Abstract
Oral microbiota ecology is influenced by environmental and host conditions, but few studies have evaluated associations between untargeted measures of the entire oral microbiome and potentially relevant environmental and host factors. This study aimed to identify salivary microbiota cluster groups using hierarchical cluster analyses (Wards method) based on 16S rRNA gene amplicon sequencing, and identify lifestyle and host factors which were associated with these groups. Group members (n = 175) were distinctly separated by microbiota profiles and differed in reported sucrose intake and allelic variation in the taste-preference-associated genes TAS1R1 (rs731024) and GNAT3 (rs2074673). Groups with higher sucrose intake were either characterized by a wide panel of species or phylotypes with fewer aciduric species, or by a narrower profile that included documented aciduric- and caries-associated species. The inferred functional profiles of the latter type were dominated by metabolic pathways associated with the carbohydrate metabolism with enrichment of glycosidase functions. In conclusion, this study supported in vivo associations between sugar intake and oral microbiota ecology, but it also found evidence for a variable microbiota response to sugar, highlighting the importance of modifying host factors and microbes beyond the commonly targeted acidogenic and acid-tolerant species. The results should be confirmed under controlled settings with comprehensive phenotypic and genotypic data.
1. Introduction
The gastro-intestinal canal is heavily colonized, with over 700 bacterial species or unnamed phylotypes in the oral cavity alone [1]. The bacteria form niche-specific ecosystems with patterns of bacterial cohabitation due to influences from factors like transmission, host receptor availability, pH, access of nutrients, oxygen and symbiosis or communication with nearby species [2,3]. Generally, these ecosystems are in equilibrium but shifts may occur due to medical or lifestyle changes [4,5,6].
Many studies report associations between host factors, including saliva quality, pH regulation, and genetics, and single or panels of bacteria [7,8,9], but few studies have evaluated the associations between host traits and the oral microbiota in an untargeted manner [10,11,12]. Carbohydrate, especially high sugar, intake is reported to correlate with the enrichment of acidogenic and acid-tolerant caries-associated species. As an example, frequent glucose pulses (low pH) on a tooth-mimicking biofilm with nine bacterial species enumerated species that thrive at a low pH, whereas others were relatively reduced [13]. This shift mirrors the tooth biofilm dysbiosis in dental caries [14]. Sucrose exposure in vivo and assessments of targeted species support these experimental findings [15,16,17]. However, associations between sugar intake and untargeted characterization of the oral microbiota remain sparsely studied [4]. That genetic variation plays a fundamental role in individual differences in food preference and thereby food selection has been described from studies targeting candidate genes and in genome wide studies (GWAS) as comprehensively reviewed [18,19]. However, non-genetic and non-lifestyle linked factors have also been indicated to influence food habits, i.e., oral microbiota as well as gut microbiota have been suggested to modulate taste perception and eating behaviours [20,21,22,23]. We, and others, have shown that the intake of and preference for sweet foods are associated with polymorphisms in sweet and bitter taste receptor encoding genes, such as TAS1R2, TAS1R3 and TAS2R38, but also glucose transporter genes (SCL2 and SCL4) and the gustducin-encoding (GNAT3) gene [24,25].
The aim of the study was to explore the global saliva microbiota structure, identify groups of subjects defined by similar oral microbiota profiles in Swedish young adults, and search for potentially associated lifestyle and host factors.
2. Materials and Methods
2.1. Study Subjects
Teenagers and young adults attending one public dental health care clinic in the city of Umeå, Sweden were recruited consecutively as they attended the dentist’s office for their regular dental health control. Those who had received antibiotic treatment in the preceding 6 months, had a systemic disease or were taking regular medication, or were unable to communicate in Swedish or English, were not approached. In total, 176 participants in the age range 17–21 years were eligible and consented to participate.
The project received ethical approval by the Swedish Ethical Review Authority (Dnr 2012-111-31M) with an addendum (Dnr 2015-389-32M), and it adhered to the Helsinki Declaration and the General Data Protection Regulation (GDPR). The project is reported in accordance with Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for cohort studies.
2.2. Saliva Sampling, Bacteria Culturing and DNA Extraction
Approximately 3 mL of chewing stimulated saliva was collected into ice-chilled test tubes. Colony-forming units (CFUs) of mutans streptococci (Streptococcus mutans and Streptococcus sobrinus) and lactobacilli were assessed per ml of fresh saliva by cultivation of 100 μL on mitis salivarius sucrose agar supplemented with 0.2 U of bacitracin (MSB) and lactobacillus selective (LBS) agar, respectively. The plates were incubated at 37°C in 5% CO2 for 48 h. The remaining saliva was stored at −80°C.
Genomic DNA was extracted from saliva samples, positive and negative controls using the GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA). Briefly, the samples were centrifuged for 5 min at 13,000 rpm, lysed in buffer with lysozyme and mutanolysin, treated with RNase and Proteinase K, and purified and washed. All DNA extractions were done at the laboratory at the Dental School, Umeå University, Sweden by the same person and with kits from the same batch. The quality of the extracted DNA was estimated using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Uppsala, Sweden) and the quantity by the Qubit 4 Fluorometer (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). The mean yield from 200 μL saliva was 32 ng/μL (range 6–92 ng/μL) and the ratio of the absorbances at 260 and 280 nm was 1.8 or higher. Mixtures of known mock bacteria were used as positive controls and sterile water as negative control.
