The Causal Effects of Blood Iron and Copper on Lipid Metabolism Diseases: Evidence from Phenome-Wide Mendelian Randomization Study
Abstract
1. Introduction
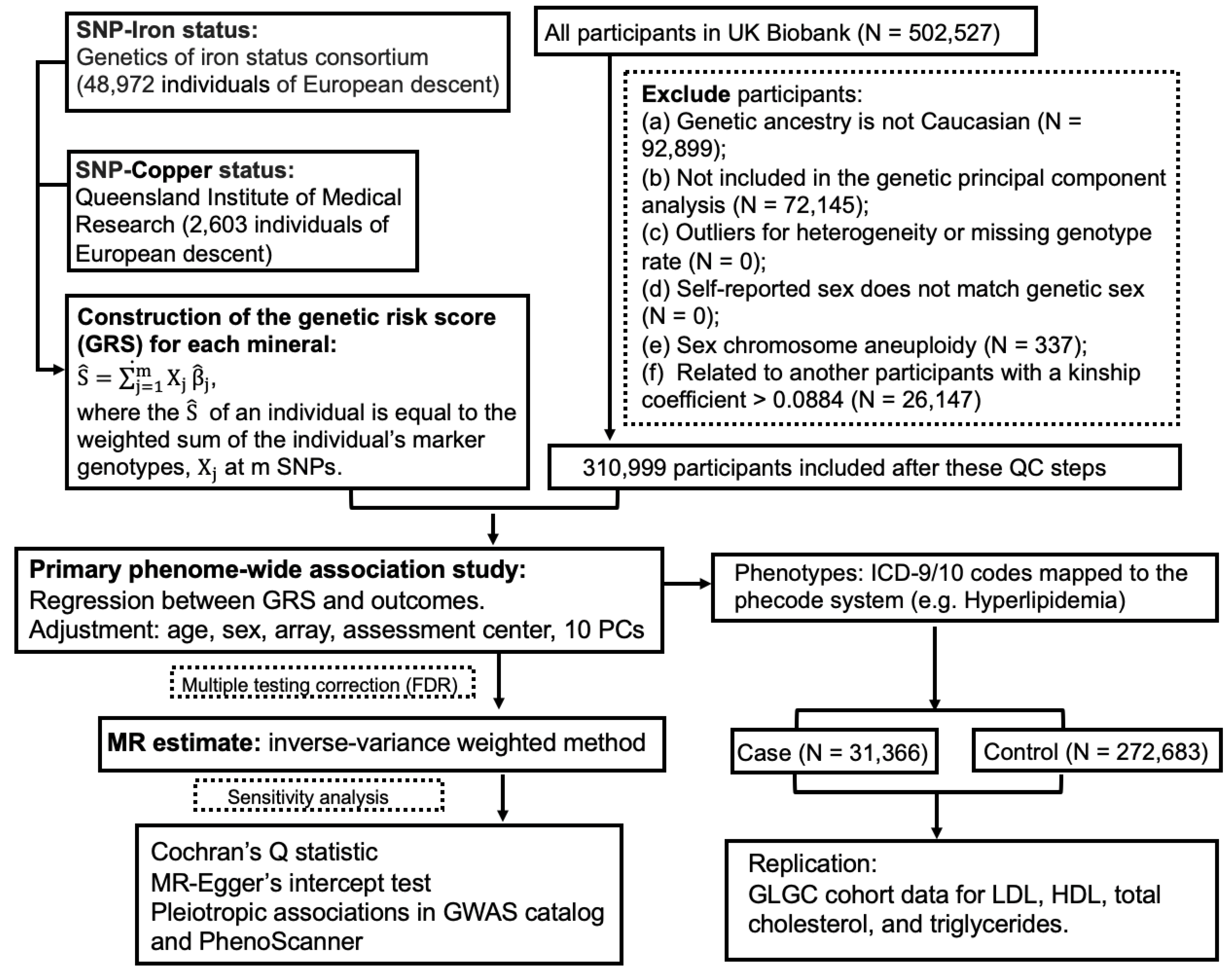
2. Methods
2.1. Genetic Instruments for Blood Iron and Copper
2.2. Study Population
2.3. Phenome-Wide Association Study
2.4. MR Analyses
2.5. Sensitivity Analyses
3. Results
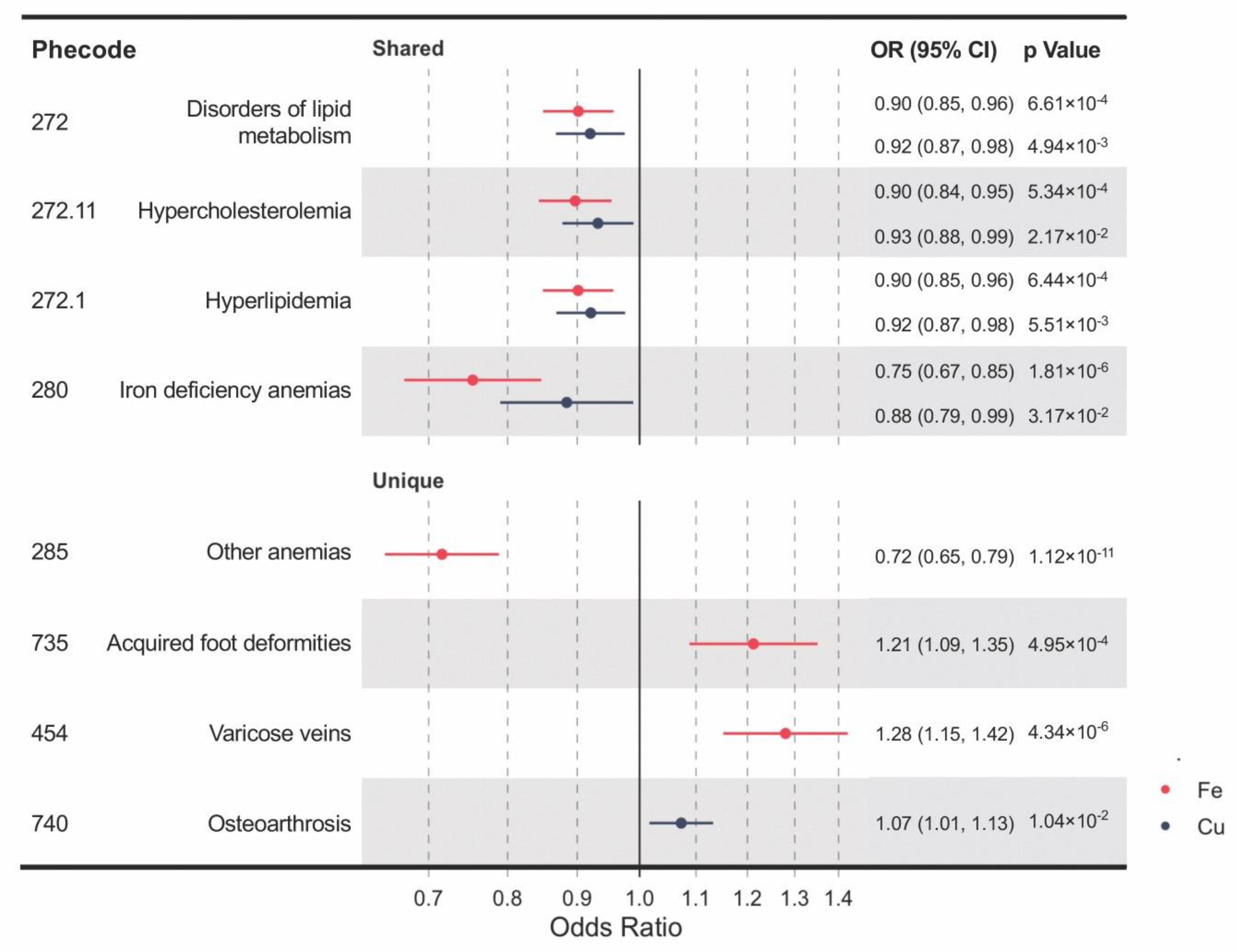
3.1. Shared and Unique Causal Clinical Effects of Blood Iron and Copper
3.2. Causality of Both Iron and Copper on Lipid Metabolism Traits
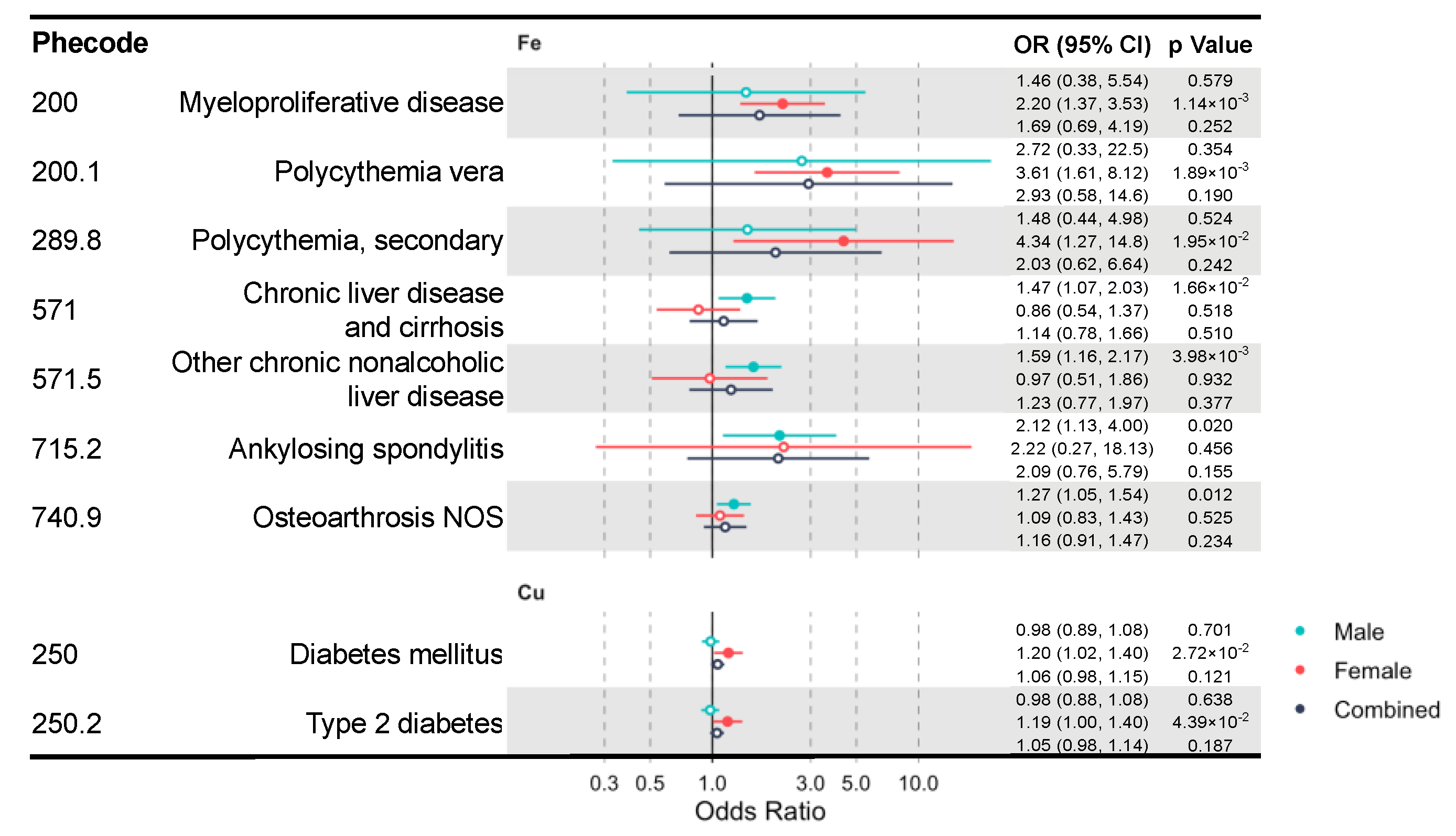
3.3. Interpretation of Potential Pleiotropy in MR Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hunsaker, E.W.; Franz, K.J. Emerging Opportunities To Manipulate Metal Trafficking for Therapeutic Benefit. Inorg. Chem. 2019, 58, 13528–13545. [Google Scholar] [CrossRef]
- Waldron, K.J.; Rutherford, J.C.; Ford, D.; Robinson, N.J. Metalloproteins and metal sensing. Nat. Cell Biol. 2009, 460, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.R.; Gahl, W.A. Disorders of metal metabolism. Transl. Sci. Rare Dis. 2017, 2, 101–139. [Google Scholar] [CrossRef] [PubMed]
- Camaschella, C.; Pagani, A.; Nai, A.; Silvestri, L. The mutual control of iron and erythropoiesis. Int. J. Lab. Hematol. 2016, 38, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Pietrangelo, A. Mechanisms of iron hepatotoxicity. J. Hepatol. 2016, 65, 226–227. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.F.; Prohaska, J.R.; Knutson, M.D. Metabolic crossroads of iron and copper. Nutr. Rev. 2010, 68, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Myint, Z.W.; Oo, T.H.; Thein, K.Z.; Tun, A.M.; Saeed, H. Copper deficiency anemia: Review article. Ann. Hematol. 2018, 97, 1527–1534. [Google Scholar] [CrossRef]
- Gulec, S.; Collins, J.F. Molecular Mediators Governing Iron-Copper Interactions. Annu. Rev. Nutr. 2014, 34, 95–116. [Google Scholar] [CrossRef]
- Vashchenko, G.; MacGillivray, R.T.A. Multi-Copper Oxidases and Human Iron Metabolism. Nutrients 2013, 5, 2289–2313. [Google Scholar] [CrossRef]
