Abstract
The intestinal microbiome plays an important role in maintaining health throughout life. The microbiota develops progressively after birth and is influenced by many factors, including the mode of delivery, antibiotics, and diet. Maternal milk is critically important to the development of the neonatal intestinal microbiota. Different bioactive components of milk, such as human milk oligosaccharides, lactoferrin, and secretory immunoglobulins, modify the composition of the neonatal microbiota. In this article, we review the role of each of these maternal milk-derived bioactive factors on the microbiota and how this modulation of intestinal bacteria shapes health, and disease.
1. Introduction
Mammals feed their infants milk for the period before they are capable of acquiring food on their own. Milk is also a mechanism by which a mother can protect the mucosal surfaces of their immunologically inexperienced infants from infection. In addition, maternal milk shapes the acquisition and development of the bacteria, archaea, viruses, protists, and fungi, collectively termed the microbiome, that colonize our barrier surfaces. The composition of the microbiome is tailored to particular tissue sites and functions [1]. While the initial colonization of the infant gastrointestinal (GI) tract with the microbiome can be volatile, there is a characteristic path for the development of the nascent microbiome after birth that is shaped by components of maternal milk. While breast-feeding likely has effects on microbial communities outside the GI tract, the most obvious and deeply studied effects are on the intestinal microbiota. Here, we will review the nutritional and bioactive components of milk and their potential effects on the developing microbiome of the infant intestine.
2. Nutritional Components of Milk
The World Health Organization recommends exclusive breastfeeding to infants, as it provides all the nutrients required for the growth of the infant for the first 6 months of life and breastfeeding with food supplementation up to 2 years of age [2]. The nutritional components of human milk can be categorized into macronutrients (Table 1) and micronutrients (vitamins and minerals). Studies have observed that human milk changes in nutrition, composition, and quantity over time, with respect to different gestational periods (pre term vs. term), the diet of the mother, sex of the infant, and other environmental factors [3,4]. Human milk in the first few days of life is called colostrum, which is high in protein, whereas fats and carbohydrates are at lower concentrations. The importance of colostrum for the health of the newborn infant is illustrated by increased mortality rates in piglets who do not receive colostrum in the days directly post-delivery [5,6]. Maternal milk proteins can be classified into two main groups: antimicrobial/immune-stimulatory and nutritional. The proteins in the nutritional group assist in the absorption of vitamins and micronutrients in the intestine of the neonate and are a source of amino acids for the developing infant [7]. Colostrum, in particular, contains high concentrations of proteins that provide protection for neonates, including antibodies and other antimicrobials [3]. The anti-microbial proteins, listed in Table 2, will be discussed at length. Within each feed, the nutritional composition of milk also varies. For example, the initial milk or foremilk, contains large amounts of water and carbohydrates, whereas the milk that comes later, called the hind milk, is richer in lipids and proteins [8].

Table 1.
Composition of term and preterm human milk.

Table 2.
Proteins present in maternal milk.
The fat concentration in human milk is highly variable and mainly consists of high concentrations of triglycerides of oleic and palmitic acids. The concentration of these lipids is influenced by the diet, especially the fat intake of the mother, thus explaining variation in the lipid concentration of the milk [3,10]. How fat and in particular milk-derived fat, directly affects intestinal bacteria is not well understood.
Lactose is the main source of nutritive carbohydrates in maternal milk and can be used for energy by some bacteria, particularly Bifidobacteria spp. and Lactobacillus spp. [11]. However, lactose is unlikely to be a significant driver of microbiome heterogeneity in infants because it does not reach the terminal ileum and colon where most intestinal bacteria live and its concentration is relatively conserved between different mothers [11]. Other carbohydrates secreted by the mammary gland include milk oligosaccharides (HMOs), [9]. Though non-nutritive to the infant, HMOs are digested by intestinal bacteria and their effects on the microbiome will be discussed below in detail [12,13].
The micronutrients in the human milk include vitamins A, B1, B2, B6, B12, and D, non-protein nitrogen-containing compounds and minerals, such as sodium, potassium, magnesium, and zinc [9]. The concentrations of these micronutrients vary significantly with availability of these components in the mother, which is highly dependent upon the diet. Many of these minerals are also requirements for various bacterial members of the microbiota. For example, zinc is a limiting reagent for the proliferation of multiple types of intestinal bacteria [14].
While all of these nutritional components are directly taken up by the host (except for HMOs), many of them will also affect members of the microbiota, either directly or via effects on host mucosal health. Malnutrition (both over and under delivery of nutrients) can have enormous effects on both the pediatric and adult microbiome [15,16,17,18], so it follows that if milk is enriched or deficient in a particular nutrient it could have important effects on the nascent infant microbiome. Further research will be necessary to determine how the different concentrations of the nutritive components of maternal milk shape the microbiome.
3. The Intestinal Microbiome
The intestinal microbiome is comprised of various bacteria, viruses, archaea, and fungi. Functionally, the intestinal microbiota provides the host with the enzymes necessary to digest complex carbohydrates and is therefore necessary for the enzymatic function of the intestine [19,20]. Specifically, the microbiome is required for the production of short chain fatty acids (SCFAs) from dietary fiber, which are then absorbed in the intestine both as an energy source and to modulate the immune system [21,22,23]. In adults, SCFAs (acetate, propionate, and butyrate) are largely produced by strict anaerobic bacteria, such as Bacteroides and Firmicutes, that together form a healthy and diverse microbiome. Thus, the presence of SCFAs is a biomarker of health of the microbiota [20]. Germ-free mice, which are bred to completely lack a microbiome, also have deficits in vitamin B and K synthesis, which can affect prothrombin levels [24]. Beyond this core digestive function, the microbiome is important to many other processes. For instance, many orally administered drugs are actually pro-drugs whose efficacy is dependent upon modification by the intestinal microbiota [25].
4. Development of the Intestinal Microbiota after Birth
The intestinal microbiota must develop from an initial state of low diversity, characterized by colonization with a limited set of microorganisms, dominated by facultative anaerobes (Enterobacteriaceae, Enterococcaceae) to a diverse and interconnected anaerobic community [26,27]. It takes about 3 years for a child to develop an intestinal microbiome composition similar to adults, i.e., dominated by Bacteroides and Firmicutes [26,27]. The infant stage of microbiome development is affected by factors such as prenatal exposure to antibiotics or toxins, the mode of delivery, antibiotic exposure after birth, diet (breast feeding versus formula feeding), the introduction of solid food, and environmental factors, such as geography or climate [26,27,28,29,30,31]. The colonization of the infant microbiome begins at delivery when infants can acquire many different types of bacteria from the environment and the vagina, skin, and intestine of their mothers [32]. It has been hypothesized that the early stage microbiome is also affected by a putative placental microbiota [33,34]. However, given the trace numbers of bacteria measured in placental studies, it is difficult to judge their importance in the face of colonization with millions of rapidly proliferating bacteria post-delivery. The microbial composition of infants changes substantially in the days directly post-delivery and is shaped by a variety of factors. Within the first hour of birth, infants delivered vaginally had mostly Bacteroides and Lactobacillus while infants born through caesarean section had increased levels of facultative anaerobes such as Enterobacteriaceae, Enterococcaceae, and other bacteria derived from the skin in their microbiota [32,35]. These differences, however, are short lived, and by one month, vaginal and C-section infants cannot be separated based upon the composition of their microbiota [36]. Infants exposed to antibiotics at birth also showed increases in facultative anaerobes, mainly Enterobacteriaceae, and a decrease in Bacteroidetes, Bifidobacterium, Lactobacillus, and Clostridium species as compared to unexposed infants [28,37,38]. Perhaps the most important modifier of the intestinal microbiome, regardless of the developmental stage, is diet. Accordingly, the diet of the infant has significant effects on the developing microbiota. Breast-milk-fed infants have an increased abundance of Bifidobacterium and Bacteroides while those fed formula maintain facultative anaerobes (Enterobacteriaceae) for longer periods of time [31,39,40]. The known bioactive factors (HMOs, antibodies, etc.) of maternal milk that contribute to these differences will be discussed below.