2.3. 16S rRNA Gene Amplicon Generation and Sequencing
Bacterial 16S rRNA gene amplicons were generated from the variable regions v1-v3 (27F-forward AGAGTTTGATCATGGCTCAG and 530R-reverse GTATTACCGCGGCTGCTG primers), and v3-v4 (357F-forward TACGGGAGGCAGCAG and 800R-reverse CCAGGGTATCTAATCC primers) from saliva and a mock community DNA. Library preparation, sequencing on the Illumina Miseq platform, and sequence demultiplexing were done at Eurofins Genomics (Ebersberg, Germany) according to their standard protocols. The company provided demultiplexed FASTQ-files, which were imported to QIIME2 [26], and DADA2 was used for denoising, pair end read fusion, chimeric sequence removal [27], and the identification of 100% identical amplicon sequence variants (ASVs) per sample [28]. Default parameters in DADA2 were used with left trimming of 13 bp for both forward and reversed reads, right trimming at 230 bp for the reversed sequences and 268 bp for the forward sequences. ASVs with >1 read were blasted against the extended Human Oral Microbiome Database (eHOMD, http://www.homd.org) for taxonomic annotation [29]. In the eHOMD blast, only ASVs with at least 98.5% identity with a named species or unnamed phylotype in eHOMD were retained, and those with the same HMT number were aggregated. The negative control contained <50 sequences and the positive control mock species were correct for representative sequences with 25 or more reads. Therefore, all comparisons were based on taxa with at least 25 reads. For simplicity, all taxa are referred to as species in the text. Sequencing failed for one saliva sample, leaving 175 samples in the final analyses.
2.4. Diet Recording
The participants reported their diet intake in a food frequency questionnaire (FFQ, http://www.matval.se). The FFQ is a semi-quantitative questionnaire, with questions on 93 food items/food aggregates selected to represent the habitual intake in Sweden and includes questions on alcoholic beverages. Participants were asked to report their typical intake in the last year. Intakes were reported on an increasing, nine-level scale, from never to four or more times a day. Portion sizes were estimated from photographs showing four portion sizes of staple foods (potatoes, rice, and pasta), meat/fish and vegetables, or standard food weights, such as for an egg or apple. Sucrose intake was estimated from nine questions, i.e., (i) fruit soup with or without a thickening agent, (ii) buns and biscuits, (iii) cookies and cakes, (iv) marmalade and jam, (v) ice cream, (vi) sodas, (vii) syrups, (viii) sugar, (ix) sweets including candies and chocolate. For sugar, the sum of mono and disaccharides (excluding lactose) from fruits, berries, vegetables, juices and honey was added. The FFQ included one question on how often the respondent ate or drank sweet products without sugar, i.e., with a sugar substitute.
Intake of energy and energy-providing nutrients was calculated by multiplying intake frequencies by portion sizes and weighting by the energy/nutrient contents in the food composition database at the National Food Administration (https://www.livsmedelsverket.se/en/food-and-content/naringsamnen).
A Healthy Diet Score that reflects healthy eating habits was calculated as previously described [30]. Briefly, frequency of intake per day was calculated for eight food/beverage groups. Favourable food groups included fish, fruits (except juices), vegetables (except potatoes) and whole grains. Unfavourable food/beverage groups included red or processed meats, desserts and sweets, sugar-sweetened beverages and fried potatoes. Intake frequencies were ranked within each sex in ascending quartile ranks for favourable foods/beverage groups, and in descending quartile ranks for unfavourable foods/beverage groups. The sum of all quartile ranks represents the Healthy Diet Score, with a minimum of zero and a maximum of 24, and with higher ranks indicating healthier food and beverage choices.
The relative validity of FFQ-derived intakes has been estimated against 24 h dietary records and/or biological markers [31,32,33,34]. For sucrose reliability, measured as correlations between registrations done one year apart, results were 0.80 for men and 0.75 for women, and the relative validity against 10 repeated 24 h recalls were 0.69 and 0.62 for sweet foods for men and women, respectively [31].
To reduce potential recording bias, energy-providing nutrients were expressed as energy standardized values (E%) and 11 participants with unrealistic reported energy intakes were excluded from analyses involving diet assessments. This was based on food intake level (FIL) scores calculated to estimate energy intake relative to minimal energy needs [35].
2.5. Recording of Medical and Other Lifestyle Conditions
Information on health status, oral hygiene, tobacco use, alcohol intake and most recent antibiotic exposure was obtained from a questionnaire. Dental caries was scored from visual and radiographic examinations in the dentist’s office with optimal lightning. Tooth surfaces that were sound according to ICDAS [36], score 0, or had caries in the enamel (e) according to ICDAS scores =1 and 2, or had caries in the enamel with a localized breakdown with or without dentine involvement (D) according to ICDAS score ≥3, or with a filling (F), were recorded. The total numbers of decayed and filled tooth surfaces (DeFS) were calculated. The M component was not considered because tooth loss occurred for orthodontic reasons or severe hypomineralization in this study group.
2.6. Genotyping of Single Nucleotide Polymorphism in Taste Associated Genes
Genotyping of 121 Single Nucleotide Polymorphisms (SNPs) in the TAS1R1, TAS1R2, TAS1R3, TAS2R16, TAS2R38, TAS2R50, SLC2A2, SLC2A4, GNAT3, CA6, SCN1B and TRPV1 taste-associated genes was performed at SciLife, Uppsala as described previously [24]. One SNP marker received a call rate of 0%. None of the remaining 120 SNPs deviated from Hardy–Weinberg equilibrium (p > 0.001), and they had an average call rate per sample of 99.8% and overall call rate of 99.8%. Genotyping data are uploaded at figshare (https://figshare.com/s/e292568e15c601e67a03).
2.7. Prediction of Functional Potential from the 16S rRNA Gene Information
Obtained representative ASVs were used to search for the potential molecular functions of the saliva microbiome using the 16S rRNA gene as marker gene, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) [37] and the Molecular Functions by orthology annotation (Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology database, KO, https://www.genome.jp/kegg/kegg1.html) [38,39]. The steps included (i) creating a closed reference feature table in QIIME2 using the trained Greengenes dataset gg-13-8-99-nb-classifier.qza (Greengenes http://greengenes.lbl.gov, [40], (ii) qiime diversity core-metrics analysis in QIIME2, and (iii) export of pathway abundances and the feature table for down-stream analyses in KO and multivariate modelling. Group separation was tested by Euclidean distances in permutational multivariate analysis of variance (PERMANOVA), Bonferroni-corrected p-values and 9999 permutations. Follow-up functional enrichment analyses were done using the STRING database (version 10.5, https://string-db.org/) [41]. The same procedure was also done for eHOMD-defined species.