- Bao, W.; Rong, Y.; Rong, S.; Liu, L. Dietary iron intake, body iron stores, and the risk of type 2 diabetes: A systematic review and meta-analysis. BMC Med. 2012, 10, 119. [Google Scholar] [CrossRef]
- Kennish, L.; Straub, R.H.; Oh, C.; Krasnokutsky, S.; Samuels, J.; Greenberg, J.D.; Huang, X.; Abramson, S.B. Age-dependent ferritin elevations and HFE C282Y mutation as risk factors for symptomatic knee osteoarthritis in males: A longitudinal cohort study. BMC Musculoskelet. Disord. 2014, 15, 8. [Google Scholar] [CrossRef]
- Lima, S.C.V.C.; Arrais, R.F.; Sales, C.H.; Almeida, M.D.G.; De Sena, K.C.M.; Oliveira, V.T.L.; De Andrade, A.S.; Pedrosa, L.F.C. Assessment of Copper and Lipid Profile in Obese Children and Adolescents. Biol. Trace Element Res. 2006, 114, 19–30. [Google Scholar] [CrossRef]
- Yu, L.; Yan, J.; Zhang, Q.; Lin, H.; Zhu, L.; Liu, Q.; Zhao, C. Association between Serum Ferritin and Blood Lipids: Influence of Diabetes and hs-CRP Levels. J. Diabetes Res. 2020, 2020, 4138696. [Google Scholar] [CrossRef]
- Li, J.; Bao, W.; Zhang, T.; Zhou, Y.; Yang, H.; Jia, H.; Wang, R.; Cao, Y.; Xiao, C. Independent relationship between serum ferritin levels and dyslipidemia in Chinese adults: A population study. PLoS ONE 2017, 12, e0190310. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.D.; Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014, 23, R89–R98. [Google Scholar] [CrossRef]
- Gill, D.; Greco, F.D.G.; Walker, A.P.; Srai, S.K.; Laffan, M.A.; Minelli, C. The Effect of Iron Status on Risk of Coronary Artery Disease. Arter. Thromb. Vasc. Biol. 2017, 37, 1788–1792. [Google Scholar] [CrossRef]
- Gill, D.; Monori, G.; Tzoulaki, I.; Dehghan, A. Iron Status and Risk of Stroke. Stroke 2018, 49, 2815–2821. [Google Scholar] [CrossRef]
- Pichler, I.; Del Greco, M.F.; Gogele, M.; Lill, C.M.; Bertram, L.; Do, C.B.; Eriksson, N.; Foroud, T.; Myers, R.H.; Nalls, M.; et al. Serum iron levels and the risk of Parkinson disease: A Mendelian randomization study. PLoS Med. 2013, 10, e1001462. [Google Scholar] [CrossRef]
