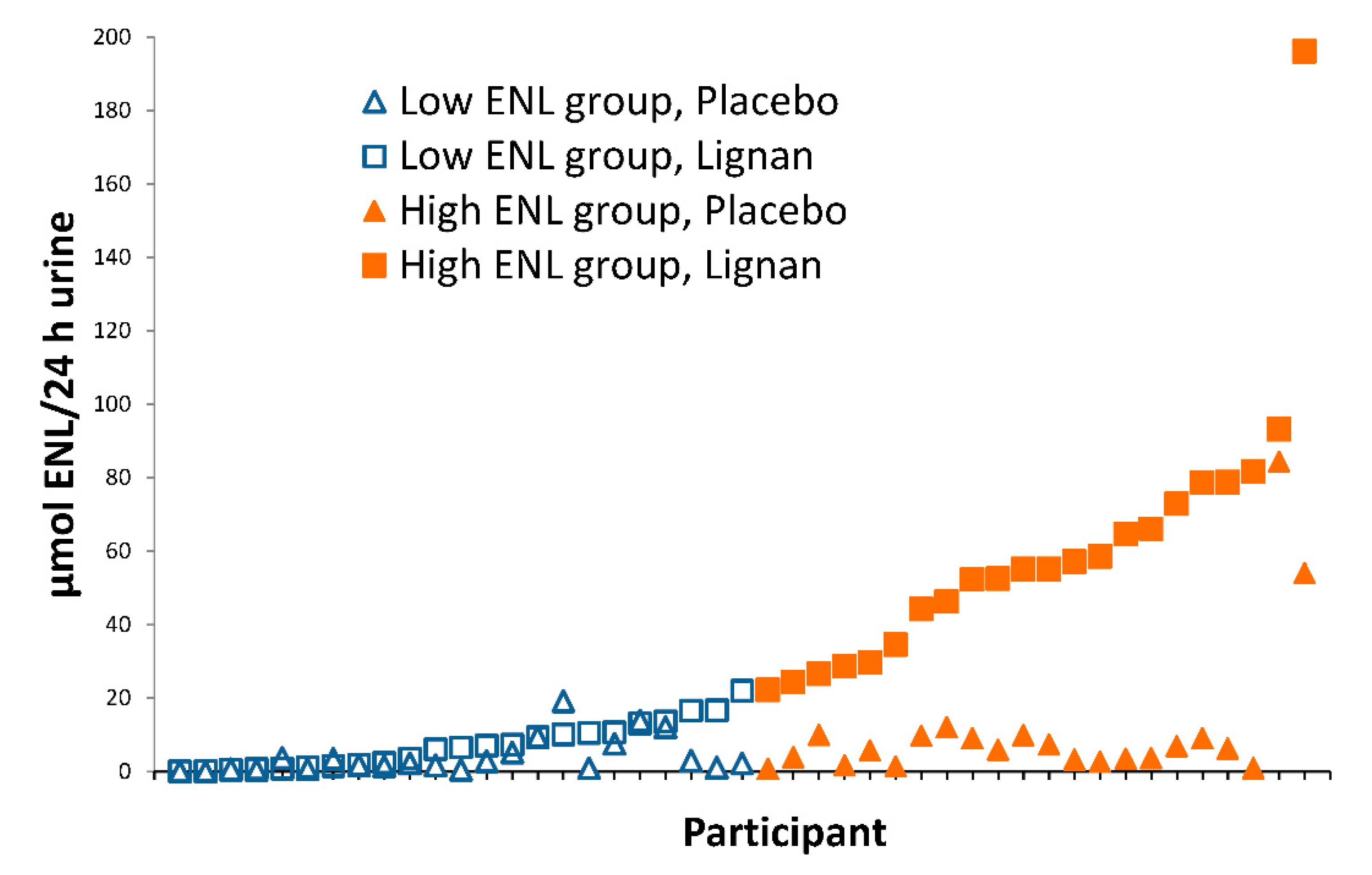
2.2. Participants
2.3. Flaxseed Lignan Extract Supplement
2.4. Specimen Collection
2.5. Laboratory Analyses
2.5.1. Plasma Bile Acids
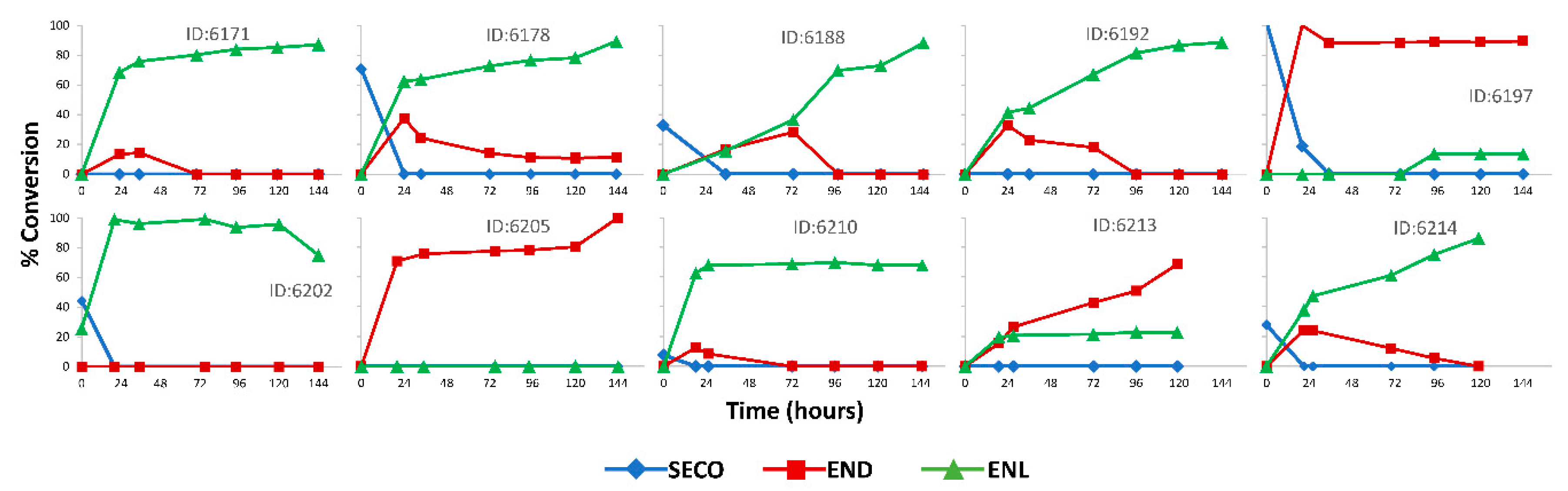
2.5.2. In Vitro Incubations
2.5.3. Lignans
2.5.4. Fecal Microbiome Nucleic Acid Extraction
2.5.5. 16S rRNA Gene Sequencing
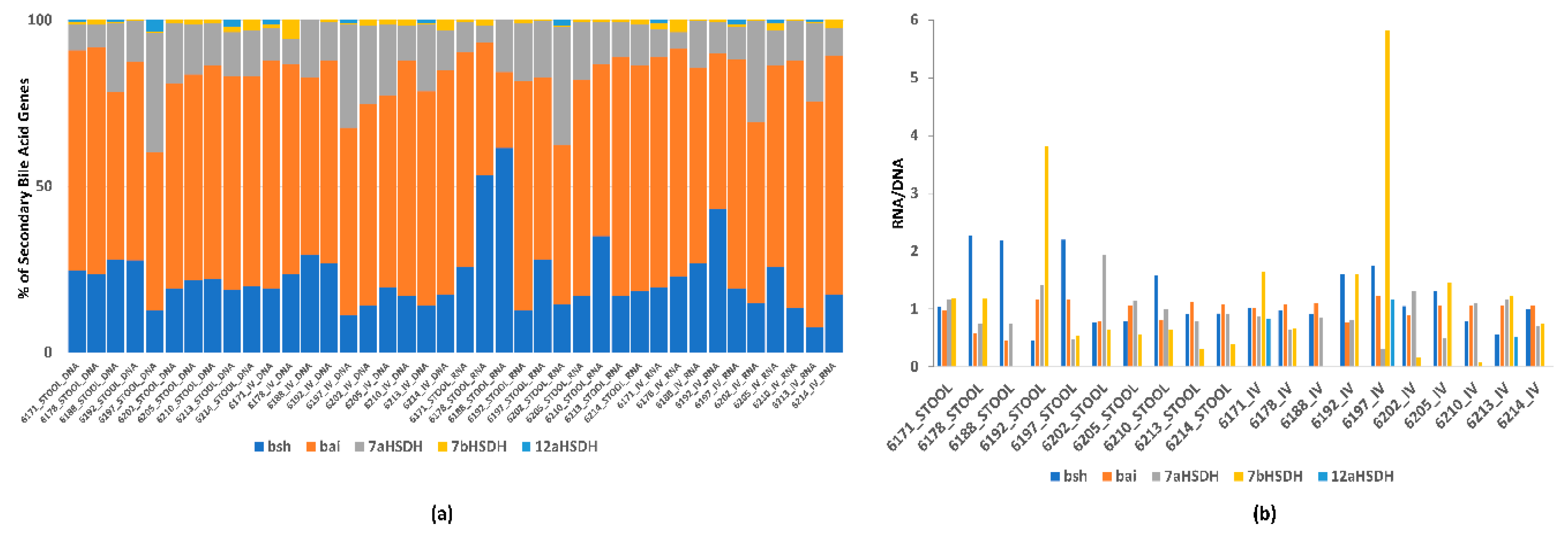
2.5.6. Stool and In Vitro Incubation Metagenomic and Metatranscriptomic Bioinformatic Analysis
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McRae, M.P. Health benefits of dietary whole grains: An umbrella review of meta-analyses. J. Chiropr. Med. 2017, 16, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, K.E.; Appleby, P.N.; Key, T.J. Fruit, vegetable, and fiber intake in relation to cancer risk: Findings from the European Prospective Investigation into Cancer and Nutrition (EPIC). Am. J. Clin. Nutr. 2014, 100, 394S–398S. [Google Scholar] [CrossRef] [PubMed]
- Kahleova, H.; Levin, S.; Barnard, N. Cardio-metabolic benefits of plant-based diets. Nutrients 2017, 9, 848. [Google Scholar] [CrossRef] [PubMed]
- Bamia, C. Dietary patterns in association to cancer incidence and survival: Concept, current evidence, and suggestions for future research. Eur. J. Clin. Nutr. 2018, 72, 818–825. [Google Scholar] [CrossRef] [PubMed]
- Tangestani, H.; Emamat, H.; Ghalandari, H.; Shab-Bidar, S. Whole grains, dietary fibers and the human gut microbiota: A systematic review of existing literature. Recent Pat Food Nutr. Agric. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Meselhy, M.R.; LI, Y.; Qin, G.; Hattori, M. Human intestinal bacteria capable of transforming secoisolariciresinol diglucoside to mammalian lignans, enterodiol and enterolactone. Chem. Pharmol. Bull. 2000, 48, 1606–1610. [Google Scholar] [CrossRef]
- Webb, A.L.; McCullough, M.L. Dietary lignans: Potential role in cancer prevention. Nutr. Cancer 2005, 51, 117–131. [Google Scholar] [CrossRef]
- Kuijsten, A.; Arts, I.C.; Vree, T.B.; Hollman, P.C. Pharmacokinetics of enterolignans in healthy men and women consuming a single dose of secoisolariciresinol diglucoside. J. Nutr. 2005, 135, 795–801. [Google Scholar] [CrossRef]
- Miles, F.L.; Navarro, S.L.; Schwarz, Y.; Gu, H.; Djukovic, D.; Randolph, T.W.; Shojaie, A.; Kratz, M.; Hullar, M.A.J.; Lampe, P.D.; et al. Plasma metabolite abundances are associated with urinary enterolactone excretion in healthy participants on controlled diets. Food Funct. 2017. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Targeting bile acids and lipotoxicity for NASH treatment. Hepatol. Commun. 2017, 1, 1002–1004. [Google Scholar] [CrossRef]
- Ajouz, H.; Mukherji, D.; Shamseddine, A. Secondary bile acids: An underrecognized cause of colon cancer. World J. Surg. Oncol. 2014, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile acid nuclear receptor FXR and digestive system diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Pang, S.; Sun, Y.; Tian, Y.; Yu, L.; Dang, N. Coordinated actions of FXR and LXR in metabolism: From pathogenesis to pharmacological targets for Type 2 Diabetes. Int. J. Endocrinol. 2014, 2014, 751859. [Google Scholar] [CrossRef] [PubMed]