2.8. Data Handling and Statistical Analyses
Unsupervised hierarchical clustering (Ward’s method) was used to classify the 175 participants by the presence (or not) of ASVs and presence (or not) of species from eHOMD identification. The number of ASVs were standardized to the level of the sample with the fewest reads after DADA2 filtering (38,293 reads), and lowest per-sample abundance of eHOMD taxa, and transformed by inverse hyperbolic sine transformation, which defines log values, including for zero-values, which are prevalent for many ASVs and some species.
Continuous phenotypic variables were presented as means with 95% confidence interval limits (CI), and when adjusted for sex, age and body mass index (BMI) using generalized linear modelling. Differences were tested with non-parametric tests. For discrete measures, the percentages in groups were estimated and proportion differences tested with Chi2 test. SPSS version 25 (IBM Corporation, Armonk, NY, USA) was used for these analyses. All tests were controlled by the Benjamini and Hochberg procedure, and those with p-values <0.05 yielding an FDR of 5% are presented.
Alpha- and beta-diversities with associated PERMANOVA tests and ASV proportions for bar charts were calculated in QIIME 2.
Multivariate modelling was performed by partial least-square regressions (PLS) (SIMCA P+ version 15.0, Sartrius Stedim Data Analytics AB, Malmö, Sweden). PLS identifies directions in an X-swarm that characterize X well and are related to Y. The software scales all variables to unit variance, and performs a K-fold cross-validation where 1/7th of the data are systematically kept out to fit a model and predict it from the remaining data (Q2-values). The results are displayed in scatter plots illustrating the separation of observations, and loading column plots displaying the mean correlation coefficient, with 95% CI between each predictor and the outcome variable. CIs that do not include zero are considered statistically significant.
The linear discriminant analysis effect size (LEfSe) method [42] was used to identify taxa effect size. Species that were shared between groups were identified in a Venn diagram [43].
3. Results
3.1. Study Group
The study included a similar number of males and females. The mean BMI was in the normal range but with variation, and approximately one in five participants had a BMI >25. Sex-adjusted mean energy intake (after exclusion of the 11 participants with unrealistic energy intake) was 1856 kcal/day, with 40.3% coming from carbohydrates (E% COH) (Table 1). Starch was the dominant type of dietary carbohydrate (117 g/day) followed by sucrose (27 g/day), monosaccharides (19 g/day), and other sugars (23 g/day) (Figure 1A). Reported median intake of sucrose corresponded to 5.8% of the total energy intake (Figure 1B).

Table 1.
Characteristics of the 175 participants in the study group.
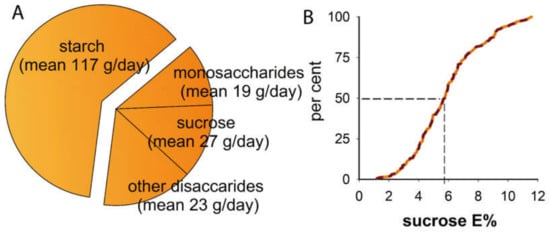
Figure 1.
Total carbohydrate and sucrose intake in the study group. Proportions of various types of carbohydrates in the reported diet (A) and cumulative percentages for sucrose intake (B).
3.2. Overall Microbiota Assessment
For the 175 saliva samples, 14,317,039 sequences passed denoising and the removal of potential chimera with 1,685,662 reads referring to the v1–v3 and 12,631,377 sequences to the v3–v4 section. Under the present conditions, the v3–v4 sequencing yielded significantly more sequences per sample, better recognition of the mock species and higher diversity (Table S1). Further analyses were based on v3–v4 sequences.
The retained 12,631,377 v3–v4 sequences corresponded to 6171 representative amplicon sequence variants (ASVs) with ≥2 reads. These were in 13 phyla and 127 genera.
3.3. Cluster Classifications Based on ASV Pattern
Four groups were classified from the hierarchical clustering of dichotomous ASVs (clusters ASV1-ASV4 (Figure 2A). The relative proportions of the 13 identified phyla and top 40 genera in the respective cluster group are presented in Figure 2B,C, respectively.
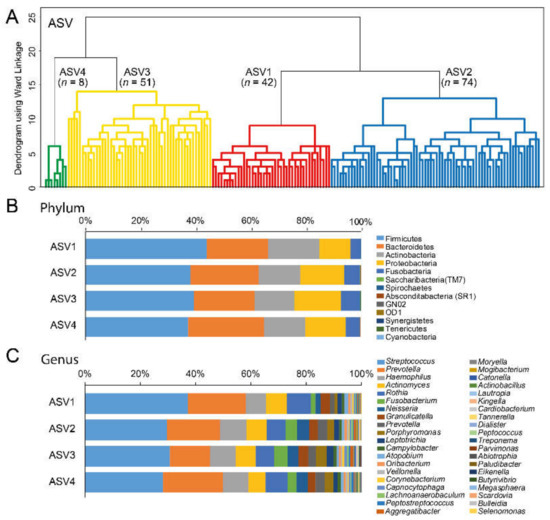
Figure 2.
(A) Dendrogram from the unsupervised hierarchical cluster analysis with Ward’s method. The clusters in red and blue refer to the groups with the highest and lowest sucrose intake, respectively, when intake in the defined cluster groups was compared, (B) bar chart for 13 identified phyla, and (C) bar chart for the top 40 identified genera out of 127.
The ASV cluster groups differed significantly in reported sugar (p = 0.001) and sucrose (p = 0.008) intake and saliva flow rate (p = 0.031) (Table 2). Further, allelic variation in the TAS1R1 (rs731024, p = 0.003), and the GNAT3 (rs2074673, p = 0.007 and rs11760281, p = 0.010) genes differed significantly between the cluster groups (Table 2). None of the other gene variants or BMI or caries scores differed between cluster groups.

