The Effects of Aronia melanocarpa Juice Consumption on the mRNA Expression Profile in Peripheral Blood Mononuclear Cells in Subjects at Cardiovascular Risk
Abstract
1. Introduction
2. Materials and Methods
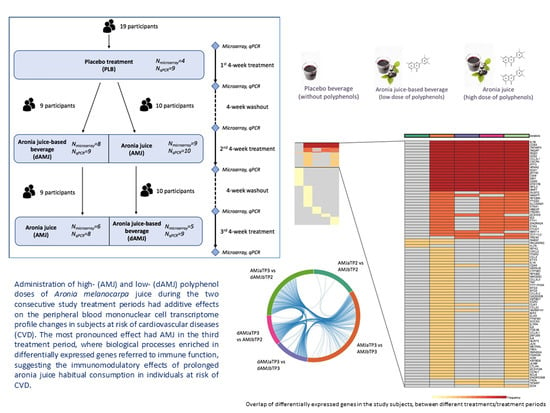
2.1. Study Design and Intervention Treatments
2.2. Sample Collection
2.3. Assessment of Study Variables
2.4. Isolation of PBMC and Preparation of the Total RNA
2.5. Samples for Microarray Analysis
2.6. Microarray PBMC mRNA Expression Profiling and Determination of Differentially Expressed Genes
2.7. Bioinformatic Analysis
2.8. Validation of PBMC Gene Expression by Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.9. Statistical Analysis
3. Results
3.1. Determination of PBMC DEGs for the Applied Treatments
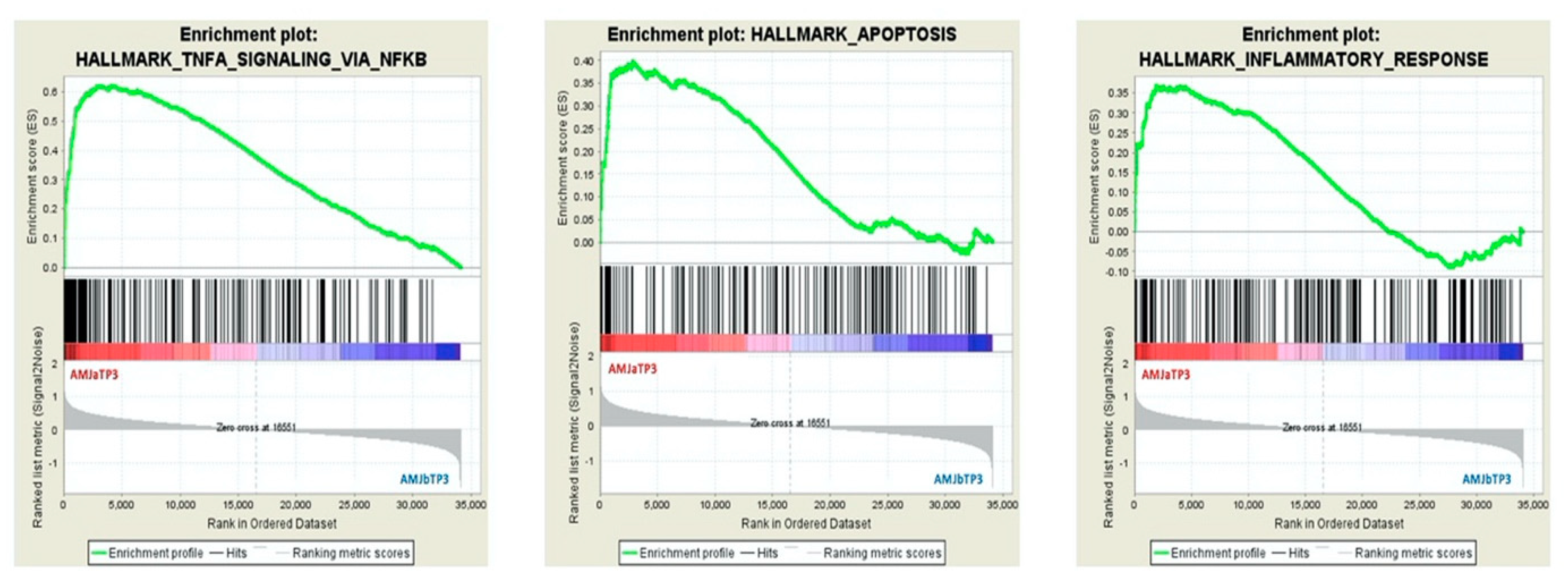
3.2. GSEA Analysis of PBMC Transcriptome
3.3. Enrichment Analysis of DEGs
3.4. RT-qPCR Validation of Selected DEGs
3.5. Effects of the Applied Treatments on Cardio-Metabolic Parameters and Leukocyte Count
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Piepoli, M.F.; Hoes, A.W.; Agewall, S.; Albus, C.; Brotons, C.; Catapano, A.L.; Cooney, M.T.; Corrà, U.; Cosyns, B.; Deaton, C.; et al. European Guidelines on cardiovascular disease prevention in clinical practice: The sixth joint task force of the European Society of Cardiology and other societies on cardiovascular disease prevention in clinical practice (constituted by representatives of 10 societies and by invited experts) developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Atherosclerosis 2016, 252, 207–274. [Google Scholar] [CrossRef] [PubMed]
- Menezes, R.; Rodriguez-Mateos, A.; Kaltsatou, A.; González-Sarrías, A.; Greyling, A.; Giannaki, C.; Andres-Lacueva, C.; Milenković, D.; Gibney, E.R.; Dumont, J.; et al. Impact of flavonols on cardiometabolic biomarkers: A meta-analysis of randomized controlled human trials to explore the role of inter-individual variability. Nutrients 2017, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- González-Sarrías, A.; García-Villalba, R.; Romo-Vaquero, M.; Alasalvar, C.; Örem, A.; Zafrilla, P.; Tomás-Barberán, F.A.; Selma, M.V.; Espín, J.C. Clustering according to urolithin metabotype explains the interindividual variability in the improvement of cardiovascular risk biomarkers in overweight-obese individuals consuming pomegranate: A randomized clinical trial. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- García-Conesa, M.T.; Chambers, K.; Combet, E.; Pinto, P.; Garcia-Aloy, M.; Andrés-Lacueva, C.; de Pascual-Teresa, S.; Mena, P.; Konic Ristic, A.; Hollands, W.J.; et al. Meta-analysis of the effects of foods and derived products containing ellagitannins and anthocyanins on cardiometabolic biomarkers: Analysis of factors influencing variability of the individual responses. Int. J. Mol. Sci. 2018, 19, 694. [Google Scholar] [CrossRef]