- Batta, A.K.; Aggarwal, S.K.; Salen, G.; Shefer, S. Selective reduction of oxo bile acids: Synthesis of 3 beta-, 7 beta-, and 12 beta-hydroxy bile acids. J. Lipid Res. 1991, 32, 977–983. [Google Scholar] [PubMed]
- Lampe, J.W.; Kim, E.; Levy, L.; Davidson, L.A.; Goldsby, J.S.; Miles, F.L.; Navarro, S.L.; Randolph, T.W.; Zhao, N.; Ivanov, I.; et al. Colonic mucosal and exfoliome transcriptomic profiling and fecal microbiome response to a flaxseed lignan extract intervention in humans. Am. J. Clin. Nutr. 2019, 110, 377–390. [Google Scholar] [CrossRef]
- Block, G.; Gillespie, C.; Rosenbaum, E.H.; Jenson, C. A rapid food screener to assess fat and fruit and vegetable intake. Am. J. Prev. Med. 2000, 18, 284–288. [Google Scholar] [CrossRef]
- Patterson, R.E.; Kristal, A.R.; Tinker, L.F.; Carter, R.A.; Bolton, M.P.; Agurs-Collins, T. Measurement characteristics of the Women’s Health Initiative food frequency questionnaire. Ann. Epidemiol. 1999, 9, 178–187. [Google Scholar] [CrossRef]
- Peñalvo, J.L.; Haajanen, K.M.; Botting, N.; Adlercreutz, H. Quantification of lignans in food using isotope dilution gas chromatography/mass spectrometry. J. Agric. Food Chem. 2005, 53, 9342–9347. [Google Scholar] [CrossRef]
- Ginos, B.N.R.; Navarro, S.L.; Schwarz, Y.; Gu, H.; Wang, D.; Randolph, T.W.; Shojaie, A.; Hullar, M.A.J.; Lampe, P.D.; Kratz, M.; et al. Circulating bile acids in healthy adults respond differently to a dietary pattern characterized by whole grains, legumes and fruits and vegetables compared to a diet high in refined grains and added sugars: A randomized, controlled, crossover feeding study. Metabolism 2018, 83, 197–204. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Raftery, D. Mass Spectrometry in Metabolomics: Methods and Protocols; Humana Press: New York, NY, USA, 2014. [Google Scholar]
- Sarafian, M.H.; Lewis, M.R.; Pechlivanis, A.; Ralphs, S.; McPhail, M.J.; Patel, V.C.; Dumas, M.E.; Holmes, E.; Nicholson, J.K. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal. Chem. 2015, 87, 9662–9670. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.C.; Randolph, T.W.; Lim, U.; Monroe, K.R.; Cheng, I.; Wilkens, L.R.; Le Marchand, L.; Lampe, J.W.; Hullar, M.A.J. Temporal variability and stability of the fecal microbiome: The Multiethnic Cohort Study. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 154–162. [Google Scholar] [CrossRef]
- Satinsky, B.M.; Gifford, S.M.; Crump, B.C.; Moran, M.A. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol. 2013, 531, 237–250. [Google Scholar] [PubMed]
- Stewart, F.J.; Ottesen, E.A.; DeLong, E.F. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010, 4, 896–907. [Google Scholar] [CrossRef]
