A Two-Sample Mendelian Randomization Analysis Investigates Associations Between Gut Microbiota and Celiac Disease
Abstract
1. Introduction
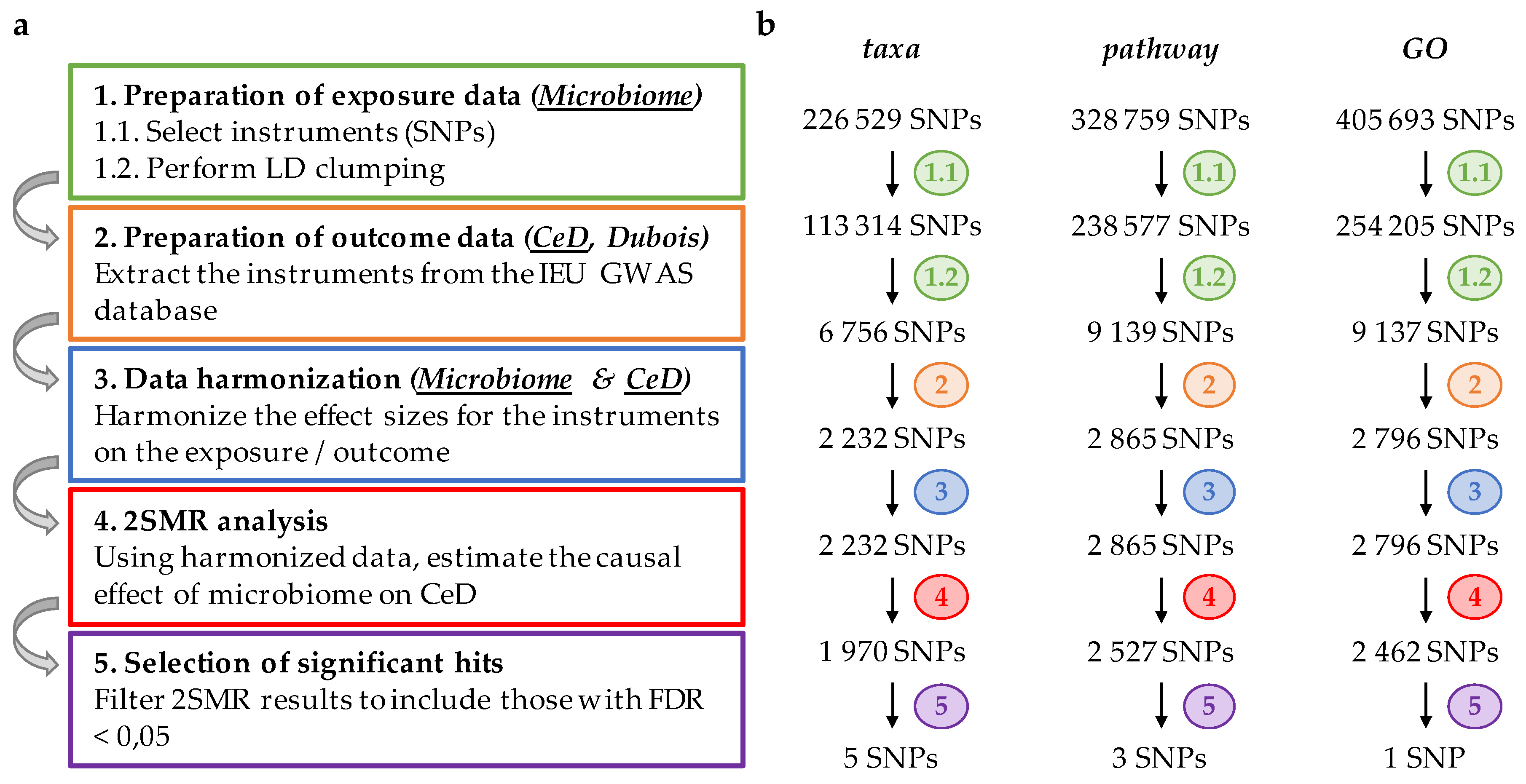
2. Materials and Methods
2.1. Preparation of Exposure Data
2.2. Preparation of Outcome Data
2.3. Harmonization of Exposure and Outcome Data
2.4. Two-Sample Mendelian Randomization (2SMR) and Statistical Analysis
3. Results
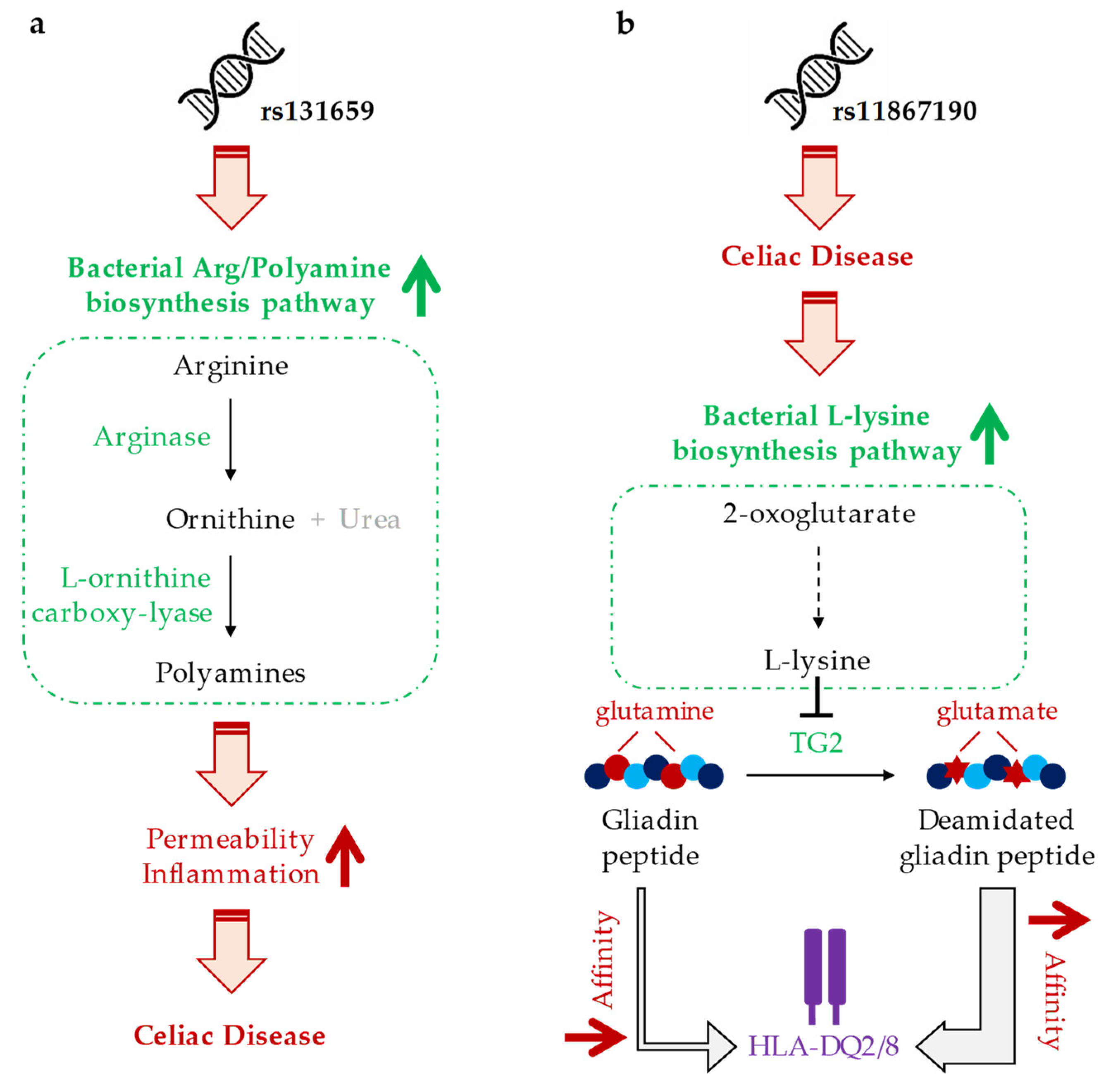
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lindfors, K.; Ciacci, C.; Kurppa, K.; Lundin, K.E.A.; Makharia, G.K.; Mearin, M.L.; Murray, J.A.; Verdu, E.F.; Kaukinen, K. Coeliac disease. Nat. Rev. Dis. Prim. 2019, 5, 1–18. [Google Scholar] [CrossRef]
- Bevan, S.; Popat, S.; Braegger, C.P.; Busch, A.; O’Donoghue, D.; Falth-Magnusson, K.; Ferguson, A.; Godkin, A.; Hogberg, L.; Holmes, G.; et al. Contribution of the MHC region to the familial risk of coeliac disease. J. Med. Genet. 1999, 36, 687–690. [Google Scholar]
- Trynka, G.; Hunt, K.A.; Bockett, N.A.; Romanos, J.; Mistry, V.; Szperl, A.; Bakker, S.F.; Bardella, M.T.; Bhaw-Rosun, L.; Castillejo, G.; et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat. Genet. 2011, 43, 1193–1201. [Google Scholar] [CrossRef]
- Dubois, P.C.A.; Trynka, G.; Franke, L.; Hunt, K.A.; Romanos, J.; Curtotti, A.; Zhernakova, A.; Heap, G.A.R.; Ádány, R.; Aromaa, A.; et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010, 42, 295–302. [Google Scholar] [CrossRef]
- Tamburini, S.; Shen, N.; Wu, H.C.; Clemente, J.C. The microbiome in early life: Implications for health outcomes. Nat. Med. 2016, 22, 713–722. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Wang, J.; Thingholm, L.B.; Skiecevičie, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.A.; Rühlemann, M.C.; Szymczak, S.; et al. Genome-wide association analysis identifies variation in Vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef]