- Milenković, D.; Morand, C.; Cassidy, A.; Konic-Ristic, A.; Tomás-Barberán, F.; Ordovas, J.M.; Kroon, P.; De Caterina, R.; Rodriguez-Mateos, A. Interindividual variability in biomarkers of cardiometabolic health after consumption of major plant-food bioactive compounds and the determinants involved. Adv. Nutr. 2017, 8, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Williamson, G. The role of polyphenols in modern nutrition. Nutr. Bull. 2017, 42, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Milenković, D.; Van de Wiele, T.; Rodriguez-Mateos, A.; de Roos, B.; Garcia-Conesa, M.T.; Landberg, R.; Gibney, E.R.; Heinonen, M.; Tomás-Barberán, F.; et al. Addressing the inter-individual variation in response to consumption of plant food bioactives: Towards a better understanding of their role in healthy aging and cardiometabolic risk reduction. Mol. Nutr. Food. Res. 2017, 61, 1600557. [Google Scholar] [CrossRef]
- Borowska, S.; Brzóska, M.M. Chokeberries (Aronia melanocarpa) and their products as a possible means for the prevention and treatment of noncommunicable diseases and unfavorable health effects due to exposure to xenobiotics. Compr. Rev. Food Sci. Food Saf. 2016, 15, 982–1017. [Google Scholar] [CrossRef]
- Parzonko, A.; Naruszewicz, M. Cardioprotective effects of Aronia melanocarpa anthocynanins. From laboratory experiments to clinical practice. Curr. Pharm. Des. 2016, 22, 174–179. [Google Scholar] [CrossRef]
- Appel, K.; Meiser, P.; Millán, E.; Collado, J.A.; Rose, T.; Gras, C.C.; Muñoz, E. Chokeberry (Aronia melanocarpa (Michx.) Elliot) concentrate inhibits NF-kB and synergizes with selenium to inhibit the release of pro-inflammatory mediators in macrophages. Fitoterapia 2015, 105, 73–82. [Google Scholar] [CrossRef]
- Ho, G.T.; Bräunlich, M.; Austarheim, I.; Wangensteen, H.; Malterud, K.E.; Slimestad, R.; Barsett, H. Immunomodulating activity of Aronia melanocarpa polyphenols. Int. J. Mol. Sci. 2014, 15, 11626–11636. [Google Scholar] [CrossRef] [PubMed]