- Kodali, H.P.; Pavilonis, B.T.; Schooling, C.M. Effects of copper and zinc on ischemic heart disease and myocardial infarction: A Mendelian randomization study. Am. J. Clin. Nutr. 2018, 108, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.; Benyamin, B.; Moore, L.; Monori, G.; Zhou, A.; Koskeridis, F.; Evangelou, E.; Laffan, M.; Walker, A.P.; Tsilidis, K.K.; et al. Associations of genetically determined iron status across the phenome: A mendelian randomization study. PLoS Med. 2019, 16, e1002833. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-W.; Zhu, Q.; Zhang, H.-Y. Mineral Nutrition and the Risk of Chronic Diseases: A Mendelian Randomization Study. Nutrients 2019, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Benyamin, B.; InterAct Consortium; Esko, T.; Ried, J.; Radhakrishnan, A.; Vermeulen, S.; Traglia, M.; Gögele, M.; Anderson, D.; Broer, L.; et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 2014, 5, 4926. [Google Scholar] [CrossRef]
- Evans, D.M.; Zhu, G.; Dy, V.; Heath, A.C.; Madden, P.A.F.; Kemp, J.P.; McMahon, G.; Pourcain, B.S.; Timpson, N.J.; Golding, J.; et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum. Mol. Genet. 2013, 22, 3998–4006. [Google Scholar] [CrossRef]
- Wu, P.; Gifford, A.; Meng, X.; Li, X.; Campbell, H.; Varley, T.; Zhao, J.; Carroll, R.; Bastarache, L.; Denny, J.C.; et al. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med. Informatics 2019, 7, e14325. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Global Lipids Genetics Consortium; Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef]
- Sanderson, E.; Smith, G.D.; Windmeijer, F.; Bowden, J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int. J. Epidemiol. 2018, 48, 713–727. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Smith, G.D.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Kamat, M.A.; Blackshaw, J.A.; Young, R.; Surendran, P.; Burgess, S.; Danesh, J.; Butterworth, A.S.; Staley, J.R. PhenoScanner V2: An expanded tool for searching human genotype–phenotype associations. Bioinformatics 2019, 35, 4851–4853. [Google Scholar] [CrossRef] [PubMed]