- Li, F.; Hullar, M.A.; Lampe, J.W. Optimization of terminal restriction fragment polymorphism (TRFLP) analysis of human gut microbiota. J. Microbiol. Methods 2007, 68, 303–311. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Heilig, H.G.; Klaassens, E.S.; Booijink, C.C.; Kleerebezem, M.; Smidt, H.; de Vos, W.M. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 2006, 1, 870–873. [Google Scholar] [CrossRef]
- El Fantroussi, S.; Urakawa, H.; Bernhard, A.E.; Kelly, J.J.; Noble, P.A.; Smidt, H.; Yershov, G.M.; Stahl, D.A. Direct profiling of environmental microbial populations by thermal dissociation analysis of native rRNAs hybridized to oligonucleotide microarrays. Appl. Environ. Microb. 2003, 69, 2377–2382. [Google Scholar] [CrossRef][Green Version]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Abubucker, S.; Segata, N.; Goll, J.; Schubert, A.M.; Izard, J.; Cantarel, B.L.; Rodriguez-Mueller, B.; Zucker, J.; Thiagarajan, M.; Henrissat, B.; et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 2012, 8, e1002358. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.Q.; Huang, H.Z.; McGarvey, P.B.; Wu, C.H.; Consortium, U. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Ferrer, L.; Foerster, H.; Fulcher, C.A.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Mueller, L.A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016, 44, D623–D631. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Heinken, A.; Ravcheev, D.A.; Baldini, F.; Heirendt, L.; Fleming, R.M.T.; Thiele, I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome 2019, 7, 75. [Google Scholar] [CrossRef]
- Touré, A.; Xueming, X. Flaxseed lignans: Source, biosynthesis, metabolism, antioxidant activity, bio-active components, and health benefits. Compr. Rev. Food Sci. Food Saf. 2010, 9, 261–269. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Zeng, H.; Jiang, Y.; Chen, P.; Fan, X.; Li, D.; Liu, A.; Ma, X.; Xie, W.; Liu, P.; Gonzalez, F.J.; et al. Schisandrol B protects against cholestatic liver injury through pregnane X receptors. Br. J. Pharmacol. 2017, 174, 672–688. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Liu, C.; Jiang, Y.; Gao, Y.; Chen, Y.; Fu, K.; Yao, X.; Huang, M.; Bi, H. Lignans from Schisandra sphenanthera protect against lithocholic acid-induced cholestasis by pregnane X receptor activation in mice. J. Ethnopharmacol. 2019, 245, 112103. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.R.; Ge, J.Q.; Xie, X.; Fan, M.L.; Fan, X.D.; Wang, H.; Dong, Z.Y.; Liao, Z.H.; Lan, X.Z.; Chen, M. Protective effects of petroleum ether extracts of Herpetospermum caudigerum against alpha-naphthylisothiocyanate-induced acute cholestasis of rats. J. Ethnopharmacol. 2017, 198, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Bohmdorfer, M.; Maier-Salamon, A.; Taferner, B.; Reznicek, G.; Thalhammer, T.; Hering, S.; Hufner, A.; Schuhly, W.; Jager, W. In Vitro metabolism and disposition of honokiol in rat and human livers. J. Pharm. Sci. 2011, 100, 3506–3516. [Google Scholar] [CrossRef]