- Gualdoni, G.A.; Kovarik, J.J.; Hofer, J.; Dose, F.; Pignitter, M.; Doberer, D.; Steinberger, P.; Somoza, V.; Wolzt, M.; Zlabinger, G.J. Resveratrol enhances TNF-α production in human monocytes upon bacterial stimulation. Biochim. Biophys. Acta 2014, 1840, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.K.; van de Water, J.; Keen, C.L.; Schmitz, H.H.; Gershwin, M.E. Modulation of TNF-alpha secretion in peripheral blood mononuclear cells by cocoa flavanols and procyanidins. Dev. Immunol. 2002, 9, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.K.; Powell, J.; Van de Water, J.; Keen, C.L.; Schmitz, H.H.; Hammerstone, J.F.; Gershwin, M.E. The effect of cocoa procyanidins on the transcription and secretion of interleukin 1 beta in peripheral blood mononuclear cells. Life Sci. 2000, 66, 1377–1386. [Google Scholar] [CrossRef]
- Zunino, S.J.; Peerson, J.M.; Freytag, T.L.; Breksa, A.P.; Bonnel, E.L.; Woodhouse, L.R.; Storms, D.H. Dietary grape powder increases IL-1β and IL-6 production by lipopolysaccharide-activated monocytes and reduces plasma concentrations of large LDL and large LDL-cholesterol particles in obese humans. Br. J. Nutr. 2014, 112, 369–380. [Google Scholar] [CrossRef]
- Pastore, S.; Lulli, D.; Maurelli, R.; Dellambra, E.; De Luca, C.; Korkina, L.G. Resveratrol induces long-lasting IL-8 expression and peculiar EGFR activation/distribution in human keratinocytes: Mechanisms and implications for skin administration. PLoS ONE 2013, 8, e59632. [Google Scholar] [CrossRef]
- Jakobek, L.; Šeruga, M.; Medvidović-Kosanović, M.; Novak, I. Antioxidant activity and polyphenols of Aronia in comparison to other berry species. Agric. Consp. Sci. 2007, 72, 301–306. [Google Scholar]
- Hwang, S.J.; Yoon, W.B.; Lee, O.-H.; Cha, S.J.; Dai Kim, J. Radical-scavenging-linked antioxidant activities of extracts from black chokeberry and blueberry cultivated in Korea. Food Chem. 2014, 146, 71–77. [Google Scholar] [CrossRef]
- Kardum, N.; Konic Ristic, A.; Zec, M.; Kojadinovic, M.; Petrovic-Oggiano, G.; Zekovic, M.; Kroon, P.A.; Glibetić, M. Design, formulation and sensory evaluation of a polyphenol-rich food placebo: An example of aronia juice for food intervention studies. Int. J. Food Sci. Nutr. 2017, 68, 742–749. [Google Scholar] [CrossRef]
- Wolfe, K.; Wu, X.; Liu, R.H. Antioxidant activity of apple peels. J. Agric. Food Chem. 2003, 51, 609–614. [Google Scholar] [CrossRef]
- Gurinović, M.; Milešević, J.; Kadvan, A.; Nikolić, M.; Zeković, M.; Djekić-Ivanković, M.; Dupouy, E.; Finglas, P.; Glibetić, M. Development, features and application of DIET ASSESS & PLAN (DAP) software in supporting public health nutrition research in Central Eastern European Countries (CEEC). Food Chem. 2018, 238, 186–194. [Google Scholar] [CrossRef]