- Turpin, W.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Kevans, D.; Smith, M.I.; Guttman, D.S.; Griffiths, A.; Panaccione, R.; Otley, A.; et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 2016, 48, 1413–1417. [Google Scholar] [CrossRef]
- Bonder, M.J.; Kurilshikov, A.; Tigchelaar, E.F.; Mujagic, Z.; Imhann, F.; Vila, A.V.; Deelen, P.; Vatanen, T.; Schirmer, M.; Smeekens, S.P.; et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016, 48, 1407–1412. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Davenport, E.R.; Beaumont, M.; Jackson, M.A.; Knight, R.; Ober, C.; Spector, T.D.; Bell, J.T.; Clark, A.G.; Ley, R.E. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe 2016, 19, 731–743. [Google Scholar] [CrossRef]
- Jackson, M.A.; Verdi, S.; Maxan, M.E.; Shin, C.M.; Zierer, J.; Bowyer, R.C.E.; Martin, T.; Williams, F.M.K.; Menni, C.; Bell, J.T.; et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; Del Vecchio Blanco, G.; et al. Metagenomics reveals dysbiosis and a potentially pathogenic N. flavescens strain in duodenum of adult celiac patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef]
- Bodkhe, R.; Shetty, S.A.; Dhotre, D.P.; Verma, A.K.; Bhatia, K.; Mishra, A.; Kaur, G.; Pande, P.; Bangarusamy, D.K.; Santosh, B.P.; et al. Comparison of small gut and whole gut microbiota of first-degree relatives with adult celiac disease patients and controls. Front. Microbiol. 2019, 10, 137–140. [Google Scholar] [CrossRef]
- De Palma, G.; Nadal, I.; Medina, M.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010, 10, 1–7. [Google Scholar] [CrossRef]
- Nadal, I.; Donant, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 2007, 56, 1669–1674. [Google Scholar] [CrossRef]
- Sellitto, M.; Bai, G.; Serena, G.; Fricke, W.F.; Sturgeon, C.; Gajer, P.; White, J.R.; Koenig, S.S.K.; Sakamoto, J.; Boothe, D.; et al. Proof of concept of microbiome-metabolome analysis and delayed gluten exposure on celiac disease autoimmunity in genetically at-risk infants. PLoS ONE 2012, 7, 33387. [Google Scholar] [CrossRef]
