Statistical and Machine-Learning Analyses in Nutritional Genomics Studies
Abstract
1. Introduction
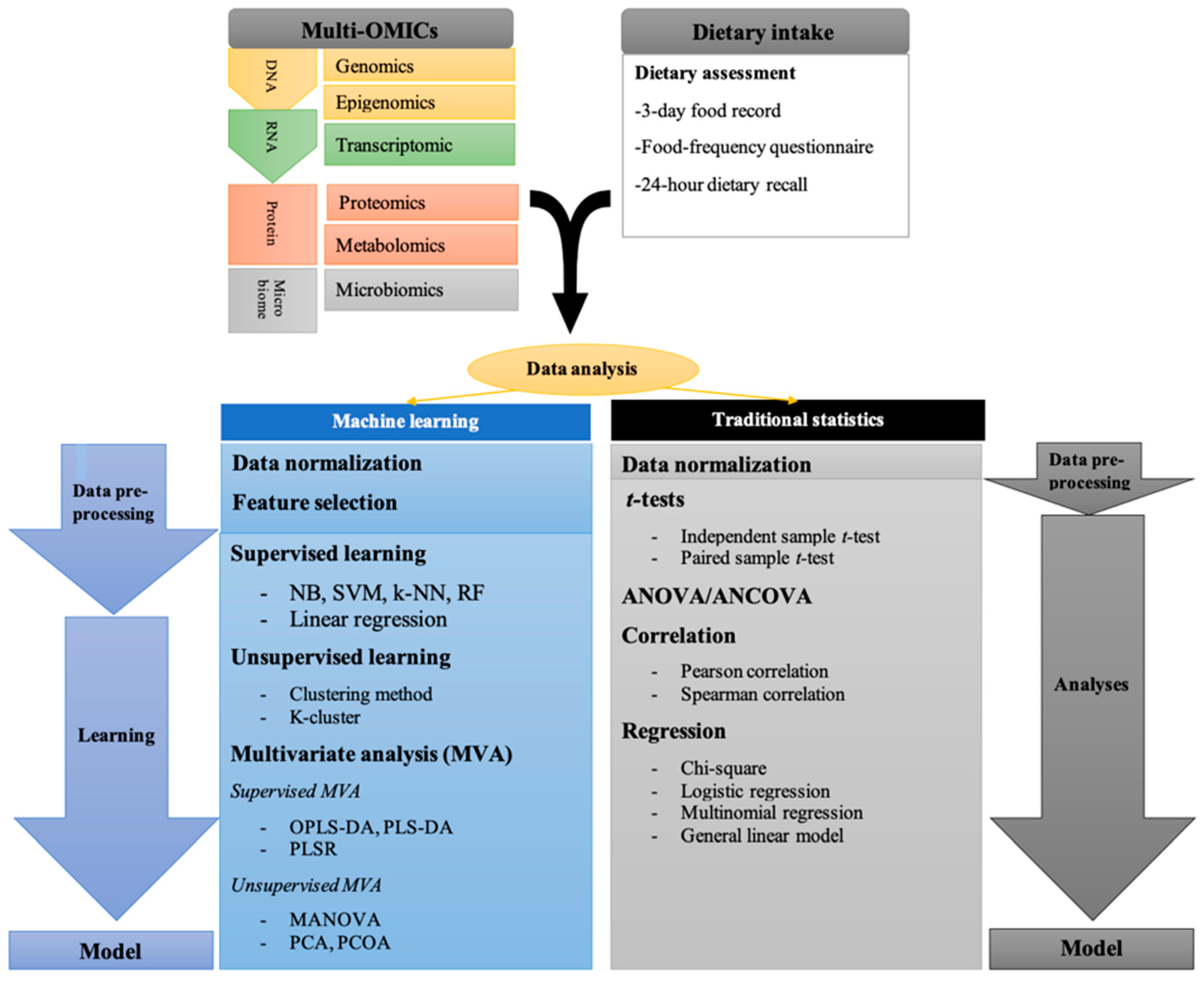
2. OMICs in Nutrition Research
3. Traditional Statistical Analysis in Nutrition Studies
4. Machine Learning in Nutrition Studies
4.1. Supervised Machine Learning
4.1.1. Data Preparation
4.1.2. Classification Methods in Supervised Machine Learning
4.1.3. Regression Method in Supervised Machine Learning
4.2. Unsupervised Machine Learning
4.3. Multivariate Analysis
4.3.1. Supervised Multivariate Analysis
4.3.2. Unsupervised Multivariate Analysis
5. Multi-OMICs Studies in Nutrition Research
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Murgia, C.; Adamski, M.M. Translation of nutritional genomics into nutrition practice: The next step. Nutrients 2017, 9, 366. [Google Scholar] [CrossRef]
- Bouchard-Mercier, A.; Paradis, A.-M.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.-C. Associations between dietary patterns and gene expression profiles of healthy men and women: A cross-sectional study. Nutr. J. 2013, 12, 24. [Google Scholar] [CrossRef]
- Tewari, A.K.; Mohanty, S.; Roy, S. Proteomics and Nutrition Research: An Overview. Genom. Proteom. Metab. Nutraceuticals Funct. Foods 2015, 245. [Google Scholar] [CrossRef]
- Tang, Z.-Z.; Chen, G.; Hong, Q.; Huang, S.; Smith, H.M.; Shah, R.D.; Scholz, M.B.; Ferguson, J.F. Multi-omic analysis of the microbiome and metabolome in healthy subjects reveals microbiome-dependent relationships between diet and metabolites. Front. Genet. 2019, 10, 454. [Google Scholar] [CrossRef]
- Paoloni-Giacobino, A.; Grimble, R.; Pichard, C. Genetics and nutrition. Clin. Nutr. 2003, 22, 429–435. [Google Scholar] [CrossRef]
- Rudkowska, I.; Guénard, F.; Julien, P.; Couture, P.; Lemieux, S.; Barbier, O.; Calder, P.C.; Minihane, A.M.; Vohl, M.-C. Genome-wide association study of the plasma triglyceride response to an n-3 polyunsaturated fatty acid supplementation. J. Lipid Res. 2014, 55, 1245–1253. [Google Scholar] [CrossRef]