Table 2.
Pheno- and genotypical characteristics for hierarchical cluster groups based on amplicon sequence variants (ASVs), and taxa aggregates based on identification in the eHOMD database. Underlying dendrograms are shown in Figure 2 and Figure 4, respectively. For eHOMD, only variables that differed significantly between the clusters are shown.
Alpha diversity was lowest for the group with the highest reported sugar intake (ASV1) when estimated as ASVs in rarefaction curves (Figure 3A), Shannon index (Figure 3B), and evenness diversities (Figure 3C), with statistically significant differences among (p-values from 1.5 × 10-3 to 1.2x10-13) and between groups (q-values from 1.3 × 10-2 to 1.7 × 10-10). Faith phylogenetic diversity index was less divergent between groups but still differed significantly (7.8 × 10-5, Figure 3D). Beta diversity differed significantly between all cluster groups (PERMANOVA p-value among groups and q-values between groups 0.001) with associated separation displayed in the Jaccard principal coordinates analysis (PCoA) plots (Figure 3E).
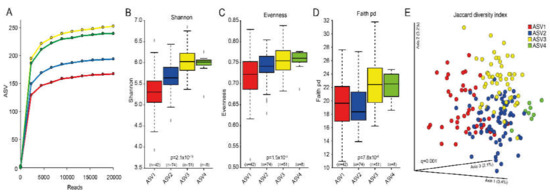
Figure 3.
Composite figure of various aspects related to cluster groups based on dichotomous ASVs. (A) rarefaction curves showing number of observed ASVs by sequencing depth (reads); (B–D) box plots of alpha diversities by the Shannon, Evenness and Faith phylogenetic diversity (pd) indexes; (E) Jaccard Principal Coordinates Analysis (PCoA) plot illustrating separation of the cluster groups based on dichotomous measures. The red colour refers to the cluster with highest sucrose intake and blue refers to the lowest.
3.4. Predicted Functions in ASV Cluster Groups
Linkage of the 6171 retained 16S rRNA ASVs to pathways by KEGG functional orthology annotation predicted 10,543 KEGG orthologies. Multivariate analysis suggested a separation of the predicted function of cluster ASV1 (highest sucrose intake) from cluster ASV2 (lowest sucrose intake) and ASV3 (both p < 0.0006). The microbiota of cluster ASV1 (compared to both cluster ASV2 and ASV3) displayed several KEGG orthology related to starch and sucrose metabolism, and a functional enrichment of glycosidase capacity according to the STRING processing, indicating the ability to hydrolase glyosidic bonds in carbohydrates (Table S2).
3.5. Taxa Determination from eHOMD and Their Cluster Classification
The 6171 representative sequence variants matched 372 named species or unnamed phylotype gene sequences in eHOMD by the set criteria. These were in nine phyla (Figure 4A) and 85 genera. The most prevalent genera were Streptococcus (28.1%), and Prevotella (16.1%), Haemophilus (8.6%), Rothia (6.9%), and Actinomyces (6.5%) (Figure 4B). The dominant species included a Streptococcus mitis/Streptococcus infantis/Streptococcus oralis complex (12.9%), followed by Prevotella melaninogenica, Haemophilus parainfluenzae, Streptococcus salivarius, and Rothia mucilaginosa, all of which represented >5% of total reads Table S3.
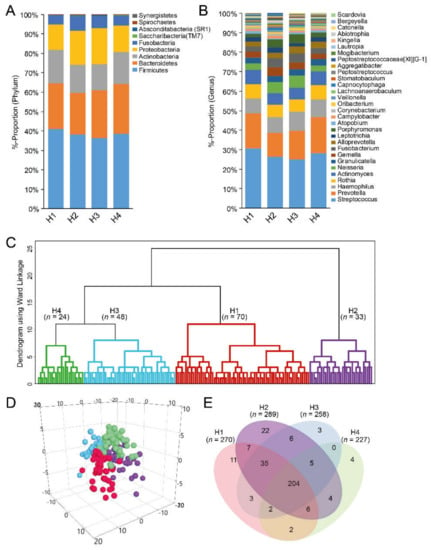
Figure 4.
Composite figure of various aspects related to cluster groups based on eHOMD-identified taxonomic names and aggregation by species. (A,B) Bar charts showing overall relative proportions in the nine represented phyla (A) and top 30 genera; (B), as classified by the eHOMD database; (C) dendrogram from hierarchical cluster analysis with Ward´s method. The red section refers to the cluster with the highest sucrose intake and blue to the lowest; (D) PLS 3D scatter plot illustrating separation of the four cluster groups; (E) Venn diagrams showing the number of species detected in all four cluster groups.
The 175 participants were classified into four groups by hierarchical clustering based on dichotomous taxa presence (clusters H1–H4, Figure 4C) with group separation, confirmed in multidimensional-PLS scatter plots (Figure 4D). Approximately half of the 372 sequences were shared among the four cluster groups (Figure 4E). Sugar (p = 0.006) and sucrose (p < 0.001) intake, and allelic variation in the GNAT3 (rs2074673 and rs11760281) gene, differed among the groups based on dichotomous measures (Table 2).
3.6. Factors Associated with Belonging to the Species Level (eHOMD) Cluster Groups
PLS identified eHOMD species, host-related traits and lifestyle factors, and mutans streptococci and lactobacilli by influential culture, and classified them into the four cluster groups. Data for H1, H2 and H4 compared to H3 are shown in Table 3 and H3 from the PLS comparison including all four cluster groups simultaneously in Table S4.

Table 3.
Species significantly influential for being classified into cluster H1, H2 or H4, respectively, when compared with cluster H3. Species in bold are known to be acidogenic and acid-tolerant species. Data for cluster H3 (lowest sucrose intake) against the three other clusters simultaneously are found in Table S4.