- Yavorska, O.; Burgess, S. MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 2017, 46, 1734–1739. [Google Scholar] [CrossRef]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Looker, A.C.; Dallman, P.R.; Carroll, M.D.; Gunter, E.W.; Johnson, C.L. Prevalence of iron deficiency in the United States. JAMA 1997, 277, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Domellöf, M.; Lönnerdal, B.; Dewey, K.G.; Cohen, R.J.; Rivera, L.L.; Hernell, O. Sex differences in iron status during infancy. Pediatrics 2002, 110, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.F.; Harris, C.; Kaye, J.; Lind, B.; Carter, R.; Anekonda, T.; Ralle, M. Gender Effects on Plasma and Brain Copper. Int. J. Alzheimer’s Dis. 2011, 2011, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Lopes, P.A.; Santos, M.C.; Vicente, L.; Rodrigues, M.O.; Pavão, M.L.; Nève, J.; Viegas-Crespo, A.M. Trace Element Status (Se, Cu, Zn) in Healthy Portuguese Subjects of Lisbon Population: A Reference Study. Biol. Trace Element Res. 2004, 101, 1–18. [Google Scholar] [CrossRef]
- Bo, S.; Durazzo, M.; Gambino, R.; Berutti, C.; Milanesio, N.; Caropreso, A.; Gentile, L.; Cassader, M.; Cavallo-Perin, P.; Pagano, G. Associations of Dietary and Serum Copper with Inflammation, Oxidative Stress, and Metabolic Variables in Adults. J. Nutr. 2008, 138, 305–310. [Google Scholar] [CrossRef]
- Zang, X.; Huang, H.; Zhuang, Z.; Chen, R.; Xie, Z.; Xu, C.; Xuming, M. The association between serum copper concentrations and cardiovascular disease risk factors in children and adolescents in NHANES. Environ. Sci. Pollut. Res. 2018, 25, 16951–16958. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; He, B.; Xiao, Y.; Chen, Y.-J. Iron metabolism and its association with dyslipidemia risk in children and adolescents: A cross-sectional study. Lipids Health Dis. 2019, 18, 1–8. [Google Scholar] [CrossRef]
- Das De, S.; Krishna, S.; Jethwa, A. Iron status and its association with coronary heart disease: Systematic review and meta-analysis of prospective studies. Atherosclerosis 2015, 238, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Gill, D.; Brewer, C.F.; Monori, G.; Trégouët, D.; Franceschini, N.; Giambartolomei, C.; Tzoulaki, I.; Dehghan, A. INVENT Consortium Effects of Genetically Determined Iron Status on Risk of Venous Thromboembolism and Carotid Atherosclerotic Disease: A Mendelian Randomization Study. J. Am. Hear. Assoc. 2019, 8, e012994. [Google Scholar] [CrossRef]