- Takeda, S.; Arai, I.; Hasegawa, M.; Tatsugi, A.; Aburada, M.; Hosoya, E. [Effect of gomisin A (TJN-101), a lignan compound isolated from Schisandra fruits, on liver function in rats]. Nihon Yakurigaku Zasshi 1988, 91, 237–244. [Google Scholar] [CrossRef]
- Ohtaki, Y.; Hida, T.; Hiramatsu, K.; Kanitani, M.; Ohshima, T.; Nomura, M.; Wakita, H.; Aburada, M.; Miyamoto, K.I. Deoxycholic acid as an endogenous risk factor for hepatocarcinogenesis and effects of gomisin A, a lignan component of Schizandra fruits. Anticancer Res. 1996, 16, 751–755. [Google Scholar]
- Miyamoto, K.; Hiramatsu, K.; Ohtaki, Y.; Kanitani, M.; Nomura, M.; Aburada, M. Effects of gomisin A on the promotor action and serum bile acid concentration in hepatocarcinogenesis induced by 3′-methyl-4-dimethylamino-azobenzene. Biol. Pharm. Bull. 1995, 18, 1443–1445. [Google Scholar] [CrossRef]
- Sanghvi, A.; Diven, W.F.; Seltman, H.J.; Paul, R.; Rizk, M. Properties of cholesterol 7 alpha-hydroxylase in rat liver microsomal acetone powder. Metabolism 1984, 33, 443–446. [Google Scholar] [CrossRef]
- Tai, T.S.; Tien, N.; Shen, H.Y.; Chu, F.Y.; Wang, C.C.N.; Lu, C.H.; Yu, H.I.; Kung, F.P.; Chuang, H.H.; Lee, Y.R.; et al. Sesamin, a Naturally Occurring Lignan, Inhibits Ligand-Induced Lipogenesis through Interaction with Liver X Receptor Alpha (LXRalpha) and Pregnane X Receptor (PXR). Evid. Based Complement. Altern. Med. 2019, 2019, 9401648. [Google Scholar] [CrossRef]
- Hirose, N.; Inoue, T.; Nishihara, K.; Sugano, M.; Akimoto, K.; Shimizu, S.; Yamada, H. Inhibition of cholesterol absorption and synthesis in rats by sesamin. J. Lipid Res. 1991, 32, 629–638. [Google Scholar]
- Jacobs, M.N.; Nolan, G.T.; Hood, S.R. Lignans, bacteriocides and organochlorine compounds activate the human pregnane X receptor (PXR). Toxicol. Appl. Pharmacol. 2005, 209, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, K.N.; Chen, C. The role of CYP3A4 in the biotransformation of bile acids and therapeutic implication for cholestasis. Ann. Transl. Med. 2014, 2, 7. [Google Scholar] [PubMed]
- Kim, H.; Fang, S. Crosstalk between FXR and TGR5 controls glucagon-like peptide 1 secretion to maintain glycemic homeostasis. Lab. Anim. Res. 2018, 34, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.J.; Briz, O. Bile-acid-induced cell injury and protection. World J. Gastroenterol. 2009, 15, 1677–1689. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Ege, T.; Gençler-Özkan, A.M.; Şen, A.; Adalı, O. Effects of folk medicinal plant epilobium hirsutum l. And its ingredient ellagic acid on rat liver bile acid synthesizing cyps in rats. PharmacologyOnline 2018, 3, 200–215. [Google Scholar]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, L.Z.; Chen, Y.J.; Zhu, P.P.; Yin, S.S.; Su, M.M.; Ni, Y.; Miao, J.; Wu, W.L.; Chen, H.; et al. Continuum of host-gut microbial Co-metabolism: Host CYP3A4/3A7 are responsible for tertiary Oxidations of deoxycholate species. Drug Metab. Dispos. 2019, 47, 283–294. [Google Scholar] [CrossRef]