In cases where the acquisition of the microbiota is perturbed, a dysbiotic intestinal microbiota can colonize the infant and lead to diseases such as necrotizing enterocolitis (NEC) and bacterial sepsis [41,42,43]. Disruption of the microbiome early-in-life is also associated with a higher incidence of chronic disease, such as obesity and atopy/asthma, in animal models [44,45,46,47], and is widely hypothesized to predispose human disease. Indeed, since the intestine has evolved to carry a microbiota, it uses that microbiota for developmental cues and to direct various functionalities. Many of the important functions of the microbiota have been exposed by germ-free mice, which have immune related defects in effector CD4 T cell differentiation, macrophage-activated motility and intestinal epithelial regeneration [48,49,50,51]. Therefore, the shaping of the microbiota by maternal milk may affect lifelong health. Accordingly, the benefits of breast milk on development are well documented, and associations can be made between feeding infants a primary diet of formula and the development of acute and chronic disease [52,53,54]. For example, human milk reduces the incidence of NEC and sepsis in preterm infants, a disease that is almost certainly associated with shifts in the microbiome [55,56]. Breast-feeding also decreases the incidence of allergy, asthma and obesity [57]. The transfer of milk-derived immune factors from mother to child has direct effects on the development of the immune system, the prevention of neonatal infection (such as respiratory syncytial virus) and, by extension, chronic inflammatory disease [58]. However, many of these same factors have important roles in determining the composition of the microbiome and so determining mechanistic explanations for bioactive factors has been a challenge. Importantly, cessation of maternal milk feeding also has benefits for immune development. In mice, the microbiome of the weaning period is critical to the development of regulatory T cells that can dampen intestinal inflammatory responses later in life [59]. Interestingly, epidermal growth factor in maternal milk, which reduces disease in neonatal mice, prevents this weaning response [59,60]. Thus, while maternal milk establishes microbiome health, it must also cease to allow proper immune function into adulthood. Below, we will discuss the specific components of maternal milk that directly affect the microbiome and how this relates to disease.
5. Human Milk Microbiota
Human milk is inhabited by its own microbiome that has been well characterized by culture and NextGen sequencing approaches [61]. While results have varied somewhat between studies, there has been a clear concordance where the milk microbiome is dominated by Staphylococcaceae and Streptococcaceae, with lesser amounts of Lactobacilliaceae, Corynebacteriaceae, and other organisms [61,62]. This structure is similar to the composition of the skin microbiome, and human milk is likely the source of the relatively large amounts of these bacteria that inhabit the neonatal intestine in the first month of life [63]. However, consistent breast feeding actually reduces the prevalence of these bacteria (in favor of Bifidobacteria and Bacteroides), so the importance of the milk microbiota in the development of the infant beyond the first few weeks of life is difficult to comprehend [31,39,40]. Further study is necessary to understand whether the human milk microbiome is necessary for establishing a healthy host/microbiome interaction in the infant.
6. Bioactive Components in Developing the Microbiome
The bioactive factors of maternal milk are non-nutritional but have a significant role in preventing infections and maintaining the mucosal epithelium and the development of the microbiome. Bioactive factors primarily consist of anti-microbials, growth factors, and white blood cells, all produced by the mother and transferred to the neonate through milk. Their source can vary, as some are produced by the maternal mammary gland epithelium and others extracted from the maternal serum. The major bioactive factors are listed in Table 3.

Table 3.
Bioactive factors in the maternal milk.
While, undoubtedly, many of these factors have direct functions in the development of the infant, here we will focus on those that are known to mediate their effects via modification of the microbiota.
7. Human Milk Oligosaccharides
Human milk oligosaccharides (HMOs) are a group of glycans with a lactose backbone at the reducing end that can be elongated by linear or branching sugars (glucose, galactose, fucose, N-acetylglucosamine, N-acetylneuraminic acid, or sialic acid) varying in length from 3 to 15 carbohydrates [65,66]. The lactose backbone of the HMOs in the maternal milk can be further fucosylated, glycosylated, or sialylated, thus forming different structural isomers [66]. There is a high number of HMOs in the maternal milk, among which about 200 have been identified through mass-spectrometry [66]. HMOs are produced by the mammary gland and are present in large amounts in human milk, about 1g/dL [9,67]. They are not broken down by the digestive enzymes of the infant and thus can reach to the distal ileum and colon [68]. HMOs are variable in each mother depending on the genetic composition, diet, body weight of the mother during pregnancy, and the stages of lactation [69,70,71,72]. Differences in the concentration of HMOs have also been observed between mothers of preterm and term infants [73,74]. Variations in HMOs have been proposed to contribute to variability in the protective effects of milk from different women against neonatal disease [67]. For instance, glycosylated or fucosylated HMOs have a similar structure to the glycosylated moieties found on the surface of the intestinal epithelium [75]. In fact, prior to the identification of their structure, HMOs were thought to be glycoproteins or glycolipids secreted from the mammary epithelium [65]. The similarity of HMO glycosylated motifs to cell surface receptors allows them to interfere with the binding of certain bacteria to the epithelium [76]. Specifically, the presence of HMOs inhibits the growth of pathogenic bacteria like Streptococcus pneumoniae and Campylobacter jejuni by interfering with the adhesion of bacteria to epithelial cells [76,77,78]. HMOs have also been demonstrated to inhibit the growth of stable toxin-expressing E. coli by binding to the stable toxin [79]. Glycosylated HMOs also prevent NEC and rotavirus infection in experimental murine models [13,80,81,82].