- Ozdemir, A.; Sevinc, C.; Selamet, U.; Turkmen, F. The Relationship Between Iron Deficiency Anemia and Lipid Metabolism in Premenopausal Women. Am. J. Med. Sci. 2007, 334, 331–333. [Google Scholar] [CrossRef]
- Wrede, C.E.; Buettner, R.; Bollheimer, L.C.; Schölmerich, J.; Palitzsch, K.-D.; Hellerbrand, C. Association between serum ferritin and the insulin resistance syndrome in a representative population. Eur. J. Endocrinol. 2006, 154, 333–340. [Google Scholar] [CrossRef]
- Campillos, M.; Cases, I.; Hentze, M.W.; Sanchez, M. SIREs: Searching for iron-responsive elements. Nucleic Acids Res. 2010, 38, W360–W367. [Google Scholar] [CrossRef]
- Davis, M.R.; Rendina, E.; Peterson, S.K.; Lucas, E.A.; Smith, B.J.; Clarke, S.L. Enhanced expression of lipogenic genes may contribute to hyperglycemia and alterations in plasma lipids in response to dietary iron deficiency. Genes Nutr. 2012, 7, 415–425. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; Mangan, D.; O’Keefe, J.H. Copper deficiency may be a leading cause of ischaemic heart disease. Open Hear. 2018, 5, e000784. [Google Scholar] [CrossRef]
- Alarcón-Corredor, O.M.; Guerrero, Y.; De Fernández, M.R.; D’Jesús, I.; Burguera, M.; Burguera, J.L.; Di Bernardo, M.L.; García, M.Y.; Alarcón, A.O. Effect of copper supplementation on lipid profile of Venezuelan hyperlipemic patients. Arch. Latinoam. Nutr. 2004, 54, 413–418. [Google Scholar]
- Kim, S.; Chao, P.Y.; Allen, K.G.D. Inhibition of elevated hepatic glutathione abolishes copper deficiency cholesterolemia. FASEB J. 1992, 6, 2467–2471. [Google Scholar] [CrossRef] [PubMed]
- Tosco, A.; Fontanella, B.; Danise, R.; Cicatiello, L.; Grober, O.M.V.; Ravo, M.; Weisz, A.; Marzullo, L. Molecular bases of copper and iron deficiency-associated dyslipidemia: A microarray analysis of the rat intestinal transcriptome. Genes Nutr. 2009, 5, 1–8. [Google Scholar] [CrossRef]