- Sánchez, E.; De Palma, G.; Capilla, A.; Nova, E.; Pozo, T.; Castillejo, G.; Varea, V.; Marcos, A.; Garrote, J.A.; Polanco, I.; et al. Influence of environmental and genetic factors linked to celiac disease risk on infant gut colonization by Bacteroides species. Appl. Environ. Microbiol. 2011, 77, 5316–5323. [Google Scholar] [CrossRef]
- Olivares, M.; Benítez-Páez, A.; de Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Marcos, A.; Garrote, J.A.; Polanco, I.; et al. Increased prevalence of pathogenic bacteria in the gut microbiota of infants at risk of developing celiac disease: The PROFICEL study. Gut Microbes 2018, 9, 551–558. [Google Scholar] [CrossRef]
- Olivares, M.; Walker, A.W.; Capilla, A.; Benítez-Páez, A.; Palau, F.; Parkhill, J.; Castillejo, G.; Sanz, Y. Gut microbiota trajectory in early life may predict development of celiac disease. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.C.; Timpson, N.; Smith, G.D. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef]
- Hartwig, F.P.; Davies, N.M.; Hemani, G.; Smith, G.D. Counterfactual causation: Avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int. J. Epidemiol. 2016, 45, 1717–1726. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-base platform supports systematic causal inference across the human phenome. Elife 2018, 7, 1–29. [Google Scholar] [CrossRef]
- Olivares, M.; Neef, A.; Castillejo, G.; De Palma, G.; Varea, V.; Capilla, A.; Palau, F.; Nova, E.; Marcos, A.; Polanco, I.; et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 2015, 64, 406–417. [Google Scholar] [CrossRef]
- Collado, M.C.; Calabuig, M.; Sanz, Y. Differences between the fecal microbiota of coeliac infants and healthy controls. Curr. Issues Intest. Microbiol. 2007, 8, 9–14. [Google Scholar]