Sugar and sucrose intakes were influential for being in clusters H1, H2 and H4 with comparably higher sucrose intake. Intake of milk was influential for clusters H1 and H4 (Table 2). A wide panel of species, including species in Actinomyces and Selenomonas, Bifidobacterium dentium and Leptotrichia sp. HMT498, were influential for being in cluster H2, whereas being in cluster H1 (with slightly higher mean sucrose intake) was determined by significantly fewer species, which included two species in Actinomyces, Bifidobacterium longum, Scardovia wiggsiae, Streptococcus mutans, two species in Veillonella and lactobacilli and mutans streptococci by culture. Cluster H4 also had fewer influential species than cluster H2 and included the cariogenic Streptococcus sobrinus.
High scores in the Healthy Diet index and for protein-related food intake were associated with membership in Cluster H3, which was the cluster with the lowest reported sucrose intake (Table S4).
3.7. Predicted Functions in Species Level (eHOMD) Cluster Groups
Predicted functions in the microbiota associated with the four HOMD-based cluster groups separated them (p < 0.0001), with cluster H3 (lowest sucrose intake) separated from the two groups with the highest sucrose intakes, i.e., H1 (p = 0.0012) and H2 (p = 0.027). Similar to the sequence variant-based analyses, cluster H1 displayed several KEGG functional orthology related to starch and sucrose metabolism (Figure 5C, Table S5) and a functional enrichment of the capacity to hydrolase glyosidic bonds (Figure 5D).
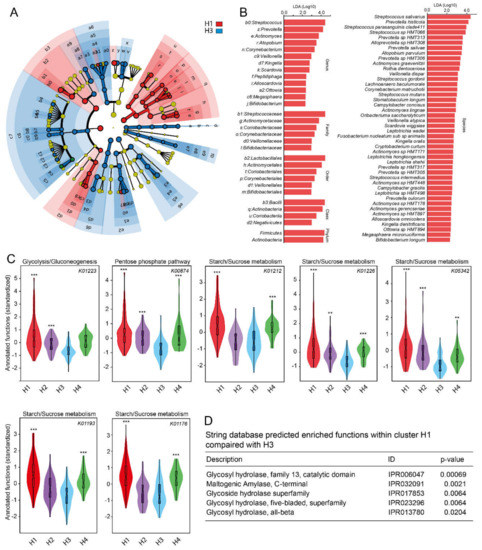
Figure 5.
LEfSe results for Cluster H1 versus H3 with highest and lowest sucrose intake, respectively. (A) Cladogram showing taxonomic representation of statistically consistent differences between the two cluster groups. The analysis was based on the abundance of the 372 species and their respective family, order, class and phylum. (B) Histogram of the linear discriminant analysis (LDA) scores in cluster groups H1 and H3. (C) Violin plots with box plots for carbohydrate metabolic pathways characteristic of all four H cluster groups, and (D) STRING database predicted enriched functions within cluster H1 (high sucrose intake) compared with cluster H3 (lowest sucrose intake). Group comparisons were done using non-parametric (Kruskal–Wallis or Mann–Whitney U test, Bonferroni corrected p values, permutation 9999). Violin labels: ***, ** and * indicate <0.001, <0.01, and <0.05, respectively.
4. Discussion
This study identified groups of people defined by similar oral microbiota profiles, targeted as dichotomized amplicon species variants (ASVs) or named species and unnamed phylotypes, and then explored lifestyle and host factors which were associated with these groups. The most striking difference between groups classified by unsupervised cluster analysis was for reported sucrose intake but associations were also found for total sugar intake, saliva flow rate and allelic variations in two taste-perception-associated genes. The group who reported the highest sucrose intake had the lowest species richness, but the microbiota in clusters with a higher sucrose intake were either defined by a pamphlet of species with no clear association with carbohydrate metabolic pathways or by a microbiota enriched for acidogenic and acid-tolerant species and carbohydrate degradation metabolic pathways. The latter species included several species that were suggested to be associated with the caries disease.
The prevailing ecological plaque hypothesis [44,45] describes an ecological shift (collapse) towards the enrichment of acidogenic and acid-tolerant species in low pH environments, such as after sugar consumption [13,16,46]. The present findings are consistent with this hypothesis, as several such species were found to be enriched in two of the three cluster groups with the highest sucrose intake. Among these were species in Actinomyces, Bifidobacterium and Veillonella, and S. wiggsiae, S. mutans, and S. sobrinus with documented relevance for dental caries in small children or adults [14,44,47]. In fact, although not statistically assured, these two cluster groups tended to have had the highest mean caries experience. Conversely, the study identified a third cluster group with similar sugar intake but not characterized by enrichment of the most acidogenic and acid-tolerant species, but rather a large number of taxa including several species in Capnocytophaga, Leptotrichia, Prevotella and Streptococcus. Thus, the effects of sugar on the salivary microbiota are potentially modified by host-related factors such as innate immunity peptides and buffer functions, but there are other possible explanations, for example, if the microbiota response to sugar is regulated by inter-bacteria communication or if there was differential measurement error in the reporting of sucrose intake in the different cluster groups. Overall, the present findings are well in line with our previous finding in the same population, where S. wiggsiae, S. mutans, B. longum, Leptotrichia sp. HMT498, and Selenomonas spp. in tooth biofilm samples discriminated caries-affected from caries-free adolescents [48], and support previous experimental findings in vitro [13] and in animals [49]. A direct comparison with the results from Anderson et al. 2018 [4] may not be appropriate as they used a different 16S rRNA segment and lower similarity requirement for taxa determination.