Bifidobacterium and Bacteroides bacteria metabolize HMOs and utilize them as an energy source [83]. The digestion of HMOs by Bifidobacteria is especially beneficial for the infant since the by-product is often SCFAs [83]. Therefore, the provision of HMOs can be seen as a way to both foster a healthy microbiome while also delivering SCFA to the intestines of infants that lack the physical capability and microbiome to digest fiber. Since the only source of HMOs is milk, studies have shown that infants who are fed with mother’s milk have increased abundance of obligate anaerobes, such as Bifidobacterium and Bacteroides, in the first few weeks of life as compared to formula-fed infants [84,85]. The duration of breastfeeding influences the presence of Bifidobacterium in the infants. Infants from countries with a higher mean breastfeeding duration had about 80%–90% colonization by Bifidobacterium longum subsp. infantis, whereas infants from countries with a low mean breastfeeding duration had about 0.7%–14% Bifidobacterium longum subsp. infantis colonization [85]. Even though the presence of HMOs in the milk facilitates the growth of certain Bifidobacterium species, the ability to digest HMOs does not always coincide with the dominance of the different species [86]. The most dominant strains of Bifidobacterium in the infant stool are Bifidobacterium longum subsp. longus and Bifidobacterium breve, but these strains have a limited capacity to digest HMOs. In contrast, the strains with a high HMO digestion capacity, Bifidobacterium bifidus and Bifidobacterium longum subsp. infantis, are found in low abundance in the infant stool. The exact mechanism behind these differences is not clear because we still do not know the source of Bifidobacterium that colonizes the infant intestine, though it is clearly not the human milk microbiome (as discussed above). Colonization, HMO digestion and the growth of different Bifidobacteria spp. may be complex and involve interactions between multiple different organisms, and further research is warranted to better understand HMO digestion and its role in the microbiome of infants [86].
8. Lactoferrin and Other Anti-Bacterial Proteins Found in Milk
Lactoferrin, referred to as the “red protein from milk”, is an iron binding protein known for its bacteriostatic properties [87]. It is produced from the glandular epithelial cells of mammals and also secreted in granules from neutrophils [88]. It is a member of the transferrin family of proteins and strongly binds iron [88]. The amount of lactoferrin in the milk varies depending on the stage of lactation as colostrum contains a high concentration of lactoferrin and it slowly decreases through the first month of lactation [89,90]. The reduced digestive capacity of the infant intestine protects milk-derived lactoferrin from degradation and maintains its functional properties, thus restricting the growth of bacteria in early life [91].
Recognition of the iron binding ability of lactoferrin led to the discovery of its anti-microbial and immunomodulatory functions [92]. Lactoferrin’s best described function is the sequestration of iron by high affinity binding, thus making it unavailable to bacteria in the gut, which require iron as an essential component for growth [93]. This bactericidal action has been demonstrated for many bacteria, including Streptococcus spp. and Vibrio spp., but some strains of bacteria, in particular the more invasive/pathogenic strains, have evolved iron-binding mechanisms to counteract lactoferrin and obtain iron [93]. Outside of its iron sequestration function, lactoferrin is also believed to have other bacteriocidal and immunomodulatory effects. For instance, it prevents the formation of biofilms in the intestinal tract and the interaction of microbes with the host epithelial cells [92,94]. Lactoferrin also interacts with the lipopolysaccharide (LPS) of gram-negative organisms and inhibits their growth [95]. In some contexts, lactoferrin supplied in maternal milk may also have effects on the host immune response. Lactoferrin induces macrophage activation to aid in the phagocytosis of gram-positive bacteria [92]. Lactoferrin also increased the activity of Myeloid-Derived Suppressor Cells (MDSCs) in both infants and mouse models, thus preventing experimental necrotizing enterocolitis in mice [96,97]. Indeed, the supplementation of lactoferrin to enteral feeds may provide protection against late-onset sepsis and NEC in preterm infants [98].
In addition to lactoferrin, there are other anti-bacterial proteins found in maternal milk that may affect the development of the infant microbiome. Lactoperoxidase is an enzyme present in the tears, saliva, and milk of mammals. Lactoperoxidase catalyzes the oxidation of thiocyanate with hydrogen peroxide to hypothiocyanite. Hypothiocyanite exhibits its anti-bacterial property by decreasing the viability of bacteria, viruses, and fungi [99]. Haptocorrin is a Vitamin B12 binding protein, previously shown to inhibit the growth of bacteria, but more recent studies have shown that the bactericidal/bacteriostatic activity could not be attributed to haptocorrin alone, so perhaps haptocorrin functions in concert with other anti-bacterial mediators [100].
9. Secretory Immunoglobulins
One of the most important anti-microbial bioactive factors in the maternal milk are the immunoglobulins, which provide passive immunity from infections to the newborn infant. The main antibodies present in the maternal milk are IgA, IgM, and IgG. IgA constitutes about 90%–95% of all antibodies, with IgM accounting for 2%–5% and IgG is less than 1% [101]. IgG can cross the placenta via the neonatal Fc Receptor (FcRn) and provide protection to the fetus in-utero and the infant for months post-delivery. In some mammals, including rodents and ruminants, FcRn is also highly expressed on the duodenum and can mediate transport of IgG from milk [102]. IgA and IgM, however, cannot cross the placenta and thus are only provided to the neonate through maternal milk to facilitate protection against mucosal infection and shape the microbiome.
Of these three antibody subtypes, the role for IgA in establishing a healthy intestinal microbiota is most clear. IgA-deficient patients have a unique intestinal microbiota, characterized by an increased relative abundance of intestinal Enterobacteriaceae [103,104,105]. Similar data have been observed in mouse models lacking in IgA (Igha−/− or Jh−/− mice), as they had equal numbers of intestinal bacteria as IgA sufficient controls but their microbiota was dominated by Enterobacteriaceae and segmented filamentous bacteria [106,107,108].
The structure of IgA consists of two IgA monomers bound together by disulphide linkages between the constant region and the J-chain (joining chain), which is produced in B cells [109]. Dimeric IgA is transported across the epithelium by the polymeric Ig receptor (pIgR). During transcytosis, a portion of the polypeptide chain of pIgR gets covalently attached to IgA after cleavage at the luminal surface of the epithelium and is called the secretory component [110]. Thus, dimeric IgA after transcytosis across the epithelial layer is known as secretory IgA (sIgA). Secretory component prevents the proteolytic degradation of IgA and is partially responsible for the high IgA/IgM ratio in the intestine since IgM is transported by pIgR at the same rate but is not irreversibly bound and protected by secretory component [109,111].
Secretory IgA is produced in the mucosal surfaces and provides anti-bacterial effects by binding to bacteria and preventing them from invading the mucosal epithelium. Unlike IgG or IgM, the anti-bacterial activity of sIgA is not mediated by complement-driven cytolysis or through opsonization and phagocytosis because these functions are blocked by binding of secretory component. Instead, IgA-based immunity against bacteria is hypothesized to be mediated by a combination of factors including: a) steric hindrance of bacterial surface molecules, b) increased uptake or sampling by the M-cells in the Peyer’s patches, c) through modification of bacterial transcription, or d) “enchaining” bacteria to prevent gene transfer [112,113,114,115]. Not all of these mechanisms are equally relevant to sIgA derived from maternal milk. For example, the increased uptake of sIgA-bound bacteria by Peyer’s patches is unlikely to be important since pups of IgA-deficient mice show increased B-cell activation and antibody production due to the lack of maternal IgA [116]. Bacterial enchainment may allow for a more efficient removal of inflammatory/invasive bacterial isolates from the microbiome and additionally prevent them from interacting with other bacteria in the intestine and acquiring new genetic material [114]. Conversely, secretory IgA can actually support the colonization of certain bacteria, such as Bacteroides, by acting as a carbohydrate source [117]. It also supports the stable colonization of Bacteroides by a distinct mucosal niche through common colonizing factor (ccf) regulation, which enhances coating by sIgA and increases the epithelial adherence of those bacteria [118]. Together, this suggests that IgA regulates the growth of a healthy microbiome in the intestine by promoting the growth of obligate anaerobes, such as Bacteroides and Firmicutes, while limiting the growth of inflammatory facultative anaerobes, such as Enterobacteriaceae.