- Ercolini, D.; Francavilla, R.; Vannini, L.; De Filippis, F.; Capriati, T.; Di Cagno, R.; Iacono, G.; De Angelis, M.; Gobbetti, M. From an imbalance to a new imbalance: Italian-style gluten-free diet alters the salivary microbiota and metabolome of African celiac children. Sci. Rep. 2015, 5, 18571. [Google Scholar] [CrossRef]
- Visconti, A.; Le Roy, C.I.; Rosa, F.; Rossi, N.; Martin, T.C.; Mohney, R.P.; Li, W.; de Rinaldis, E.; Bell, J.T.; Venter, J.C.; et al. Interplay between the human gut microbiome and host metabolism. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Wacklin, P.; Kaukinen, K.; Tuovinen, E.; Collin, P.; Lindfors, K.; Partanen, J.; Mäki, M.; Mättuö, J. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 2013, 19, 934–941. [Google Scholar] [CrossRef]
- Sánchez, E.; Donat, E.; Ribes-Koninckx, C.; Fernández-Murga, M.L.; Sanz, Y. Duodenal-mucosal bacteria associated with celiac disease in children. Appl. Environ. Microbiol. 2013, 79, 5472–5479. [Google Scholar] [CrossRef]
- Verdu, E.F.; Galipeau, H.J.; Jabri, B. Novel players in coeliac disease pathogenesis: Role of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 497–506. [Google Scholar] [CrossRef]
- Mullaney, J.A.; Stephens, J.E.; Costello, M.E.; Fong, C.; Geeling, B.E.; Gavin, P.G.; Wright, C.M.; Spector, T.D.; Brown, M.A.; Hamilton-Williams, E.E. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome 2018, 6. [Google Scholar]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Rotoli, B.M.; Visigalli, R.; Ingoglia, F.; Cirlini, M.; Prandi, B.; Dall’Asta, V. Gliadin-mediated production of polyamines by RAW264.7 macrophages modulates intestinal epithelial permeability in vitro. Biochim. Biophys. Acta—Mol. Basis Dis. 2015, 1852, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Barilli, A.; Gaiani, F.; Prandi, B.; Cirlini, M.; Ingoglia, F.; Visigalli, R.; Rotoli, B.M.; De’Angelis, N.; Sforza, S.; De’Angelis, G.L.; et al. Gluten peptides drive healthy and celiac monocytes toward an M2-like polarization. J. Nutr. Biochem. 