- Gurinović, M.; Milešević, J.; Novaković, R.; Kadvan, A.; Djekić-Ivanković, M.; Šatalić, Z.; Korošec, M.; Spiroski, I.; Ranić, M.; Dupouy, E.; et al. Improving nutrition surveillance and public health research in Central and Eastern Europe/Balkan Countries using the Balkan Food Platform and dietary tools. Food Chem. 2016, 193, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Eijssen, L.M.T.; Goelela, V.S.; Kelder, T.; Adriaens, M.E.; Evelo, C.T.; Radonjic, M. A user-friendly workflow for analysis of Illumina gene expression bead array data available at the arrayanalysis.org portal. BMC Genom. 2015, 16, 482. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef]
- Shi, W.; Oshlack, A.; Smyth, G.K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010, 38, e204. [Google Scholar] [CrossRef]
- Smyth, G.K.; Michaud, J.; Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Diyagama, D.; Neilson, J.; van Laar, R.; Dobrovic, A.; Holloway, A.; Smyth, G.K. Empirical array quality weights in the analysis of microarray data. BMC Bioinform. 2006, 7, 261. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, A.K.; Simon, P.; Seeburger, J.; Hoffmann, J.; Ziemer, G.; Aebert, H.; Wendel, H.P. Cytokine gene expression in monocytes of patients undergoing cardiopulmonary bypass surgery evaluated by real-time PCR. J. Cell. Mol. Med. 2003, 7, 146–156. [Google Scholar] [CrossRef] [PubMed][Green Version]
- D’Amore, S.; Vacca, M.; Cariello, M.; Graziano, G.; D’Orazio, A.; Salvia, R.; Sasso, R.C.; Sabbà, C.; Palasciano, G.; Moschetta, A. Genes and miRNA expression signatures in peripheral blood mononuclear cells in healthy subjects and patients with metabolic syndrome after acute intake of extra virgin olive oil. Biochim. Biophys. Acta 2016, 1861, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Schwager, J.; Richard, N.; Widmer, F.; Raederstorff, D. Resveratrol distinctively modulates the inflammatory profiles of immune and endothelial cells. BMC Complement. Altern. Med. 2017, 17, 309. [Google Scholar] [CrossRef]
- Nilsson, J.; Hansson, G.K.; Shah, P.K. Immunomodulation of atherosclerosis: Implications for vaccine development. Arterioscler. Thromb. Vasc. Biol. 2005, 25, 18–28. [Google Scholar] [CrossRef]
- Zhou, X.; Paulsson, G.; Stemme, S.; Hansson, G.K. Hypercholesterolemia is associated with a T helper (Th) 1/Th2 switch of the autoimmune response in atherosclerotic apo E-knockout mice. J. Clin. Investig. 1998, 101, 1717–1725. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Caligiuri, G.; Hamsten, A.; Lefvert, A.K.; Hansson, G.K. LDL immunization induces T-cell-dependent antibody formation and protection against atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 108–114. [Google Scholar] [CrossRef]