Studies in both mice and humans have shown that maternal milk is the only source of sIgA for pups in the first weeks of life [116,119,120]. This is likely because it takes about 3–4 weeks for the neonatal intestine to be populated by IgA-secreting B cells [120,121]. IgA in the milk is related to the maternal microbiome because during pregnancy, intestinal IgA+ B cells traffic to the mammary gland where they secrete IgA into milk, as directed by the chemokine CCL28 [122,123]. Recent studies in mice have identified the presence of B-cells with the same variable regions in both the intestine and mammary glands, confirming the enteromammary B cell circuit [124]. Thus, the sIgA secreted by mammary gland B cells is shaped by the intestinal microorganisms that drive the strongest intestinal IgA responses in the mother [125]. However, since the bacteria present in the infant are not necessarily shared with the mother, maternal IgA may not possess all of the specificities necessary to bind all of the bacteria found in an infant. This is particularly true in preterm infants whose microbiome is more shaped to their physical environment and therefore dominated by facultative anaerobes that are largely absent from the maternal intestine [126]. Additionally, bacterial invasion of the infant will not generally trigger a maternal response, and, therefore, escape from IgA binding due to mutation, horizontal gene transfer, or colonization with a new strain will proceed without modification of the dominant maternal IgA response. In accord with this hypothesis, the relative fraction of IgA-bound bacteria, specifically Enterobacteriaceae, decreases prior to the onset of NEC, even though these children are receiving human milk [120]. Therefore, the specificities of IgA present in breast milk might be as important as the amount of antibody in shaping the infant microbiota and that IgA binding is necessary to restrict the growth of Enterobacteriaceae in the intestine. Since the anti-bacterial IgA repertoire is dependent upon the intestinal B cell repertoire [124], it is likely to differ between mothers according to differences in the microbiome and infectious history. The protection provided by maternal IgA is potentially important to the development of a healthy relationship between the microbiota and the host immune system. In mouse models, a lack of maternal IgA has been associated with more rapid and more robust immune responses in pups. Specifically, the pups of IgA-deficient mothers produce their own IgA much more rapidly (prior to weaning) and also show increased T cell activation in gut-associated lymphoid tissue [116,127]. These differences in lymphocytes are potentially important to the long-term health of the infant due to the longevity of the cells, though more research on this subject is necessary.
Despite its relatively low abundance in maternal milk, IgG has important effects on host-bacterial interaction in neonates. How IgG functions with regard to the microbiota is complex since it is transferred to the infant’s circulation (either in utero or via milk) or acts directly on bacteria within the intestine. However, it is clear that milk-derived IgG is critical for controlling enteric infections, independent of the effect of a placentally transferred antibody. Enterobacteriaceae-specific IgG in maternal milk has been shown to be critical for protecting neonatal mice against enteric infections with Citrobacter rodentium and enterotoxigenic E. coli [128,129]. IgG is also important for establishing homeostasis with regard to the newly colonizing microbiota by preventing the activation of the immune system in gut-associated lymphoid tissue and shaping Innate Lymphoid Cell development in the intestine [130,131]. Interestingly, mucosal IgG1 in mice targets Akkermansia muciniphilia, a bacteria that lives close to the intestinal epithelium and consumes mucus [132]. IgA also preferentially targets bacteria that live close to the epithelium, such as segmented filamentous bacteria [107]. Thus, perhaps both maternal milk-supplied IgG and IgA are a mechanism by which mothers can protect their children from invasion by the organisms that live closest to the intestine and are most likely to be capable of invasion.
10. Conclusions
The acquisition of microbiomes that are appropriate to the various body sites is an important component of development. If infants become colonized with microorganisms that are ill-suited to the barrier tissue site or are innately invasive and inflammatory, it can lead to both immediate and long-term health consequences. As a result, mammals have developed the ability to shape the intestinal microbiome of their infants and push it towards a state of health via the provision of milk (Figure 1). Here, we have reviewed the critical components of maternal milk that we know to shape the development of the microbiome, but important questions still remain. Unlike the anti-bacterial mechanisms of the internal organs, which are largely based on the killing of invasive organisms, the components at work in maternal milk are typically bacteriostatic and often we do not know how they mediate their effects to regulate the microbiome. For instance, over time the infant intestine becomes de-oxygenated, which is critical for the acquisition of fastidious anaerobes (Bifidobacteria, Bacteroides, Clostridia) necessary for the healthy function of the microbiome [133]. It has been hypothesized that facultative anaerobes can contribute to de-oxygenation, but these organisms are also dangerous for the host as this group (Enterobacteriaceae) contains many of the most inflammatory and invasive strains. How many facultative anaerobes are present in the infant intestine, how long they colonize, and how they associate to the infant intestine is clearly regulated by the components of mother’s milk, but how this might be balanced with a necessity for these organisms for de-oxygenation and colonization resistance is not known. For this and other questions related to maternal milk and the microbiome, we argue a comprehensive approach that tries to understand how particular host mediators affect bacterial colonization by shifts in bacterial gene expression is required. This ‘strain-centric’ approach is important because the effects of any given bioactive component of milk will change in response to shifts in the microbiome, driven either by new colonizers or evolution amongst long-term members. Furthermore, many bioactive factors, such as HMOs and the anti-bacterial repertoire of antibodies, differ significantly between mothers, implying that the fitness of any given bacterial isolate will be individualized to each host. Integrating analyses of the bacterial response to milk components with the host response, particularly the immune response, will lead to a better understanding of how the infant microbiome and thus maternal milk can shape the development of disease into adulthood.
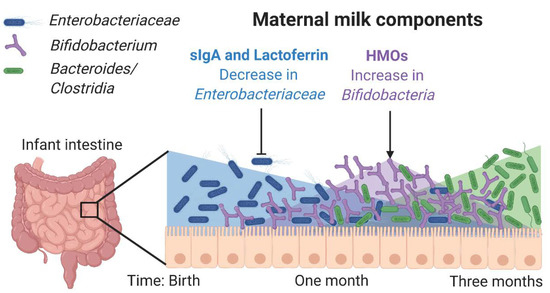
Figure 1.