- Lin, Q.; Tan, X.; Wang, W.; Zeng, W.; Gui, L.; Su, M.; Liu, C.; Jia, W.; Xu, L.; Lan, K. Species differences of bile acid redox metabolism: Tertiary oxidation of deoxycholate is conserved in preclinical animals. Drug Metab. Dispos. 2020, 48, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Yanagi, K.; Cheng, C.; Alaniz, R.C.; Lee, K.; Jayaraman, A. Interactions between gut microbiota and non-alcoholic liver disease: The role of microbiota-derived metabolites. Pharmacol. Res. 2019, 141, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Nihira, T.; Suzuki, E.; Kitaoka, M.; Nishimoto, M.; Ohtsubo, K.; Nakai, H. Discovery of beta-1,4-D-mannosyl-N-acetyl-D-glucosamine phosphorylase involved in the metabolism of N-glycans. J. Biol. Chem. 2013, 288, 27366–27374. [Google Scholar] [CrossRef]
- Yoder, S.; Lancaster, S.; Hullar, M.A.J.; Lampe, J.W. Gut microbial metabolism of plant lignans: Influence on human health. In Diet-Microbe Interactions in the Gut; Del Rio, D., Tuohy, K., Eds.; Elsevier: Oxford, UK, 2015; pp. 103–117. [Google Scholar]
- Halldin, E.; Eriksen, A.K.; Brunius, C.; da Silva, A.B.; Bronze, M.; Hanhineva, K.; Aura, A.M.; Landberg, R. Factors explaining inter-personal variation in plasma enterolactone concentrations in humans. Mol. Nutr. Food Res. 2019, 63, e1801159. [Google Scholar] [CrossRef]
- Liu, N.; Fosses, A.; Kampik, C.; Parsiegla, G.; Denis, Y.; Vita, N.; Fierobe, H.P.; Perret, S. In Vitro and In Vivo exploration of the cellobiose and cellodextrin phosphorylases panel in Ruminiclostridium cellulolyticum: Implication for cellulose catabolism. Biotechnol. Biofuels 2019, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; You, W.; Wu, S.; Poetsch, A.; Xu, C. Secretomic analyses of Ruminiclostridium papyrosolvens reveal its enzymatic basis for lignocellulose degradation. Biotechnol. Biofuels 2019, 12, 183. [Google Scholar] [CrossRef]
- Harris, S.C.; Devendran, S.; Mendez-Garcia, C.; Mythen, S.M.; Wright, C.L.; Fields, C.J.; Hernandez, A.G.; Cann, I.; Hylemon, P.B.; Ridlon, J.M. Bile acid oxidation by Eggerthella lenta strains C592 and DSM 2243(T). Gut Microbes 2018, 9, 523–539. [Google Scholar]
- Bess, E.N.; Bisanz, J.E.; Yarza, F.; Bustion, A.; Rich, B.E.; Li, X.; Kitamura, S.; Waligurski, E.; Ang, Q.Y.; Alba, D.L.; et al. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat. Microbiol. 2020, 5, 56–66. [Google Scholar] [CrossRef]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Thakare, R.; Alamoudi, J.A.; Gautam, N.; Rodrigues, A.D.; Alnouti, Y. Species differences in bile acids I. Plasma and urine bile acid composition. J. Appl. Toxicol. 2018, 38, 1323–1335. [Google Scholar] [CrossRef]
- Alnouti, Y. Bile Acid sulfation: A pathway of bile acid elimination and detoxification. Toxicol. Sci. 2009, 108, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Ghaffarzadegan, T.; Zanzer, Y.C.; Ostman, E.; Hallenius, F.; Essen, S.; Sandahl, M.; Nyman, M. Postprandial responses of serum bile acids in healthy humans after ingestion of turmeric before medium/high-fat breakfasts. Mol. Nutr. Food Res. 2019, 63, e1900672. [Google Scholar] [CrossRef] [PubMed]