The strength of the present study is the comparably large sample and that the study group likely represents the underlying population with minimal selection bias since the attendance rate to the public dental clinics is very high, as care is free in the targeted age group and that participants who were enrolled consecutively agreed to participate. The major weakness relates to the inherent difficulties in measuring diet using questionnaires [50]. Monitoring sugar intake may be even more challenging in the dentist´s office, since patients are aware that sugar is bad for the teeth. This may be a source of systematic measurement error which may be manifested in the low self-reported sugar intake and the limited variation compared to other studies in the country [51]. However, energy-adjustment, excluding participants who reported the most implausible energy intakes and use of robust statistical methods based on ranking rather than reported consumption, was performed to reduce the impact of this error, but it cannot completely exclude a nullifying bias from the under-reporting of sucrose intake.
Factors influential for being classified in a cluster group were assessed using multivariate PLS regression, which has the advantage of accepting correlated variables. These models were strong, but it should be noted that this is partly an effect of the fact that the cluster groups were formed by cluster analysis of bacterial taxa. However, since clustering was unsupervised, the association with sucrose intake and which specific bacteria were present in each cluster is not a consequence of clustering per se and does not prevent the comparison of phenotypic characteristics of people in the cluster groups or species that were influential for being in the groups.
An alternative model to the idea that sugar intake and, indirectly, gene variations, drives oral microbiota transformation would be that the oral microbiota per se influences the taste phenotype of the host and, accordingly, the individual’s food selection, as previously suggested [20,21,22,23]. In the present study, sucrose intake was the strongest explanatory diet factor for microbiota clustering, and supported by several in vitro and a few in vivo studies [4,7,52]. We suggest that the variant oral microbiota ecologies (here, the clusters) are a consequence of low pH due to sugar intake [42,45,52]. Thus, we suggest that the clinical implication of the present findings is that even a moderate difference in sugar intake may affect the oral microbiota into a more cariogenic composition, which may be reverted by sugar restriction. However, we cannot fully exclude that the alternative hypothesis is of relevance too.
We, and others, have reported that genetic variants located in or near genes related to taste perception and preference were associated with diet preference and caries [33], but studies have not evaluated variations in such genes in relation to the concerted oral microbiome. In this study, allelic variations in the TAS1R1 and GNAT3 genes were significantly associated with the pattern of oral bacteria at the group level. The TAS1R1 gene is involved in bitter taste sensations with known associations with sweet food preferences [53], whereas the GNAT3 gene, encoding the gustducin alpha-3 chain protein, can be involved in sweet, bitter or umami taste sensation depending on heterodimeric formations with various TAS proteins [54]. Besides taste perception, the gustducin protein functions as a sugar sensor in the gut with suggested effects on sugar absorption and metabolic syndrome [55]. Thus, hypothetically, the GNAT3 gene may affect the oral microbiota via dietary habits or secondary effects from diet-related metabolic disorders [56].
The present study also found that under the present conditions sequencing of the v1-v3 amplicons of the 16S rDNA gene yielded significantly fewer sequences and poorer recognition of the mock species than the v3-v4 amplicons. This is in line with previous reports [57,58] and stresses the importance of taking the sequencing protocol into consideration when data are compared from different studies.
5. Conclusions
In conclusion, this study supported in vivo associations between sugar intake and oral microbiota ecology, but it also confirmed variations in the apparent microbiota response to sugar, highlighting the importance of considering microbes beyond the commonly studied acidogenic and acid-tolerant species and modifying host factors. The result, need to be confirmed, preferentially under controlled forms, such as in a randomized clinical trial design and with the monitoring of host pheno- and genotypical characteristics and a potential biomarker for diet, such as untargeted metabolomics.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6643/12/3/681/s1. Table S1: Taxa detected by Illumina MISeq sequencing of the v1–v3 versus the v3–v4 variable regions of 16S rDNA. Identities are from eHOMD blasting at 98.5% identity and cut-off at 25 reads per species. Table S2: Partial least square regression analysis (PLS-DA) was used to identify bacterial functions differentiating between ASV cluster 2, with the lowest sucrose intake (sugar 13.6 E%, sucrose 5.3E%), to that of cluster ASV 1, 3, and 4 with higher sucrose intakes. Bacterial functions were predicted by Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt 2) in QIIME 2 and Kyoto Encyclopedia of Genes and Genomes (KEGG orthology, KO). Bold labelled functions are linked to Carbohydrate metabolism (e.g., starch, sucrose, and glycolysis). Table S3: Prevalence of 372 species identified from the eHOMD database at 98.5% identity and represented by at least 25 reads. Data are for cluster groups from hierarchical clustering of dichotomous variables. p-values for the prevalences are from Chi-square analyses. Species and p-values in bold are statistically significant at FDR 0.05. Table S4: Bacteria and other host-associated variables with a statistically significant association (PLS correlation coefficients with 95% CI that does not include 0) in PLS modelling. Cluster H3 (the cluster with the lowest reported sucrose intake) was modelled against cluster H1, H2, and H4 individually or together. Sucrose intake (E%) and caries (DeFS) are group mean values standardized for sex, age and BMI and sex and age, respectively. Variables are ordered by taxa names and other host factors at the end. Species in bold are species that commonly are referred to as acidophilic species. Table S5: Multivariate analysis (PLS-DA) was used to identify bacterial functions differentiating between cluster H3, with the lowest sucrose intake) to that of clusters H1, H2, and H4 with higher sucrose intake. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt 2) in QIIME 2 and Kyoto Encyclopedia of Genes and Genomes (KEGG orthology, KO) predicted bacterial functions. Functions in bold are linked to carbohydrate metabolism (e.g., starch, sucrose, and glycolysis).
Author Contributions
Conceptualization, A.E., P.L.H. and I.J.; methodology, A.E.; software, A.E., S.H. and I.J.; validation, A.E., P.L.H., S.H, P.H. and I.J.; formal analysis, A.E. and I.J.; investigation, P.L.H; resources, I.J and P.L.H.; data curation, A.E., S.H. and I.J.; writing—original draft preparation, A.E., P.L.H., S.H, P.H. and I.J; writing—review and editing, A.E., P.L.H., S.H, P.H. and I.J; visualization, A.E and I.J.; supervision, I.J.; project administration, I.J.; funding acquisition, I.J and P.L.H. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by The Swedish Patent Revenue Fund under Grant I 2017-019; TUA, the County Council of Västerbotten, Sweden; The Swedish Dental Association; and the County Council of Västerbotten internal research foundation under Grant VLL-663251.