Maternal milk components shape the microbiota. Enterobacteriaceae is one of the first colonizers of the infant intestine and is controlled by secretory IgA (sIgA) from milk, prior to the infant’s own production of sIgA. Lactoferrin inhibits many types of Enterobacteriaceae by binding iron and preventing epithelial adhesion. Human milk oligosaccharides (HMOs) support the outgrowth of Bifidobacteriaceae, which can convert HMOs into Short Chain Fatty Acids, which are an important energy source for the intestinal epithelium and also contribute to immunoregulation. HMOs also serve as decoy receptors limiting bacterial association with the intestinal epithelium. Together these components induce an environment conducive to the colonization of the strict anaerobes that will compose the healthy adult intestinal microbiome.
Author Contributions
K.P.G. and T.W.H. wrote the review. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the R.K. Mellon Foundation Institute for Pediatric Research and the National Institutes of Health (R01DK120697).
Acknowledgments
The authors would like to apologize that due to length requirements, not all work in this growing field could be discussed and properly cited. We would like to thank J. Tometich for critical reading of the manuscript.
Conflicts of Interest
The authors, K.P.G. and T.W.H., share a patent on a methodology to determine the specificity of antibodies from maternal milk samples.
References
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Eidelman, A.I.; Schanler, R. Breastfeeding and the use of human milk. Pediatrics 2012, 115, 496–506. [Google Scholar]
- Nommsen, L.A.; Lovelady, C.A.; Heinig, M.J.; Lönnerdal, B.; Dewey, K.G. Determinants of energy, protein, lipid, and lactose concentrations in human milk during the first 12 mo of lactation: The DARLING Study. Am. J. Clin. Nutr. 1991, 53, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Powe, C.E.; Knott, C.D.; Conklin-Brittain, N. Infant sex predicts breast milk energy content. Am. J. Hum. Biol. 2010, 22, 50–54. [Google Scholar] [CrossRef]
- Quesnel, H.; Farmer, C.; Devillers, N. Colostrum intake: Influence on piglet performance and factors of variation. Livest. Sci. 2012, 146, 105–114. [Google Scholar] [CrossRef]
- Devillers, N.; Le Dividich, J.; Prunier, A. Influence of colostrum intake on piglet survival and immunity. Animal 2011, 5, 1605–1612. [Google Scholar] [CrossRef]
- Lönnerdal, B. Human milk proteins. In Protecting Infants through Human Milk; Springer: Berlin/Heidelberg, Germany, 2004; pp. 11–25. [Google Scholar]
- Saarela, T.; Kokkonen, J.; Koivisto, M. Macronutrient and energy contents of human milk fractions during the first six months of lactation. Acta Paediatr. 2005, 94, 1176–1181. [Google Scholar] [CrossRef]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar]
- Martin, M.A.; Lassek, W.D.; Gaulin, S.J.; Evans, R.W.; Woo, J.G.; Geraghty, S.R.; Davidson, B.S.; Morrow, A.L.; Kaplan, H.S.; Gurven, M.D. Fatty acid composition in the mature milk of Bolivian forager-horticulturalists: Controlled comparisons with a US sample. Matern. Child Nutr. 2012, 8, 404–418. [Google Scholar] [CrossRef]
- Forsgard, R.A. Lactose digestion in humans: Intestinal lactase appears to be constitutive whereas the colonic microbiome is adaptable. Am. J. Clin. Nutr. 2019, 110, 273–279. [Google Scholar] [CrossRef]
- Newburg, D.S.; Ruiz-Palacios, G.M.; Morrow, A.L. Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 2005, 25, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Morrow, A.L.; Ruiz-Palacios, G.M.; Jiang, X.; Newburg, D.S. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 2005, 135, 1304–1307. [Google Scholar] [CrossRef] [PubMed]
- Lonergan, Z.R.; Skaar, E.P. Nutrient Zinc at the Host-Pathogen Interface. Trends Biochem. Sci. 2019, 44, 1041–1056. [Google Scholar] [CrossRef] [PubMed]
- Blanton, L.V.; Barratt, M.J.; Charbonneau, M.R.; Ahmed, T.; Gordon, J.I. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 2016, 352, 1533. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Hand, T.W. Role of nutrition, infection, and the microbiota in the efficacy of oral vaccines. Clin. Sci. 2018, 132, 1169–1177. [Google Scholar] [CrossRef]
- Backhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Tan, J.; McKenzie, C.; Potamitis, M.; Thorburn, A.N.; Mackay, C.R.; Macia, L. The role of short-chain fatty acids in health and disease. Adv. Immunol. 2014, 121, 91–119. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Haiser, H.J.; Van Treuren, W.; Garg, N.; Reddivari, L.; Vanamala, J.; Dorrestein, P.C.; Turnbaugh, P.J.; Knight, R. The intestinal metabolome: An intersection between microbiota and host. Gastroenterology 2014, 146, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Zimmermann-Kogadeeva, M.; Wegmann, R.; Goodman, A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570, 462–467. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 2014, 5, 427. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Zou, Z.-H.; Liu, D.; Li, H.-D.; Zhu, D.-P.; He, Y.; Hou, T.; Yu, J.-L. Prenatal and postnatal antibiotic exposure influences the gut microbiota of preterm infants in neonatal intensive care units. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 9. [Google Scholar] [CrossRef]
- Reyman, M.; van Houten, M.A.; van Baarle, D.; Bosch, A.A.; Man, W.H.; Chu, M.L.J.; Arp, K.; Watson, R.L.; Sanders, E.A.; Fuentes, S. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 2019, 10, 1–12. [Google Scholar]
- Chong, C.Y.L.; Bloomfield, F.H.; O’Sullivan, J.M. Factors affecting gastrointestinal microbiome development in neonates. Nutrients 2018, 10, 274. [Google Scholar] [CrossRef]
- Ho, N.T.; Li, F.; Lee-Sarwar, K.A.; Tun, H.M.; Brown, B.P.; Pannaraj, P.S.; Bender, J.M.; Azad, M.B.; Thompson, A.L.; Weiss, S.T. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat. Commun. 2018, 9, 4169. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed]
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65. [Google Scholar] [CrossRef] [PubMed]