- Heinrich, P.C.; Behrmann, I.; Haan, S.; Hermanns, H.M.; Müller-Newen, G.; Schaper, F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003, 374, 1–20. [Google Scholar] [CrossRef]
- Sun, D.; Li, S.; Wu, H.; Zhang, M.; Zhang, X.; Wei, L.; Qin, X.; Gao, E. Oncostatin M (OSM) protects against cardiac ischaemia/reperfusion injury in diabetic mice by regulating apoptosis, mitochondrial biogenesis and insulin sensitivity. J. Cell. Mol. Med. 2015, 19, 1296–1307. [Google Scholar] [CrossRef]
- Xie, Z.; Chan, E.C.; Druey, K.M. R4 regulator of G protein signaling (RGS) proteins in inflammation and immunity. AAPS J. 2016, 18, 294–304. [Google Scholar] [CrossRef]
- Tran, T.; Paz, P.; Velichko, S.; Cifrese, J.; Belur, P.; Yamaguchi, K.D.; Ku, K.; Mirshahpanah, P.; Reder, A.T.; Croze, E. Interferonβ-1b induces the expression of RGS1 a negative regulator of G-protein signaling. Int. J. Cell Biol. 2010, 2010, 529376. [Google Scholar] [CrossRef] [PubMed]
- Caunt, C.J.; Keyse, S.M. Dual-specificity MAP kinase phosphatases (MKPs): Shaping the outcome of MAP kinase signalling. FASEB J. 2013, 280, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Liu, Y.X.; Jin, Y.J.; Hall, E.J.; Barrett, J.C. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature 2003, 422, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wu, X.; Chen, Q.; Zhu, S.; Liu, Y.; Pan, D.; Chen, X.; Li, D. The anti-apoptotic and cardioprotective effects of salvianolic acid a on rat cardiomyocytes following ischemia/reperfusion by DUSP-mediated regulation of the ERK1/2/JNK pathway. PLoS ONE 2014, 9, e102292. [Google Scholar] [CrossRef]
- Jeffrey, K.L.; Brummer, T.; Rolph, M.S.; Liu, S.M.; Callejas, N.A.; Grumont, R.J.; Gillieron, C.; Mackay, F.; Grey, S.; Camps, M.; et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat. Immunol. 2006, 7, 274–283. [Google Scholar] [CrossRef]
- Lu, D.; Liu, L.; Ji, X.; Gao, Y.; Chen, X.; Liu, Y.; Jin, Y. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat. Immunol. 2015, 16, 1263–1273. [Google Scholar] [CrossRef]
- Boss, A.; Kao, C.H.; Murray, P.M.; Marlow, G.; Barnett, M.P.; Ferguson, L.R. Human intervention study to assess the effects of supplementation with olive leaf extract on peripheral blood mononuclear cell gene expression. Int. J. Mol. Sci. 2016, 17, 2019. [Google Scholar] [CrossRef]
- Barrera-Reyes, P.K.; Hernández-Ramírez, N.; Cortés, J.; Poquet, L.; Redeuil, K.; Rangel-Escareño, C.; Kussmann, M.; Silva-Zolezzi, I.; Tejero, M.E. Gene expression changes by high-polyphenols cocoa powder intake: A randomized crossover clinical study. Eur. J. Nutr. 2019, 58, 1887–1898. [Google Scholar] [CrossRef]