- Story, J.A.; Furumoto, E.J.; Buhman, K.K. Dietary fiber and bile acid metabolism—An update. Adv. Exp. Med. Biol. 1997, 427, 259–266. [Google Scholar] [PubMed]
- Luo, L.; Aubrecht, J.; Li, D.; Warner, R.L.; Johnson, K.J.; Kenny, J.; Colangelo, J.L. Assessment of serum bile acid profiles as biomarkers of liver injury and liver disease in humans. PLoS ONE 2018, 13, e0193824. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Pan, L.X.; Li, H.; Forman, B.M.; Erickson, S.K.; Shefer, S.; Bollineni, J.; Batta, A.K.; Christie, J.; Wang, T.H.; et al. Regulation of the farnesoid X receptor (FXR) by bile acid flux in rabbits. J. Biol. Chem. 2002, 277, 50491–50496. [Google Scholar] [CrossRef] [PubMed]
| Abbreviation | Enzyme |
|---|---|
| BSH | Choloylglycine Hydrolase (EC 3.5.1.24) |
| 12aHSDH | 12–alpha-hydroxysteroid dehydrogenase (EC 1.1.1.176) |
| 7aHSDH | 7–alpha-hydroxysteroid dehydrogenase (EC 1.1.1.159) |
| 7bHSDH | 7–beta-hydroxysteroid dehydrogenase (EC 1.1.1.201) |
| 3aHSDH | 3–alpha-hydroxysteroid dehydrogenase (EC 1.1.1.53) |
| 3bHSDH | 3–beta-hydroxysteroid dehydrogenase (EC 1.1.1.145 |
| Bile acid inducible (Bai) Gene Cluster | |
| BaiA | NAD(P)-dependent 3–alpha-hydroxysteroid dehydrogenase (EC 1.1.1.50) |
| BaiB | Bile acid-coenzyme A ligase (EC 6.2.1.7) |
| BaiCD | NAD(H)-dependent 7–alpha-hydroxy–3–oxo-delta 4–cholenoic acid oxidoreductase |
| BaiE | Bile acid 7–alpha dehydratase (EC 4.2.1.106) |
| BaiF | Bile acid CoA-transferase (EC 2.8.3.25) |
| BaiG | Bile acids transporter, MFS family |
| BaiH | NAD(H)-dependent 7–beta-hydroxy–3–oxo-delta 4–cholenoic acid oxidoreductase |
| BaiI | Bile acid 7–beta-dehydratase (EC 4.2.1.106) |
| CHSTR | Cholesterol reductase (gene not known) |
| Characteristic | All Participants | Low ENL | High ENL (n = 23) |
|---|---|---|---|
| (n = 46) | (n = 23) | ||
| Female n (%) | 23 (50.0) | 11 (47.8) | 12 (52.2) |
| Age (years) | 32.1 (8.4) | 33.1 (9.3) | 31.1 (7.6) |
| BMI (kg/m2) | 26.7 (5.8) | 27.2 (7.1) | 26.2 (4.3) |
| Energy intake (kcal/day) b | 2067 (518) | 2289 (602) | 1865 (324) |
| Dietary fat (g/day) b | 86 (30) | 96 (34) | 77 (23) |
| Dietary fiber (g/day) b | 20.5 (7) | 19.2 (7.2) | 21.6 (6.7) |
| Dietary protein (g/day) b | 86.9 (30.3) | 99.8 (35.5) | 74.6 (17.6) |
| Race/ethnicity (n) | |||
| American Indian | 2 | 1 | 1 |
| Asian | 11 | 7 | 4 |
| Black African American | 2 | 1 | 1 |
| Caucasian | 28 | 12 | 16 |
| Other (more than 1 race) | 3 | 2 | 1 |
| Urinary Enterolignans (µmol/24 h) c | |||
| SECO | 0.4 (0.5) | 0.5 (0.6) | 0.3 (0.4) |
| END | 1.1 (2.7) | 1.2 (3) | 1.1 (2.4) |
| ENL | 7.6 (14.3) | 4.1 (5) | 11.1 (19.2) |
| Baseline plasma bile acid groups (nM) d | |||
| Σ Total bile acids | 2630 (2515) | 2623 (2652) | 2639 (2428) |
| Σ Primary bile acids | 1511 (1747) | 1411 (1633) | 1617 (1890) |
| Σ Secondary bile acids | 1119 (984) | 1212 (1173) | 1022 (737) |
| Σ Total taurine-conjugated | 293 (666) | 193.6 (200) | 397 (926) |
| Σ Total glycine-conjugated | 1309 (1274) | 1284 (1103) | 1336 (1455) |