- De Goffau, M.C.; Lager, S.; Sovio, U.; Gaccioli, F.; Cook, E.; Peacock, S.J.; Parkhill, J.; Charnock-Jones, D.S.; Smith, G.C. Human placenta has no microbiome but can contain potential pathogens. Nature 2019, 572, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Forster, S.C.; Tsaliki, E.; Vervier, K.; Strang, A.; Simpson, N.; Kumar, N.; Stares, M.D.; Rodger, A.; Brocklehurst, P. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019, 574, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef]
- Tapiainen, T.; Koivusaari, P.; Brinkac, L.; Lorenzi, H.A.; Salo, J.; Renko, M.; Pruikkonen, H.; Pokka, T.; Li, W.; Nelson, K. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Mazzola, G.; Murphy, K.; Ross, R.P.; Di Gioia, D.; Biavati, B.; Corvaglia, L.T.; Faldella, G.; Stanton, C. Early gut microbiota perturbations following intrapartum antibiotic prophylaxis to prevent group B streptococcal disease. PLoS ONE 2016, 11, e0157527. [Google Scholar] [CrossRef]
- Lee, S.A.; Lim, J.Y.; Kim, B.-S.; Cho, S.J.; Kim, N.Y.; Kim, O.B.; Kim, Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr. Res. Pract. 2015, 9, 242–248. [Google Scholar] [CrossRef]
- Madan, J.C.; Hoen, A.G.; Lundgren, S.N.; Farzan, S.F.; Cottingham, K.L.; Morrison, H.G.; Sogin, M.L.; Li, H.; Moore, J.H.; Karagas, M.R. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016, 170, 212–219. [Google Scholar] [CrossRef]
- Pammi, M.; Cope, J.; Tarr, P.I.; Warner, B.B.; Morrow, A.L.; Mai, V.; Gregory, K.E.; Kroll, J.S.; McMurtry, V.; Ferris, M.J. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome 2017, 5, 31. [Google Scholar] [CrossRef]
- Graham III, P.L.; Della-Latta, P.; Wu, F.; Zhou, J.; Saiman, L. The gastrointestinal tract serves as the reservoir for Gram-negative pathogens in very low birth weight infants. Pediatr. Infect. Dis. J. 2007, 26, 1153–1156. [Google Scholar] [CrossRef]
- Carl, M.A.; Ndao, I.M.; Springman, A.C.; Manning, S.D.; Johnson, J.R.; Johnston, B.D.; Burnham, C.-A.D.; Weinstock, E.S.; Weinstock, G.M.; Wylie, T.N. Sepsis from the gut: The enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis. 2014, 58, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Yamanishi, S.; Cox, L.; Methe, B.A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012, 488, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; An, D.; Zeissig, S.; Vera, M.P.; Richter, J.; Franke, A.; Glickman, J.N.; Siebert, R.; Baron, R.M.; Kasper, D.L.; et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012, 336, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Muller, P.A.; Koscso, B.; Rajani, G.M.; Stevanovic, K.; Berres, M.L.; Hashimoto, D.; Mortha, A.; Leboeuf, M.; Li, X.M.; Mucida, D.; et al. Crosstalk between Muscularis Macrophages and Enteric Neurons Regulates Gastrointestinal Motility. Cell 2014, 158, 1210. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef]
- Greer, F.R.; Sicherer, S.H.; Burks, A.W.; Committee On, N.; Section On, A. The Effects of Early Nutritional Interventions on the Development of Atopic Disease in Infants and Children: The Role of Maternal Dietary Restriction, Breastfeeding, Hydrolyzed Formulas, and Timing of Introduction of Allergenic Complementary Foods. Pediatrics 2019, 143. [Google Scholar] [CrossRef] [PubMed]
- Oddy, W.H. Breastfeeding, Childhood Asthma, and Allergic Disease. Ann. Nutr. Metab. 2017, 70, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Singhal, A. The role of infant nutrition in the global epidemic of non-communicable disease. Proc. Nutr. Soc. 2016, 75, 162–168. [Google Scholar] [CrossRef]
- Quigley, M.; Embleton, N.D.; McGuire, W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst. Rev. 2018, 6, CD002971. [Google Scholar] [CrossRef] [PubMed]
- Cortez, J.; Makker, K.; Kraemer, D.F.; Neu, J.; Sharma, R.; Hudak, M.L. Maternal milk feedings reduce sepsis, necrotizing enterocolitis and improve outcomes of premature infants. J. Perinatol. 2018, 38, 71–74. [Google Scholar] [CrossRef]
- Walker, W.A. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatr. Res. 2017, 82, 387–395. [Google Scholar] [CrossRef]
- Dixon, D.L. The Role of Human Milk Immunomodulators in Protecting Against Viral Bronchiolitis and Development of Chronic Wheezing Illness. Child 2015, 2, 289–304. [Google Scholar] [CrossRef]
- Al Nabhani, Z.; Dulauroy, S.; Marques, R.; Cousu, C.; Al Bounny, S.; Dejardin, F.; Sparwasser, T.; Berard, M.; Cerf-Bensussan, N.; Eberl, G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity 2019, 50, 1276–1288. [Google Scholar] [CrossRef]
- Good, M.; Sodhi, C.P.; Egan, C.E.; Afrazi, A.; Jia, H.; Yamaguchi, Y.; Lu, P.; Branca, M.F.; Ma, C.; Prindle, T., Jr.; et al. Breast milk protects against the development of necrotizing enterocolitis through inhibition of Toll-like receptor 4 in the intestinal epithelium via activation of the epidermal growth factor receptor. Mucosal Immunol. 2015, 8, 1166–1179. [Google Scholar] [CrossRef]
- Gomez-Gallego, C.; Garcia-Mantrana, I.; Salminen, S.; Collado, M.C. The human milk microbiome and factors influencing its composition and activity. In Seminars in Fetal and Neonatal Medicine; Elsevier: Amsterdam, The Netherlands, 2016; pp. 400–405. [Google Scholar]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Cacho, N.T.; Lawrence, R.M. Innate immunity and breast milk. Front. Immunol. 2017, 8, 584. [Google Scholar] [CrossRef]
- Newburg, D.S. Oligosaccharides in human milk and bacterial colonization. J. Pediatr. Gastroenterol. Nutr. 2000, 30, S8–S17. [Google Scholar] [CrossRef] [PubMed]
- Thomson, P.; Medina, D.A.; Garrido, D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018, 75, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef]
- Michaelsen, K.F.; Skafte, L.; Badsberg, J.H.; Jørgensen, M.J.J. Variation in macronutrients in human bank milk: Influencing factors and implications for human milk banking. J. Pediatr. Gastroenterol. Nutr. 1990, 11, 229–239. [Google Scholar] [CrossRef]
- Coppa, G.V.; Gabrielli, O.; Pierani, P.; Catassi, C.; Carlucci, A.; Giorgi, P.L. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics 1993, 91, 637–641. [Google Scholar]
- Sprenger, N.; De Castro, C.A.; Steenhout, P.; Thakkar, S.K. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS ONE 2017, 12, e0171814. [Google Scholar] [CrossRef]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef]