- Muredda, L.; Kępczyńska, M.A.; Zaibi, M.S.; Alomar, S.Y.; Trayhurn, P. IL-1β and TNFα inhibit GPR120 (FFAR4) and stimulate GPR84 (EX33) and GPR41 (FFAR3) fatty acid receptor expression in human adipocytes: Implications for the anti-inflammatory action of n-3 fatty acids. Arch. Physiol. Biochem. 2018, 124, 97–108. [Google Scholar] [CrossRef]
- Pokimica, B.; García-Conesa, M.T.; Zec, M.; Debeljak-Martačić, J.; Ranković, S.; Vidović, N.; Petrović-Oggiano, G.; Konić-Ristić, A.; Glibetić, M. Chokeberry juice containing polyphenols does not affect cholesterol or blood pressure but modifies the composition of plasma phospholipids fatty acids in individuals at cardiovascular risk. Nutrients 2019, 11, 850. [Google Scholar] [CrossRef]
- Gibney, E.R.; Milenković, D.; Combet, E.; Ruskovska, T.; Greyling, A.; González-Sarrías, A.; de Roos, B.; Tomás-Barberán, F.; Morand, C.; Rodriguez-Mateos, A. Factors influencing the cardiometabolic response to (poly)phenols and phytosterols: A review of the COST Action POSITIVe activities. Eur. J. Nutr. 2019, 58 (Suppl. 2), 37–47. [Google Scholar] [CrossRef] [PubMed]
- Chareonrungrueangchai, K.; Wongkawinwoot, K.; Anothaisintawee, T.; Reutrakul, S. Dietary Factors and Risks of Cardiovascular Diseases: An Umbrella Review. Nutrients 2020, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Cespedes, E.M.; Hu, F.B. Dietary patterns: From nutritional epidemiologic analysis to national guidelines. Am. J. Clin. Nutr. 2015, 101, 899–900. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jiménez, J.; Fezeu, L.; Touvier, M.; Arnault, N.; Manach, C.; Hercberg, S.; Galan, P.; Scalbert, A. Dietary intake of 337 polyphenols in French adults. Am. J. Clin. Nutr. 2011, 93, 1220–1228. [Google Scholar] [CrossRef]

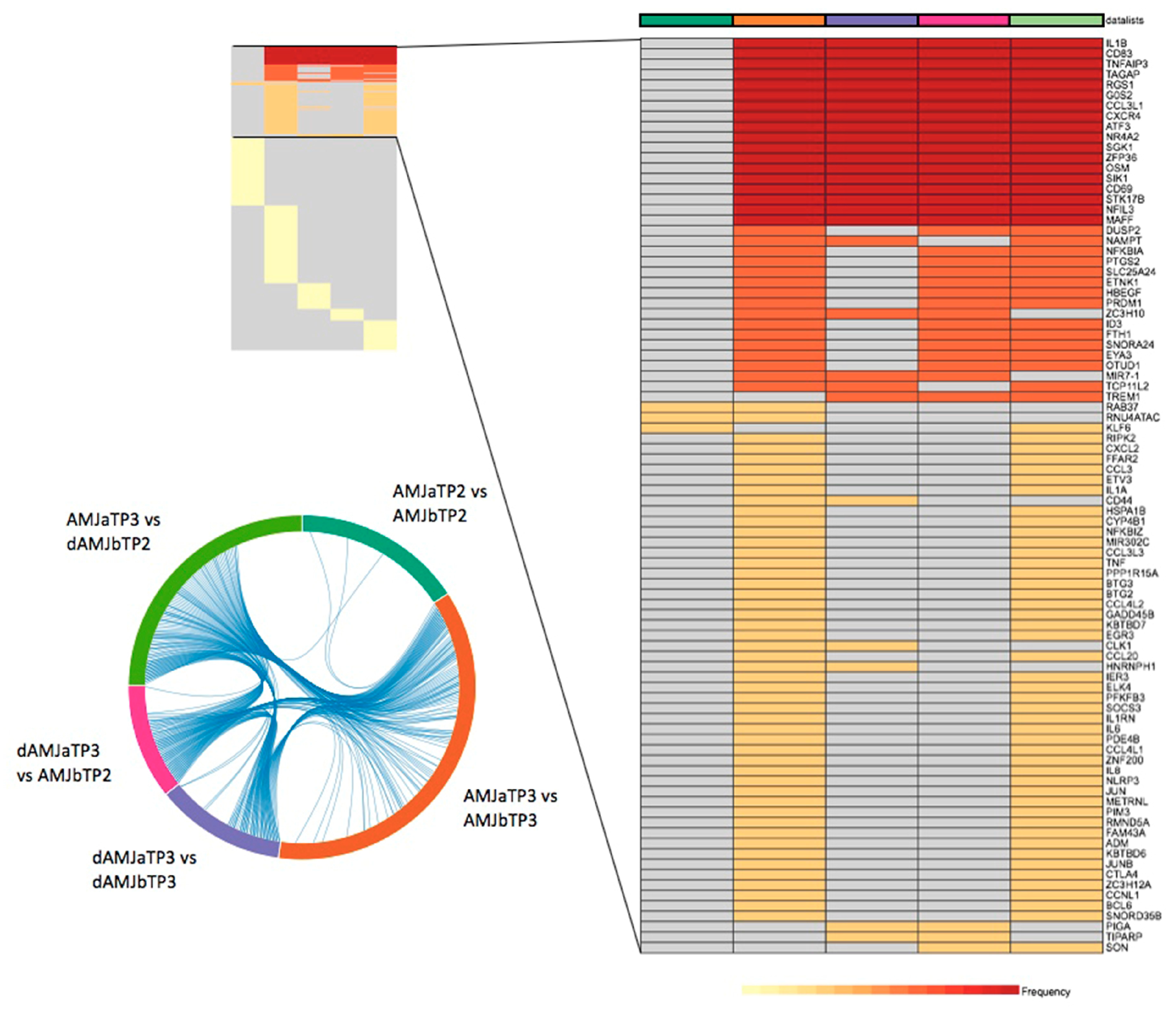
| Comparison | Number of DEGs * |
|---|---|
| PLB aTP1 vs. PLB bTP1 | 0 |
| AMJ aTP2 vs. AMJ bTP2 | 78 |
| dAMJ aTP2 vs. dAMJ bTP2 | 0 |
| AMJ aTP3 vs. AMJ bTP3 | 172 |
| dAMJ aTP3 vs. dAMJ bTP3 | 62 |
| AMJ aTP3 vs. dAMJ bTP2 | 115 |
| dAMJ aTP3 vs. AMJ bTP2 | 51 |
| Comparison | Enriched HALLMARK GENE SET | Size | ES | NES | FDR q-val |
|---|---|---|---|---|---|
| AMJ aTP3 vs. AMJ bTP3 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 199 | 0.622 | 3.05 | 0.000 |
| HALLMARK_APOPTOSIS | 161 | 0.399 | 1.8 | 0.002 | |
| HALLMARK_INFLAMMATORY_RESPONSE | 200 | 0.370 | 1.82 | 0.003 | |
| HALLMARK_INTERFERON_GAMMA_RESPONSE | 199 | 0.370 | 1.80 | 0.002 | |
| HALLMARK_INTERFERON_ALPHA_RESPONSE | 96 | 0.354 | 1.54 | 0.034 | |
| HALLMARK_P53_PATHWAY | 196 | 0.306 | 1.49 | 0.047 | |
| HALLMARK_HYPOXIA | 197 | 0.304 | 1.48 | 0.045 | |
| HALLMARK_UV_RESPONSE_UP | 158 | 0.314 | 1.48 | 0.041 | |