2018, 54, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Elli, L.; Roncoroni, L.; Hils, M.; Pasternack, R.; Barisani, D.; Terrani, C.; Vaira, V.; Ferrero, S.; Bardella, M.T. Immunological effects of transglutaminase-treated gluten in coeliac disease. Hum. Immunol. 2012, 73, 992–997. [Google Scholar] [CrossRef]
- Wang, J.; Kurilshikov, A.; Radjabzadeh, D.; Turpin, W.; Croitoru, K.; Bonder, M.J.; Jackson, M.A.; Medina-Gomez, C.; Frost, F.; Homuth, G.; et al. Meta-analysis of human genome-microbiome association studies: The MiBioGen consortium initiative. Microbiome 2018, 6. [Google Scholar] [CrossRef]
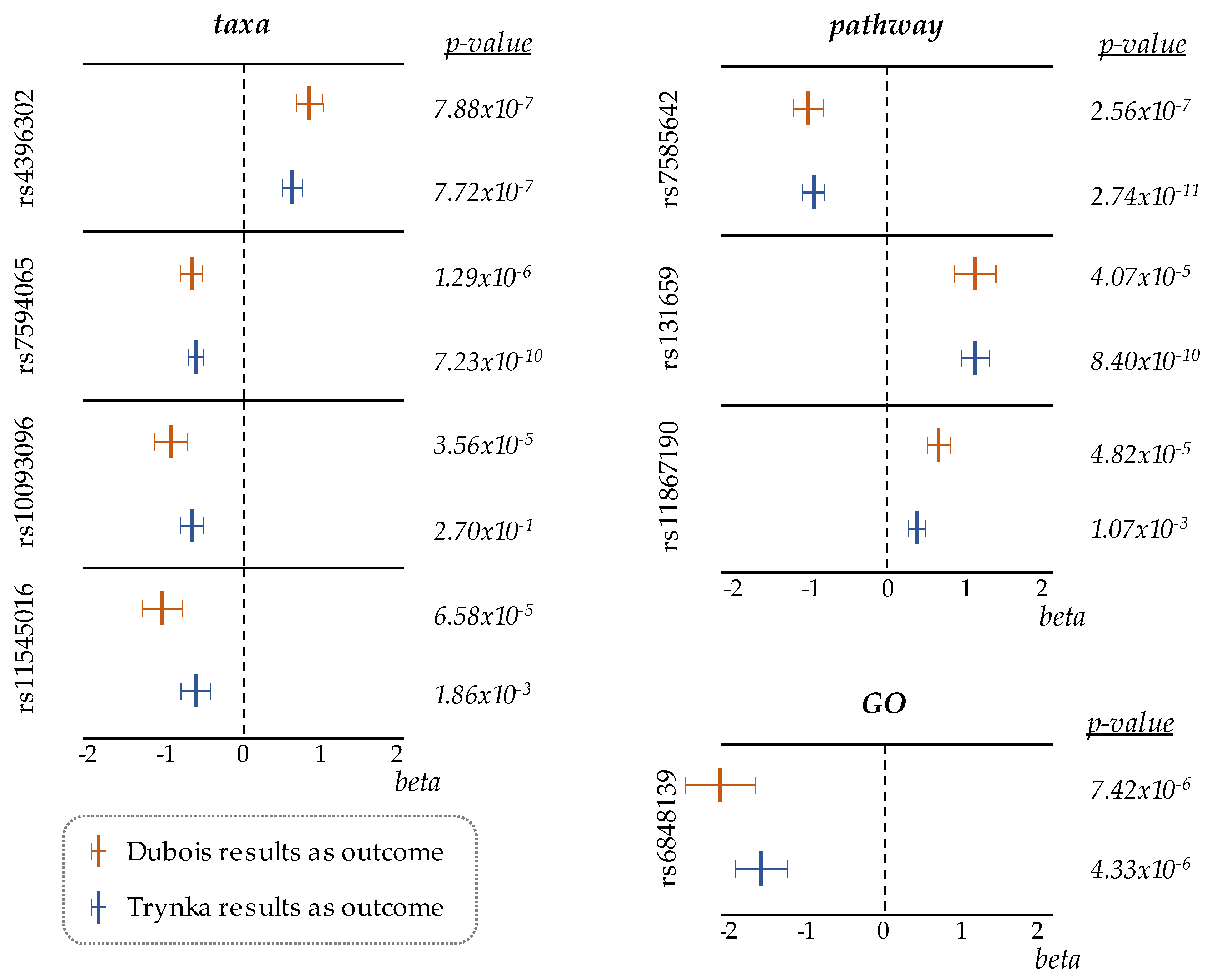
| SNP | Effect/Other Alleles | Chr. | Position | p-Value | Effect size ± SE | FDR |
|---|---|---|---|---|---|---|
| taxa | ||||||
| rs4396302 | A/G | 11 | 128420926 | 7.88 × 10−7 | 0.80 ± 0.16 | 0.001 |
| rs7594065 | T/C | 2 | 204814676 | 1.29 × 10−6 | −0.61 ± 0.13 | 0.001 |
| rs10093096 | C/T | 8 | 64907701 | 3.56 × 10−5 | −0.84 ± 0.20 | 0.027 |
| rs11545016 | T/C | 8 | 22438313 | 6.58 × 10−5 | −0.96 ± 0.24 | 0.037 |
| rs12913063 | T/C | 15 | 75424593 | 9.97 × 10−5 | 1.09 ± 0.28 | 0.044 |
| pathway | ||||||
| rs7585642 | A/C | 2 | 61217542 | 2.56 × 10−7 | −0.92 ± 0.18 | 0.001 |
| rs131659 | G/A | 22 | 21964761 | 4.07 × 10−5 | 1.04 ± 0.25 | 0.046 |
| rs11867190 | A/G | 17 | 5261220 | 4.82 × 10−5 | 0.59 ± 0.14 | 0.046 |
| GO | ||||||
| rs6848139 | C/A | 4 | 123395041 | 7.42 × 10−6 | −1.95 ± 0.43 | 0.021 |
| SNP | Associated Microbiota Trait |
|---|---|
| taxa | |
| rs4396302 | Firmicutes (p), Clostridia (c), Clostridiales (o), Peptostreptococcaceae (f), Peptostreptococcaceae (g), Peptostreptococcaceae unclassified (s) |
| rs7594065 | Firmicutes (p), Clostridia (c), Clostridiales (o), Clostridiales noname (f), Pseudoflavonifractor (g) |