- Soliai, M.M.; Kato, A.; Stanhope, C.T.; Norton, J.E.; Naughton, K.A.; Klinger, A.I.; Kern, R.C.; Tan, B.K.; Schleimer, R.P.; Nicolae, D.L.; et al. Multi-omics co-localization with genome-wide association studies reveals context-specific mechanisms of asthma risk variants. bioRxiv 2019, 593558. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, R.; Cheng, F.; Wei, Q.; Ji, Y.; Yang, H.; Zhong, X.; Tao, R.; Wen, Z.; Sutcliffe, J.S.; et al. A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nat. Neurosci. 2019, 22, 691. [Google Scholar] [CrossRef]
- Dubourg-Felonneau, G.; Cannings, T.; Cotter, F.; Thompson, H.; Patel, N.; Cassidy, J.W.; Clifford, H.W. Machine Learning for Health (ML4H) Workshop at NeurIPS. arXiv 2018, arXiv:1811.10455. [Google Scholar]
- Drabsch, T.; Gatzemeier, J.; Pfadenhauer, L.; Hauner, H.; Holzapfel, C. Associations between single nucleotide polymorphisms and total energy, carbohydrate, and fat intakes: A systematic review. Adv. Nutr. 2018, 9, 425–453. [Google Scholar] [CrossRef]
- Park, S.; Daily, J.W.; Zhang, X.; Jin, H.S.; Lee, H.J.; Lee, Y.H. Interactions with the MC4R rs17782313 variant, mental stress and energy intake and the risk of obesity in Genome Epidemiology Study. Nutr. Metab. 2016, 13, 38. [Google Scholar] [CrossRef]
- Grimaldi, K.A.; van Ommen, B.; Ordovas, J.M.; Parnell, L.D.; Mathers, J.C.; Bendik, I.; Brennan, L.; Celis-Morales, C.; Cirillo, E.; Daniel, H. Proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice. Genes Nutr. 2017, 12, 35. [Google Scholar] [CrossRef]
- Zhang, D.; Li, Z.; Wang, H.; Yang, M.; Liang, L.; Fu, J.; Wang, C.; Ling, J.; Zhang, Y.; Zhang, S. Interactions between obesity-related copy number variants and dietary behaviors in childhood obesity. Nutrients 2015, 7, 3054–3066. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Baier, S.R.; Nguyen, C.; Xie, F.; Wood, J.R.; Zempleni, J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J. Nutr. 2014, 144, 1495–1500. [Google Scholar] [CrossRef]
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA methylation and DNA methyltransferases. Epigenet. Chromatin 2017, 10, 23. [Google Scholar] [CrossRef]
- Karlić, R.; Chung, H.-R.; Lasserre, J.; Vlahoviček, K.; Vingron, M. Histone modification levels are predictive for gene expression. Proc. Natl. Acad. Sci. USA 2010, 107, 2926–2931. [Google Scholar] [CrossRef]
- Druesne-Pecollo, N.; Latino-Martel, P. Modulation of histone acetylation by garlic sulfur compounds. Anti Cancer Agent 2011, 11, 254–259. [Google Scholar] [CrossRef]