- Carr, T.P.; Lei, K.Y. In Vivo Apoprotein Catabolism of High Density Lipoproteins in Copper-Deficient, Hypercholesterolemic Rats. Exp. Biol. Med. 1989, 191, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Lutsenko, S.; Burkhead, J.L. The Role of Copper as a Modifier of Lipid Metabolism; IntechOpen: London, UK, 2013. [Google Scholar]
- Spivak, J.L. Myeloproliferative Neoplasms. N. Engl. J. Med. 2017, 377, 895–896. [Google Scholar] [CrossRef]
- Ginzburg, Y.Z.; Feola, M.; Zimran, E.; Varkonyi, J.; Ganz, T.; Hoffman, R. Dysregulated iron metabolism in polycythemia vera: Etiology and consequences. Leukemia 2018, 32, 2105–2116. [Google Scholar] [CrossRef]
- Kucukguven, A.; Khalil, R.A. Matrix metalloproteinases as potential targets in the venous dilation associated with varicose veins. Curr. Drug Targets 2013, 14, 287–324. [Google Scholar] [PubMed]
- Fukaya, E.; Flores, A.M.; Lindholm, D.; Gustafsson, S.; Zanetti, D.; Ingelsson, E.; Leeper, N.J. Clinical and Genetic Determinants of Varicose Veins. Circulation 2018, 138, 2869–2880. [Google Scholar] [CrossRef]
- Zamboni, P.; Scapoli, G.; Lanzara, V.; Izzo, M.; Fortini, P.; Legnaro, R.; Palazzo, A.; Tognazzo, S.; Gemmati, D. Serum Iron and Matrix Metalloproteinase-9 Variations in Limbs Affected by Chronic Venous Disease and Venous Leg Ulcers. Dermatol. Surg. 2005, 31, 644–649. [Google Scholar] [CrossRef]
- Chiang, C.-P.; Chang, J.Y.-F.; Wang, Y.-P.; Wu, Y.-H.; Wu, Y.-C.; Sun, A. Atrophic glossitis: Etiology, serum autoantibodies, anemia, hematinic deficiencies, hyperhomocysteinemia, and management. J. Formos. Med Assoc. 2020, 119, 774–780. [Google Scholar] [CrossRef]
- Smith, G.D. Mendelian randomization: Prospects, potentials, and limitations. Int. J. Epidemiol. 2004, 33, 30–42. [Google Scholar] [CrossRef]
- Pierce, B.L.; Burgess, S. Efficient Design for Mendelian Randomization Studies: Subsample and 2-Sample Instrumental Variable Estimators. Am. J. Epidemiol. 2013, 178, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Smith, G.D.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
| SNP | Effect Allele | Baseline Allele | Chr | Closest Gene | % Variance Explained | F-Statistic | EAF | Beta a | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 SNPs for Fe from Benyamin et al. (N b = 48,972) | ||||||||||