| rs10093096 | Proteobacteria (p) |
| rs11545016 | Firmicutes (p), Clostridia (c), Clostridiales (o), Lachnospiraceae (f), Lachnospiraceae noname (g) |
| pathway | |
| rs7585642 | PWY-6060 (malonate degradation II, biotin-dependent) |
| rs131659 | ARG+POLYAMINE-SYN (super pathway of arginine and polyamine biosynthesis) |
| rs11867190 | PWY-3081 (L-lysine biosynthesis V) |
| GO | |
| rs6848139 | GO:0016831 (MF, carboxy-lyase activity) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Santisteban, I.; Cilleros-Portet, A.; Moyua-Ormazabal, E.; Kurilshikov, A.; Zhernakova, A.; Garcia-Etxebarria, K.; Fernandez-Jimenez, N.; Bilbao, J.R. A Two-Sample Mendelian Randomization Analysis Investigates Associations Between Gut Microbiota and Celiac Disease. Nutrients 2020, 12, 1420. https://doi.org/10.3390/nu12051420
García-Santisteban I, Cilleros-Portet A, Moyua-Ormazabal E, Kurilshikov A, Zhernakova A, Garcia-Etxebarria K, Fernandez-Jimenez N, Bilbao JR. A Two-Sample Mendelian Randomization Analysis Investigates Associations Between Gut Microbiota and Celiac Disease. Nutrients. 2020; 12(5):1420. https://doi.org/10.3390/nu12051420
Chicago/Turabian StyleGarcía-Santisteban, Iraia, Ariadna Cilleros-Portet, Elisabet Moyua-Ormazabal, Alexander Kurilshikov, Alexandra Zhernakova, Koldo Garcia-Etxebarria, Nora Fernandez-Jimenez, and Jose Ramon Bilbao. 2020. "A Two-Sample Mendelian Randomization Analysis Investigates Associations Between Gut Microbiota and Celiac Disease" Nutrients 12, no. 5: 1420. https://doi.org/10.3390/nu12051420
APA StyleGarcía-Santisteban, I., Cilleros-Portet, A., Moyua-Ormazabal, E., Kurilshikov, A., Zhernakova, A., Garcia-Etxebarria, K., Fernandez-Jimenez, N., & Bilbao, J. R. (2020). A Two-Sample Mendelian Randomization Analysis Investigates Associations Between Gut Microbiota and Celiac Disease. Nutrients, 12(5), 1420. https://doi.org/10.3390/nu12051420