| Bile Acid | All Participants (n = 45) | Low ENL (n = 23) | High ENL (n = 22) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Placebo | Flax | Pb | Placebo | Flax | Pb | Placebo | Flax | Pb | |
| Primary | |||||||||
| Σ Primary bile acids | 1364 (960) | 1202 (868) | 0.18 | 1415 (865) | 1337 (899) | 0.72 | 1310 (1067) | 1060 (830) | 0.25 |
| Cholic acid | 262 (313) | 142 (88) | 0.004 | 275 (379) | 134 (60) | 0.04 | 284 (233) | 149 (110) | 0.04 |
| Taurohyocholic acid | 6.5 (4.0) | 5.8 (3.9) | 0.14 | 5.9 (3.3) | 5.2 (2.4) | 0.51 | 7.3 (4.6) | 6.4 (5.1) | 0.13 |
| Glycocholic acid | 273 (306) | 269 (309) | 0.62 | 234 (169) | 286 (320) | 0.94 | 315 (403) | 252 (304) | 0.44 |
| Taurocholic acid | 123 (188) | 132 (182) | 0.64 | 88.7 (58.8) | 116 (137) | 0.92 | 159 (250) | 150 (220) | 0.52 |
| Chenodeoxycholic acid | 430 (420) | 388 (386) | 0.85 | 518 (485) | 501 (474) | 0.96 | 338 (324) | 270 (219) | 0.84 |
| Glycochenodeoxycholic acid | 269 (217) | 265 (232) | 0.96 | 293 (224) | 296 (234) | 0.94 | 243 (212) | 233 (231) | 0.99 |
| Secondary | |||||||||
| Σ Secondary bile acids | 1830 (726) | 1802 (900) | 0.34 | 1900 (819) | 1961 (990) | 0.51 | 1756 (626) | 1635 (782) | 0.91 |
| Deoxycholic acid | 480 (338) | 453 (318) | 0.86 | 486 (371) | 487 (326) | 0.49 | 473 (308) | 417 (314) | 0.23 |
| Taurodeoxycholic acid | 57.5 (62.2) | 65.5 (84.5) | 0.90 | 60.3 (71.8) | 63.5 (88.8) | 0.89 | 55.6 (51.8) | 67.5 (81.8) | 0.77 |
| Glycodeoxycholic acid | 311 (295) | 311 (304) | 0.72 | 344 (331) | 331 (324) | 0.84 | 275 (253) | 290 (288) | 0.75 |
| Lithocholic acid | 73.3 (40.1) | 72.2 (49.6) | 0.87 | 66.8 (45.2) | 72.6 (46.1) | 0.38 | 80.1 (33.5) | 71.7 (32.4) | 0.29 |
| Glycolithocholic acid | 69.4 (39.4) | 65.0 (37.3) | 0.63 | 70.1 (46.0) | 66.5 (45.1) | 0.90 | 68.6 (32.1) | 63.4 (27.8) | 0.54 |
| Taurolithocholic acid | 14.9 (7.0) | 18.3 (15.1) | 0.24 | 13.1 (6.8) | 17.0 (9.1) | 0.01 | 16.7 (6.8) | 19.6 (19.7) | 0.58 |
| Isolithocholic acid | 73.5 (33.0) | 73.8 (32.6) | 0.97 | 59.8 (24.2) | 68.7 (36.3) | 0.29 | 87.9 (35.4) | 79.2 (28.1) | 0.28 |
| Glycoursodeoxycholic acid | 146 (104) | 134 (108) | 0.17 | 169 (122) | 155 (112) | 0.82 | 122 (76) | 112 (103) | 0.05 |
| Glycohyodeoxycholic acid | 132 (91) | 122 (90) | 0.33 | 150 (105) | 139 (92) | 0.99 | 113 (69) | 105 (87) | 0.14 |
| Hyodeoxycholic acid | 106 (48) | 107 (68) | 0.46 | 108 (50) | 124 (88) | 0.96 | 104 (47) | 77.8 (31.5) | 0.16 |
| Glycohyocholic acid | 36.3 (27.4) | 29.9 (25.1) | 0.11 | 33.7 (28.6) | 30.6 (29.6) | 0.60 | 39.0 (26.4) | 29.3 (20.0 | 0.03 |
| 5β-Cholanic acid-3β, 12α-diol | 158 (81) | 167 (102) | 0.61 | 160 (79) | 191 (117) | 0.21 | 156 (85) | 142 (81) | 0.61 |
| Muricholic acid | 164 (124) | 176 (172) | 0.78 | 171 9137) | 208 (202) | 0.28 | 158 (111) | 143 (131) | 0.22 |
| Tauro-α-muricholic acid | 8.1 (9.6) | 6.9 (8.7) | 0.08 | 8.3 (10.5) | 6.6 (9.8) | 0.07 | 7.7 (8.8) | 7.3 (7.6) | 0.46 |