| rs1800562 | A | G | 6 | HFE | 1.30 | 645 | 0.067 | 0.328 | 0.016 | 2.72 × 10−97 |
| rs1799945 | G | C | 6 | HFE | 0.90 | 445 | 0.15 | 0.189 | 0.010 | 1.10 × 10−81 |
| rs855791 | G | A | 22 | TMPRSS6 | 1.60 | 796 | 0.554 | 0.181 | 0.007 | 1.32 × 10−139 |
| 2 SNPs for Cu from Evans et al. (N b = 2603) | ||||||||||
| rs1175550 | G | A | 1 | SMIM1 | 1.45 | 38 | 0.23 | 0.198 | 0.032 | 5.03 × 10−10 |
| rs2769264 | G | T | 1 | SELENBP1 | 3.15 | 85 | 0.18 | 0.313 | 0.034 | 2.63 × 10−20 |
| Exposure/Outcome | MR Method | Beta | SE | 95% CI | P-Effect | P-Pleiotropy | n_Total | Data |
|---|---|---|---|---|---|---|---|---|
| Iron | ||||||||
| HDL cholesterol | WM | −0.008 | 0.015 | (−0.037, 0.021) | 0.602 | - | 183,990 | GLGC |
| IVW | −0.003 | 0.013 | (−0.028, 0.022) | 0.801 | 0.396 | |||
| MR Egger | −0.062 | 0.055 | (−0.170, 0.045) | 0.459 | 0.430 | |||
| WM | −0.005 | 0.004 | (−0.012, 0.003) | 0.231 | - | 224,140 | UKBB | |
| IVW | −0.002 | 0.004 | (−0.011, 0.006) | 0.589 | 0.162 | |||
| MR Egger | −0.021 | 0.014 | (−0.048, 0.005) | 0.361 | 0.385 | |||
| LDL cholesterol | WM | −0.058 | 0.019 | (−0.095, −0.021) | 0.002 | - | 169,960 | GLGC |
| IVW | −0.100 | 0.043 | (−0.184, −0.015) | 0.020 | 6 × 10−5 | |||
| MR Egger | −0.351 | 0.059 | (−0.467, −0.235) | 0.106 | 0.143 | |||
| WM | −0.089 | 0.014 | (−0.116, −0.062) | 5.27 × 10−11 | - | 244,476 | UKBB | |
| IVW | −0.083 | 0.042 | (−0.165, −0.001) | 0.048 | 7.6 × 10−13 | |||
| MR Egger | −0.279 | 0.127 | (−0.528, −0.031) | 0.271 | 0.356 | |||
| Total cholesterol | WM | −0.047 | 0.019 | (−0.085, −0.010) | 0.013 | - | 184,158 | GLGC |
| IVW | −0.083 | 0.044 | (−0.169, 0.003) | 0.060 | 1.9 × 10−5 | |||
| MR Egger | −0.342 | 0.057 | (−0.454, −0.230) | 0.106 | 0.135 | |||
| WM | −0.096 | 0.018 | (−0.132, −0.061) | 8.99 × 10−8 | - | 244,950 | UKBB | |
| IVW | −0.090 | 0.056 | (−0.201, 0.020) | 0.109 | 9.8 × 10−14 | |||
| MR Egger | −0.359 | 0.162 | (−0.676, −0.042) | 0.270 | 0.336 | |||
| Triglycerides | IVW | 0.034 | 0.012 | (0.010, 0.059) | 0.006 | 0.958 | 174,687 | GLGC |
| WM | 0.033 | 0.013 | (0.007, 0.059) | 0.014 | - | |||