- Gabrielli, O.; Zampini, L.; Galeazzi, T.; Padella, L.; Santoro, L.; Peila, C.; Giuliani, F.; Bertino, E.; Fabris, C.; Coppa, G.V. Preterm milk oligosaccharides during the first month of lactation. Pediatrics 2011, 128, e1520–e1531. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.A.; Gaerlan, S.C.; Strum, J.S.; Dimapasoc, L.M.; Mirmiran, M.; Tancredi, D.J.; Smilowitz, J.T.; Kalanetra, K.M.; Mills, D.A.; German, J.B. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J. Proteome Res. 2012, 11, 4662–4672. [Google Scholar] [CrossRef] [PubMed]
- Kunz, C.; Rudloff, S.; Baier, W.; Klein, N.; Strobel, S. Oligosaccharides in human milk: Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 2000, 20, 699–722. [Google Scholar] [CrossRef] [PubMed]
- Porras, O.; Andersson, B.; Hanson, L.Å.; Lagergård, T.; Edén, C.S. Inhibition of Attachment of Streptococcus Pneumoniae and Haemophilus Influenzae by Human Milk. In Human Lactation 2; Springer: Berlin/Heidelberg, Germany, 1986; pp. 559–568. [Google Scholar]
- Andersson, B.; Porras, O.; Hanson, L.Å.; Lagergård, T.; Svanborg-Edén, C. Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J. Infect. Dis. 1986, 153, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, L.; Soto, L.; Newburg, D.; Ruiz-Palacios, G. Campylobacter jejuni receptor analogs present in human milk. Microbiol. Ecol. Health Dis. 1991, 4, S27. [Google Scholar]
- Newburg, D.S.; Pickering, L.K.; McCluer, R.H.; Cleary, T.G. Fucosylated oligosaccharides of human milk protect suckling mice from heat-stabile enterotoxin of Escherichia coli. J. Infect. Dis. 1990, 162, 1075–1080. [Google Scholar] [CrossRef]
- Good, M.; Sodhi, C.P.; Yamaguchi, Y.; Jia, H.; Lu, P.; Fulton, W.B.; Martin, L.Y.; Prindle, T.; Nino, D.F.; Zhou, Q. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br. J. Nutr. 2016, 116, 1175–1187. [Google Scholar] [CrossRef]
- Yolken, R.H.; Peterson, J.A.; Vonderfecht, S.L.; Fouts, E.T.; Midthun, K.; Newburg, D.S. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Investig. 1992, 90, 1984–1991. [Google Scholar] [CrossRef]
- Bode, L. Human Milk Oligosaccharides in the Prevention of Necrotizing Enterocolitis: A Journey From in vitro and in vivo Models to Mother-Infant Cohort Studies. Front Pediatr. 2018, 6, 385. [Google Scholar] [CrossRef]
- Kirmiz, N.; Robinson, R.C.; Shah, I.M.; Barile, D.; Mills, D.A. Milk Glycans and Their Interaction with the Infant-Gut Microbiota. Annu. Rev. Food Sci. Technol. 2018, 9, 429–450. [Google Scholar] [CrossRef]
- Lawson, M.A.; O’Neill, I.J.; Kujawska, M.; Javvadi, S.G.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2019, 14, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Taft, D.H.; Liu, J.; Maldonado-Gomez, M.X.; Akre, S.; Huda, M.N.; Ahmad, S.M.; Stephensen, C.B.; Mills, D.A. Bifidobacterial Dominance of the Gut in Early Life and Acquisition of Antimicrobial Resistance. Msphere 2018, 3, e00441-18. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, A.; Katoh, T.; Sakanaka, M.; Ling, Y.; Yamada, C.; Asakuma, S.; Urashima, T.; Tomabechi, Y.; Katayama-Ikegami, A.; Kurihara, S. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Groves, M.L. The isolation of a red protein from Milk2. J. Am. Chem. Soc. 1960, 82, 3345–3350. [Google Scholar] [CrossRef]
- Ward, P.; Paz, E.; Conneely, O. Lactoferrin. Cell. Mol. Life Sci. 2005, 62, 2540. [Google Scholar] [CrossRef]
- Rai, D.; Adelman, A.S.; Zhuang, W.; Rai, G.P.; Boettcher, J.; Lönnerdal, B. Longitudinal changes in lactoferrin concentrations in human milk: A global systematic review. Crit. Rev. Food Sci. Nutr. 2014, 54, 1539–1547. [Google Scholar] [CrossRef]
- Villavicencio, A.; Rueda, M.S.; Turin, C.G.; Ochoa, T.J. Factors affecting lactoferrin concentration in human milk: How much do we know? Biochem. Cell Biol. 2017, 95, 12–21. [Google Scholar] [CrossRef]
- Hamosh, M. Bioactive factors in human milk. Pediatr. Clin. N. Am. 2001, 48, 69–86. [Google Scholar] [CrossRef]
- Jenssen, H.; Hancock, R.E. Antimicrobial properties of lactoferrin. Biochimie 2009, 91, 19–29. [Google Scholar] [CrossRef]
- Arnold, R.R.; Cole, M.F. A bactericidal effect for human lactoferrin. Sci. Transl. Med. 1977, 197, 263–265. [Google Scholar] [CrossRef]
- García-Montoya, I.A.; Cendón, T.S.; Arévalo-Gallegos, S.; Rascón-Cruz, Q. Lactoferrin a multiple bioactive protein: An overview. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 226–236. [Google Scholar]
- Appelmelk, B.J.; An, Y.-Q.; Geerts, M.; Thijs, B.G.; De Boer, H.; MacLaren, D.M.; De Graaff, J.; Nuijens, J.H. Lactoferrin is a lipid A-binding protein. Immunity 1994, 62, 2628–2632. [Google Scholar] [CrossRef]
- He, Y.-M.; Li, X.; Perego, M.; Nefedova, Y.; Kossenkov, A.V.; Jensen, E.A.; Kagan, V.; Liu, Y.-F.; Fu, S.-Y.; Ye, Q.-J. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat. Med. 2018, 24, 224. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Perego, M.; Xiao, Q.; He, Y.; Fu, S.; He, J.; Liu, W.; Li, X.; Tang, Y.; Li, X. Lactoferrin-induced myeloid-derived suppressor cell therapy attenuates pathologic inflammatory conditions in newborn mice. J. Clin. Investig. 2019, 129. [Google Scholar] [CrossRef]
- Pammi, M.; Suresh, G. Enteral lactoferrin supplementation for prevention of sepsis and necrotizing enterocolitis in preterm infants. Cochrane Database Syst. Rev. 2017, 6, CD007137. [Google Scholar] [CrossRef]
- Welk, A.; Meller, C.; Schubert, R.; Schwahn, C.; Kramer, A.; Below, H.J.B. Effect of lactoperoxidase on the antimicrobial effectiveness of the thiocyanate hydrogen peroxide combination in a quantitative suspension test. BMC Microbiol. 2009, 9, 134. [Google Scholar] [CrossRef]
- Jensen, H.R.; Laursen, M.F.; Lildballe, D.L.; Andersen, J.B.; Nexø, E.; Licht, T.R. Effect of the vitamin B12-binding protein haptocorrin present in human milk on a panel of commensal and pathogenic bacteria. BMC Res. Notes 2011, 4, 208. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Secretory IgA: Designed for anti-microbial defense. Front. Immunol. 2013, 4, 222. [Google Scholar] [CrossRef]
- Suleiman, Y.B.; Yoshida, M.; Nishiumi, S.; Tanaka, H.; Mimura, T.; Nobutani, K.; Yamamoto, K.; Takenaka, M.; Aoganghua, A.; Miki, I. Neonatal Fc receptor for IgG (FcRn) expressed in the gastric epithelium regulates bacterial infection in mice. Mucosal Immunol. 2012, 5, 87–98. [Google Scholar] [CrossRef]