| Σ Glycine-conjugated bile acids | 1237 (925) | 1196 (925) | 0.62 | 1295 (876) | 1304 (967) | 0.95 | 1176 (860) | 1084 (887) | 0.15 |
| Σ Taurine-conjugated bile acids | 210 (231) | 229 (260) | 0.96 | 176 (134) | 208 (228) | 0.94 | 245 (301) | 251 (293) | 0.95 |
| Bile Acid | Type | β | SE | P Value a |
|---|---|---|---|---|
| Glycoursodeoxycholic acid | Secondary | −0.2144 | 0.0455 | 2.39 × 10−6 * |
| Glycohyodeoxycholic acid | Secondary | −0.1964 | 0.0439 | 7.69 × 10−6 * |
| Isolithocholic acid | Secondary | 0.1095 | 0.0298 | 0.0002 * |
| Hyodeoxycholic acid | Secondary | −0.1095 | 0.0298 | 0.0002 * |
| Muricholic acid | Secondary | −0.1715 | 0.0513 | 0.0008 * |
| Lithocholic acid | Secondary | 0.0999 | 0.0397 | 0.01 |
| Chenodeoxycholic acid | Primary | −0.1359 | 0.0628 | 0.03 |
| Glycolithocholic acid | Secondary | 0.0654 | 0.0388 | 0.09 |
| Taurolithocholic acid | Secondary | 0.0786 | 0.0434 | 0.07 |
| Cholic acid | Primary | 0.0685 | 0.0475 | 0.15 |
| 5β-Cholanic acid-3β, 12α-diol | Secondary | −0.0431 | 0.0366 | 0.24 |
| Glycochenodeoxycholic acid | Primary | −0.0701 | 0.0674 | 0.30 |
| Taurodeoxycholic acid | Secondary | 0.0746 | 0.0815 | 0.36 |
| Deoxycholic acid | Secondary | 0.0473 | 0.0537 | 0.38 |
| Tauro-α-muricholic acid | Secondary | 0.0418 | 0.0825 | 0.61 |
| Glycocholic acid | Primary | −0.0477 | 0.0759 | 0.53 |
| Glycodeoxycholic acid | Secondary | 0.0256 | 0.0745 | 0.73 |
| Glycohyocholic acid | Secondary | −0.0085 | 0.0496 | 0.86 |
| Taurocholic acid | Primary | −0.0044 | 0.0773 | 0.95 |
| Taurohyocholic acid | Primary | 0.0001 | 0.0416 | 1.00 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro, S.L.; Levy, L.; Curtis, K.R.; Elkon, I.; Kahsai, O.J.; Ammar, H.S.; Randolph, T.W.; Hong, N.N.; Carnevale Neto, F.; Raftery, D.; et al. Effect of a Flaxseed Lignan Intervention on Circulating Bile Acids in a Placebo-Controlled Randomized, Crossover Trial. Nutrients 2020, 12, 1837. https://doi.org/10.3390/nu12061837
Navarro SL, Levy L, Curtis KR, Elkon I, Kahsai OJ, Ammar HS, Randolph TW, Hong NN, Carnevale Neto F, Raftery D, et al. Effect of a Flaxseed Lignan Intervention on Circulating Bile Acids in a Placebo-Controlled Randomized, Crossover Trial. Nutrients. 2020; 12(6):1837. https://doi.org/10.3390/nu12061837
Chicago/Turabian StyleNavarro, Sandi L., Lisa Levy, Keith R. Curtis, Isaac Elkon, Orsalem J. Kahsai, Hamza S. Ammar, Timothy W. Randolph, Natalie N. Hong, Fausto Carnevale Neto, Daniel Raftery, and et al. 2020. "Effect of a Flaxseed Lignan Intervention on Circulating Bile Acids in a Placebo-Controlled Randomized, Crossover Trial" Nutrients 12, no. 6: 1837. https://doi.org/10.3390/nu12061837
APA StyleNavarro, S. L., Levy, L., Curtis, K. R., Elkon, I., Kahsai, O. J., Ammar, H. S., Randolph, T. W., Hong, N. N., Carnevale Neto, F., Raftery, D., Chapkin, R. S., Lampe, J. W., & Hullar, M. A. J. (2020). Effect of a Flaxseed Lignan Intervention on Circulating Bile Acids in a Placebo-Controlled Randomized, Crossover Trial. Nutrients, 12(6), 1837. https://doi.org/10.3390/nu12061837