| HALLMARK_IL6_JAK_STAT3_SIGNALING | 87 | 0.342 | 1.47 | 0.039 | |
| HALLMARK_KRAS_SIGNALING_UP | 200 | 0.298 | 1.46 | 0.037 | |
| dAMJ aTP3 vs. dAMJ bTP3 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 199 | 0.492 | 2.35 | 0.000 |
| AMJ aTP2 vs. AMJ bTP2 | None |
| Gene | AMJaTP2 vs. AMJbTP2 | dAMJaTP2 vs. dAMJbTP2 | AMJaTP3 vs. AMJbTP3 | dAMJaTP3 vs. dAMJbTP3 | PLBaTP1 vs. PLBbTP1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FC | p * | FC | p * | FC | p * | FC | p * | FC | p * | |
| IL1B | 0.79 | 0.799 | 1.01 | 0.948 | 26.41 † | 0.025 | 47.65 † | 0.021 | 1.50 | 0.173 |
| TNF | 0.70 | 0.284 | 0.96 | 0.594 | 23.64 † | 0.012 | 16.07 | 0.021 | 1.32 | 0.139 |
| IL8 | 0.87 | 0.508 | 3.06 | 0.021 | 9.14 † | 0.093 | 30.11 | 0.008 | 3.50 | 0.008 |
| DUSP2 | 0.93 | 0.646 | 1.04 | 0.767 | 5.34 † | 0.036 | 4.89 | 0.015 | 1.58 | 0.017 |
| OSM | 0.96 | 0.721 | 1.08 | 0.515 | 3.62 † | 0.025 | 3.89 † | 0.001 | 2.00 | 0.011 |
| RGS1 | 1.16 | 0.878 | 1.1 | 0.953 | 4.04 † | 0.036 | 4.83 † | 0.001 | 1.25 | 0.028 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stojković, L.; Jovanović, I.; Zivković, M.; Zec, M.; Djurić, T.; Zivotić, I.; Kuveljić, J.; Kolaković, A.; Kolić, I.; Djordjević, A.; et al. The Effects of Aronia melanocarpa Juice Consumption on the mRNA Expression Profile in Peripheral Blood Mononuclear Cells in Subjects at Cardiovascular Risk. Nutrients 2020, 12, 1484. https://doi.org/10.3390/nu12051484
Stojković L, Jovanović I, Zivković M, Zec M, Djurić T, Zivotić I, Kuveljić J, Kolaković A, Kolić I, Djordjević A, et al. The Effects of Aronia melanocarpa Juice Consumption on the mRNA Expression Profile in Peripheral Blood Mononuclear Cells in Subjects at Cardiovascular Risk. Nutrients. 2020; 12(5):1484. https://doi.org/10.3390/nu12051484
Chicago/Turabian StyleStojković, Ljiljana, Ivan Jovanović, Maja Zivković, Manja Zec, Tamara Djurić, Ivan Zivotić, Jovana Kuveljić, Ana Kolaković, Ivana Kolić, Ana Djordjević, and et al. 2020. "The Effects of Aronia melanocarpa Juice Consumption on the mRNA Expression Profile in Peripheral Blood Mononuclear Cells in Subjects at Cardiovascular Risk" Nutrients 12, no. 5: 1484. https://doi.org/10.3390/nu12051484
APA StyleStojković, L., Jovanović, I., Zivković, M., Zec, M., Djurić, T., Zivotić, I., Kuveljić, J., Kolaković, A., Kolić, I., Djordjević, A., Glibetić, M., Alavantić, D., & Stanković, A. (2020). The Effects of Aronia melanocarpa Juice Consumption on the mRNA Expression Profile in Peripheral Blood Mononuclear Cells in Subjects at Cardiovascular Risk. Nutrients, 12(5), 1484. https://doi.org/10.3390/nu12051484