- Catanzaro, J.R.; Strauss, J.D.; Bielecka, A.; Porto, A.F.; Lobo, F.M.; Urban, A.; Schofield, W.B.; Palm, N.W. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Fadlallah, J.; El Kafsi, H.; Sterlin, D.; Juste, C.; Parizot, C.; Dorgham, K.; Autaa, G.; Gouas, D.; Almeida, M.; Lepage, P.; et al. Microbial ecology perturbation in human IgA deficiency. Sci. Transl. Med. 2018, 10, eaan1217. [Google Scholar] [CrossRef] [PubMed]
- Planer, J.D.; Peng, Y.; Kau, A.L.; Blanton, L.V.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Gordon, J.I. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 2016, 534, 263. [Google Scholar] [CrossRef] [PubMed]
- Mirpuri, J.; Raetz, M.; Sturge, C.R.; Wilhelm, C.L.; Benson, A.; Savani, R.C.; Hooper, L.V.; Yarovinsky, F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes 2014, 5, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Meek, B.; Doi, Y.; Muramatsu, M.; Chiba, T.; Honjo, T.; Fagarasan, S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl. Acad. Sci. USA 2004, 101, 1981–1986. [Google Scholar] [CrossRef]
- Kawamoto, S.; Maruya, M.; Kato, L.M.; Suda, W.; Atarashi, K.; Doi, Y.; Tsutsui, Y.; Qin, H.; Honda, K.; Okada, T.; et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity 2014, 41, 152–165. [Google Scholar] [CrossRef]
- Woof, J.; Russell, M. Structure and function relationships in IgA. Mucosal Immunol. 2011, 4, 590. [Google Scholar] [CrossRef]
- Pabst, O.; Slack, E. IgA and the intestinal microbiota: The importance of being specific. Mucosal Immunol. 2019, 13, 12–21. [Google Scholar] [CrossRef]
- Brandtzaeg, P. Human secretory immunoglobulin M. An immunochemical and immunohistochemical study. Immunology 1975, 29, 559. [Google Scholar]
- Cullender, T.C.; Chassaing, B.; Janzon, A.; Kumar, K.; Muller, C.E.; Werner, J.J.; Angenent, L.T.; Bell, M.E.; Hay, A.G.; Peterson, D.A. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 2013, 14, 571–581. [Google Scholar] [CrossRef]
- Peterson, D.A.; McNulty, N.P.; Guruge, J.L.; Gordon, J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2007, 2, 328–339. [Google Scholar] [CrossRef]
- Moor, K.; Diard, M.; Sellin, M.E.; Felmy, B.; Wotzka, S.Y.; Toska, A.; Bakkeren, E.; Arnoldini, M.; Bansept, F.; Völler, T. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 2017, 544, 498. [Google Scholar] [CrossRef] [PubMed]
- Fransen, F.; Zagato, E.; Mazzini, E.; Fosso, B.; Manzari, C.; El Aidy, S.; Chiavelli, A.; D’Erchia, A.M.; Sethi, M.K.; Pabst, O.; et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity 2015, 43, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Harris, N.L.; Spoerri, I.; Schopfer, J.F.; Nembrini, C.; Merky, P.; Massacand, J.; Urban, J.F., Jr.; Lamarre, A.; Burki, K.; Odermatt, B.; et al. Mechanisms of neonatal mucosal antibody protection. J. Immunol. 2006, 177, 6256–6262. [Google Scholar] [CrossRef] [PubMed]
- Briliūtė, J.; Urbanowicz, P.A.; Luis, A.S.; Baslé, A.; Paterson, N.; Rebello, O.; Hendel, J.; Ndeh, D.A.; Lowe, E.C.; Martens, E.C. Complex N-glycan breakdown by gut Bacteroides involves an extensive enzymatic apparatus encoded by multiple co-regulated genetic loci. Nat. Microbiol. 2019, 4, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.; Ladinsky, M.; Yu, K.; Sanders, J.; Yoo, B.; Chou, W.-C.; Conner, M.; Earl, A.; Knight, R.; Bjorkman, P. Gut microbiota utilize immunoglobulin A for mucosal colonization. Sci. Transl. Med. 2018, 360, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Rogier, E.W.; Frantz, A.L.; Bruno, M.E.; Wedlund, L.; Cohen, D.A.; Stromberg, A.J.; Kaetzel, C.S. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl. Acad. Sci. USA 2014, 111, 3074–3079. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, K.P.; Macadangdang, B.R.; Rogers, M.B.; Tometich, J.T.; Firek, B.A.; Baker, R.; Ji, J.; Burr, A.H.; Ma, C.; Good, M. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat. Med. 2019, 25, 1110–1115. [Google Scholar] [CrossRef]
- Rognum, T.O.; Thrane, P.S.; Stoltenberg, L.; Vege, Å.; Brandtzaeg, P. Development of intestinal mucosal immunity in fetal life and the first postnatal months. Pediatr. Res. 1992, 32, 145. [Google Scholar] [CrossRef]
- Roux, M.; Mcwilliams, M.; Phillips-Quagliata, J.M.; Weisz-Carrington, P.; Lamm, M. Origin of IgA-secreting plasma cells in the mammary gland. J. Exp. Med. 1977, 146, 1311–1322. [Google Scholar] [CrossRef]
- Wilson, E.; Butcher, E.C. CCL28 controls immunoglobulin (Ig) A plasma cell accumulation in the lactating mammary gland and IgA antibody transfer to the neonate. J. Exp. Med. 2004, 200, 805–809. [Google Scholar] [CrossRef]
- Lindner, C.; Thomsen, I.; Wahl, B.; Ugur, M.; Sethi, M.K.; Friedrichsen, M.; Smoczek, A.; Ott, S.; Baumann, U.; Suerbaum, S. Diversification of memory B cells drives the continuous adaptation of secretory antibodies to gut microbiota. Nat. Immunol. 2015, 16, 880. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.; Olm, M.R.; Firek, B.A.; Baker, R.; Thomas, B.C.; Morowitz, M.J.; Banfield, J.F. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat. Commun. 2017, 8, 1814. [Google Scholar] [CrossRef] [PubMed]
- Torow, N.; Marsland, B.J.; Hornef, M.W.; Gollwitzer, E.S. Neonatal mucosal immunology. Mucosal Immunol. 2017, 10, 5–17. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Cisalpino, D.; Varadarajan, S.; Hellman, J.; Warren, H.S.; Cascalho, M.; Inohara, N.; Núñez, G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 2016, 44, 647–658. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, W.; Wu, M.; Song, X.; Caro, F.; Sun, X.; Gazzaniga, F.; Stefanetti, G.; Oh, S.; Mekalanos, J.J. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 2020, 577, 543–548. [Google Scholar] [CrossRef]
- Koch, M.A.; Reiner, G.L.; Lugo, K.A.; Kreuk, L.S.; Stanbery, A.G.; Ansaldo, E.; Seher, T.D.; Ludington, W.B.; Barton, G.M. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 2016, 165, 827–841. [Google Scholar] [CrossRef]
- Gomez de Aguero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.; et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019, 364, 1179–1184. [Google Scholar] [CrossRef]
- Friedman, E.S.; Bittinger, K.; Esipova, T.V.; Hou, L.; Chau, L.; Jiang, J.; Mesaros, C.; Lund, P.J.; Liang, X.; FitzGerald, G.A.; et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc. Natl. Acad. Sci. USA 2018, 115, 4170–4175. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).