| MR Egger | 0.049 | 0.053 | (−0.055, 0.154) | 0.524 | 0.986 | |||
| WM | 0.047 | 0.012 | (0.025, 0.070) | 4.2 × 10−5 | - | 244,754 | UKBB | |
| IVW | 0.043 | 0.016 | (0.012, 0.074) | 0.006 | 0.043 | |||
| MR Egger | −0.040 | 0.035 | (−0.109, 0.030) | 0.463 | 0.250 | |||
| Copper | ||||||||
| HDL cholesterol | IVW | 0.004 | 0.028 | (−0.050, 0.058) | 0.880 | 0.116 | 94,311 | GLGC |
| IVW | 0.002 | 0.003 | (−0.004, 0.008) | 0.448 | 0.885 | 221,738 | UKBB | |
| LDL cholesterol | IVW | −0.048 | 0.019 | (−0.085, −0.011) | 0.011 | 0.785 | 89,888 | GLGC |
| IVW | −0.008 | 0.013 | (−0.033, 0.017) | 0.543 | 0.100 | 241,831 | UKBB | |
| Total cholesterol | IVW | −0.043 | 0.018 | (−0.079, −0.007) | 0.020 | 0.823 | 94,595 | GLGC |
| IVW | −0.013 | 0.016 | (−0.044, 0.017) | 0.388 | 0.121 | 242,304 | UKBB | |
| Triglycerides | IVW | −0.024 | 0.020 | (−0.063, 0.015) | 0.233 | 0.241 | 91,013 | GLGC |
| IVW | −0.008 | 0.009 | (−0.025, 0.008) | 0.333 | 0.874 | 242,112 | UKBB |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Liu, C.; Francis, M.; Sun, Y.; Ryu, M.-S.; Grider, A.; Ye, K. The Causal Effects of Blood Iron and Copper on Lipid Metabolism Diseases: Evidence from Phenome-Wide Mendelian Randomization Study. Nutrients 2020, 12, 3174. https://doi.org/10.3390/nu12103174
Zhou J, Liu C, Francis M, Sun Y, Ryu M-S, Grider A, Ye K. The Causal Effects of Blood Iron and Copper on Lipid Metabolism Diseases: Evidence from Phenome-Wide Mendelian Randomization Study. Nutrients. 2020; 12(10):3174. https://doi.org/10.3390/nu12103174
Chicago/Turabian StyleZhou, Jingqi, Chang Liu, Michael Francis, Yitang Sun, Moon-Suhn Ryu, Arthur Grider, and Kaixiong Ye. 2020. "The Causal Effects of Blood Iron and Copper on Lipid Metabolism Diseases: Evidence from Phenome-Wide Mendelian Randomization Study" Nutrients 12, no. 10: 3174. https://doi.org/10.3390/nu12103174
APA StyleZhou, J., Liu, C., Francis, M., Sun, Y., Ryu, M.-S., Grider, A., & Ye, K. (2020). The Causal Effects of Blood Iron and Copper on Lipid Metabolism Diseases: Evidence from Phenome-Wide Mendelian Randomization Study. Nutrients, 12(10), 3174. https://doi.org/10.3390/nu12103174