Acknowledgments
DDS Linda Eriksson is acknowledged for collecting clinical data and samples and Agneta Rönnlund for skilful laboratory work.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Caufield, P.W.; Schon, C.N.; Saraithong, P.; Li, Y.; Argimon, S. Oral lactobacilli and dental caries: A model for niche adaptation in humans. J. Dent. Res. 2015, 94, 110S–118S. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S. Talk of the town: Interspecies communication in oral biofilms. Mol. Oral. Microbiol. 2010, 25, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Rothballer, M.; Altenburger, M.J.; Woelber, J.P.; Karygianni, L.; Lagkouvardos, I.; Hellwig, E.; Al-Ahmad, A. In-vivo shift of the microbiota in oral biofilm in response to frequent sucrose consumption. Sci. Rep. 2018, 8, 14202. [Google Scholar] [CrossRef]
- Bibbo, S.; Ianiro, G.; Giorgio, V.; Scaldaferri, F.; Masucci, L.; Gasbarrini, A.; Cammarota, G. The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 4742–4749. [Google Scholar]
- Romani Vestman, N.; Chen, T.; Lif Holgerson, P.; Ohman, C.; Johansson, I. Oral microbiota shift after 12-week supplementation with lactobacillus reuteri DSM 17938 and PTA 5289; A randomized control trial. PLoS ONE 2015, 10, e0125812. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Belstrom, D.; Jersie-Christensen, R.R.; Lyon, D.; Damgaard, C.; Jensen, L.J.; Holmstrup, P.; Olsen, J.V. Metaproteomics of saliva identifies human protein markers specific for individuals with periodontitis and dental caries compared to orally healthy controls. PeerJ 2016, 4, e2433. [Google Scholar] [CrossRef]
- Esberg, A.; Haworth, S.; Brunius, C.; Lif Holgerson, P.; Johansson, I. Carbonic anhydrase 6 gene variation influences oral microbiota composition and caries risk in Swedish adolescents. Sci. Rep. 2019, 24, 452. [Google Scholar] [CrossRef]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef]
- Aleti, G.; Baker, J.L.; Tang, X.; Alvarez, R.; Dinis, M.; Tran, N.C.; Melnik, A.V.; Zhong, C.; Ernst, M.; Dorrestein, P.C.; et al. Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Divaris, K. Searching deep and wide: Advances in the molecular understanding of dental caries and periodontal disease. Adv. Dent. Res. 2019, 30, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, D.J.; McKee, A.S.; Marsh, P.D. Effects of carbohydrate pulses and pH on population shifts within oral microbial communities in vitro. J. Dent. Res. 1989, 68, 1298–1302. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.R.; Kressirer, C.A.; Rothmiller, S.; Johansson, I.; Chalmers, N.I. The caries microbiome: Implications for reversing dysbiosis. Adv. Dent. Res. 2018, 29, 78–85. [Google Scholar] [CrossRef]
- Keller, M.K.; Kressirer, C.A.; Belstrom, D.; Twetman, S.; Tanner, A.C.R. Oral microbial profiles of individuals with different levels of sugar intake. J. Oral. Microbiol. 2017, 9, 1355207. [Google Scholar] [CrossRef]
- Tsutsumi, K.; Maruyama, M.; Uchiyama, A.; Shibasaki, K. Characterisation of a sucrose-independent in vitro biofilm model of supragingival plaque. Oral. Dis. 2018, 24, 465–475. [Google Scholar] [CrossRef]
- Wennerholm, K.; Birkhed, D.; Emilson, C.G. Effects of sugar restriction on Streptococcus mutans and Streptococcus sobrinus in saliva and dental plaque. Caries Res. 1995, 29, 54–61. [Google Scholar] [CrossRef]
- Hwang, L.D.; Lin, C.; Gharahkhani, P.; Cuellar-Partida, G.; Ong, J.S.; An, J.; Gordon, S.D.; Zhu, G.; MacGregor, S.; Lawlor, D.A.; et al. New insight into human sweet taste: A genome-wide association study of the perception and intake of sweet substances. Am. J. Clin. Nutr. 2019, 1, 1724–1737. [Google Scholar] [CrossRef]
- Robino, A.; Concas, M.P.; Catamo, E.; Gasparini, P. A brief review of genetic approaches to the study of food preferences: Current knowledge and future directions. Nutrients 2019, 26, 1735. [Google Scholar] [CrossRef]
- Cattaneo, C.; Gargari, G.; Koirala, R.; Laureati, M.; Riso, P.; Guglielmetti, S.; Pagliarini, E. New insights into the relationship between taste perception and oral microbiota composition. Sci. Rep. 2019, 5, 3549. [Google Scholar] [CrossRef]
- Cattaneo, C.; Riso, P.; Laureati, M.; Gargari, G.; Pagliarini, E. Exploring associations between interindividual differences in taste perception, oral microbiota composition, and reported food intake. Nutrients 2019, 24, 1167. [Google Scholar] [CrossRef] [PubMed]
- Besnard, P.; Christensen, J.E.; Brignot, H.; Bernard, A.; Passilly-Degrace, P.; Nicklaus, S.; Pais de Barros, J.P.; Collet, X.; Lelouvier, B.; Servant, F.; et al. Obese subjects with specific gustatory papillae microbiota and salivary cues display an impairment to sense lipids. Sci. Rep. 2018, 30, 6742. [Google Scholar] [CrossRef] [PubMed]
- Alcock, J.; Maley, C.C.; Aktipis, C.A. Is eating behavior manipulated by the gastrointestinal microbiota? Evolutionary pressures and potential mechanisms. Bioessays 2014, 36, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Esberg, A.; Haworth, S.; Holgerson, P.L.; Johansson, I. Allelic variation in taste genes is associated with taste and diet preferences and dental caries. Nutrients 2019, 29, 1491. [Google Scholar] [CrossRef] [PubMed]
- Fushan, A.A.; Simons, C.T.; Slack, J.P.; Drayna, D. Association between common variation in genes encoding sweet taste signaling components and human sucrose perception. Chem. Senses 2010, 35, 579–592. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The human oral microbiome database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010, 2010, 1–10. [Google Scholar] [CrossRef]
- Nettleton, J.A.; Hivert, M.F.; Lemaitre, R.N.; McKeown, N.M.; Mozaffarian, D.; Tanaka, T.; Wojczynski, M.K.; Hruby, A.; Djousse, L.; Ngwa, J.S.; et al. Meta-analysis investigating associations between healthy diet and fasting glucose and insulin levels and modification by loci associated with glucose homeostasis in data from 15 cohorts. Am. J. Epidemiol. 2013, 177, 103–115. [Google Scholar] [CrossRef]
- Johansson, I.; Hallmans, G.; Wikman, A.; Biessy, C.; Riboli, E.; Kaaks, R. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002, 5, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Wennberg, M.; Vessby, B.; Johansson, I. Evaluation of relative intake of fatty acids according to the Northern Sweden FFQ with fatty acid levels in erythrocyte membranes as biomarkers. Public Health Nutr 2009, 12, 1477–1484. [Google Scholar] [CrossRef] [PubMed]
- Klingberg, S.; Ellegard, L.; Johansson, I.; Jansson, J.H.; Hallmans, G.; Winkvist, A. Dietary intake of naturally occurring plant sterols is related to a lower risk of a first myocardial infarction in men but not in women in northern Sweden. J. Nutr. 2013, 143, 1630–1635. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Holgerson, P.L.; Kressin, N.R.; Nunn, M.E.; Tanner, A.C. Snacking habits and caries in young children. Caries Res. 2010, 44, 421–430. [Google Scholar] [CrossRef]
- Black, A.E. Critical evaluation of energy intake using the Goldberg cut-off for energy intake: Basal metabolic rate. A practical guide to its calculation, use and limitations. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 1119–1130. [Google Scholar] [CrossRef]
- Pitts, N.B.; Ekstrand, K.R.; Foundation, I. International Caries Detection and Assessment System (ICDAS) and its International Caries Classification and Management System (ICCMS) - methods for staging of the caries process and enabling dentists to manage caries. Community Dent. Oral. Epidemiol. 2013, 41, e41–e52. [Google Scholar] [CrossRef]
- Douglas, G.M.; Beiko, R.G.; Langille, M.G.I. Predicting the functional potential of the microbiome from marker genes using PICRUSt. Methods Mol. Biol. 2018, 1849, 169–177. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C.; VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. Available online: http://bioinfogpcnbcsices/tools/venny/indexhtml (accessed on 12 December 2019).
- Marsh, P.D.; Zaura, E. Dental biofilm: Ecological interactions in health and disease. J. Clin. Periodontol. 2017, 44, S12–S22. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: Ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef]
- Marsh, P.D. In sickness and in health-what does the oral microbiome mean to us? An ecological perspective. Adv. Dent. Res. 2018, 29, 60–65. [Google Scholar] [CrossRef]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef]
- Eriksson, L.; Lif Holgerson, P.; Johansson, I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. Sci. Rep. 2017, 7, 5861. [Google Scholar] [CrossRef]
- Borkent, D.; Reardon, R.J.M.; Mc, L.G.; Glendinning, L.; Dixon, P.M. A microbiome analysis of equine peripheral dental caries using next generation sequencing. Equine Vet. J. 2020, 52, 67–75. [Google Scholar] [CrossRef]
- Poslusna, K.; Ruprich, J.; de Vries, J.H.; Jakubikova, M.; van’t Veer, P. Misreporting of energy and micronutrient intake estimated by food records and 24 h recalls, control and adjustment methods in practice. Br. J. Nutr. 2009, 101, S73–S85. [Google Scholar] [CrossRef]
- Moraeus, L.; Lemming, E.W.; Hursti, U.K.; Arnemo, M.; Sipinen, J.P.; Lindroos, A.K. Riksmaten Adolescents 2016–2017: A national dietary survey in Sweden—Design, methods, and participation. Food Nutr. Res. 2018, 62. [Google Scholar] [CrossRef]
- Bowen, W.H.; Burne, R.A.; Wu, H.; Koo, H. Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 2018, 26, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Rawal, S.; Hayes, J.E.; Wallace, M.R.; Bartoshuk, L.M.; Duffy, V.B. Do polymorphisms in the TAS1R1 gene contribute to broader differences in human taste intensity? Chem. Senses 2013, 38, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Staszewski, L.; Tang, H.; Adler, E.; Zoller, M.; Li, X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 14258–14263. [Google Scholar] [CrossRef] [PubMed]
- Laffitte, A.; Neiers, F.; Briand, L. Functional roles of the sweet taste receptor in oral and extraoral tissues. Curr. Opin. Clin. Nutr. Metab. Care 2014, 17, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bailo, B.; Toguri, C.; Eny, K.M.; El-Sohemy, A. Genetic variation in taste and its influence on food selection. OMICS 2009, 13, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Castelino, M.; Eyre, S.; Moat, J.; Fox, G.; Martin, P.; Ho, P.; Upton, M.; Barton, A. Optimisation of methods for bacterial skin microbiome investigation: Primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 2017, 21, 23. [Google Scholar] [CrossRef]
- Teng, F.; Nair, S.S.D.; Zhu, P.; Li, S.; Huang, S.; Li, X.; Xu, J.; Yang, F. Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 2018, 5, 16321. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).