- Reuter, S.; Gupta, S.C.; Park, B.; Goel, A.; Aggarwal, B.B. Epigenetic changes induced by curcumin and other natural compounds. Genes Nutr. 2011, 6, 93–108. [Google Scholar] [CrossRef]
- Herrera-Marcos, L.; Lou-Bonafonte, J.; Arnal, C.; Navarro, M.; Osada, J. Transcriptomics and the mediterranean diet: A systematic review. Nutrients 2017, 9, 472. [Google Scholar] [CrossRef]
- Hsiao, L.-L.; Jensen, R.; Yoshida, T.; Clark, K.; Blumenstock, J.; Gullans, S. Correcting for signal saturation errors in the analysis of microarray data. Biotechniques 2002, 32, 330–336. [Google Scholar] [CrossRef]
- Konstantinidou, V.; Khymenets, O.; Fitó Colomer, M.; Fornell, T.; Anglada Busquets, R.; Dopazo, A.; Covas Planells, M.I. Characterization of human gene expression changes after olive oil ingestion: An exploratory approach. Folia Biol. 2009, 55, 85–91. [Google Scholar]
- Geyer, P.E.; Wewer Albrechtsen, N.J.; Tyanova, S.; Grassl, N.; Iepsen, E.W.; Lundgren, J.; Madsbad, S.; Holst, J.J.; Torekov, S.S.; Mann, M. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 2016, 12, 901. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef]
- Metabolomexchange. Available online: http://www.metabolomexchange.org/site/ (accessed on 12 October 2020).
- Ley, S.H.; Sun, Q.; Willett, W.C.; Eliassen, A.H.; Wu, K.; Pan, A.; Grodstein, F.; Hu, F.B. Associations between red meat intake and biomarkers of inflammation and glucose metabolism in women. Am. J. Clin. Nutr. 2013, 99, 352–360. [Google Scholar] [CrossRef]
- Cho, C.E.; Taesuwan, S.; Malysheva, O.V.; Bender, E.; Yan, J.; Caudill, M.A. Choline and one-carbon metabolite response to egg, beef and fish among healthy young men: A short-term randomized clinical study. Clin. Nutr. Exp. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- O’Connor, S.; Greffard, K.; Leclercq, M.; Julien, P.; Weisnagel, S.J.; Gagnon, C.; Droit, A.; Bilodeau, J.F.; Rudkowska, I. Increased Dairy Product Intake Alters Serum Metabolite Profiles in Subjects at Risk of Developing Type 2 Diabetes. Mol. Nutr. Food Res. 2019, 63, 1900126. [Google Scholar] [CrossRef]
- Abbondio, M.; Palomba, A.; Tanca, A.; Fraumene, C.; Pagnozzi, D.; Serra, M.; Marongiu, F.; Laconi, E.; Uzzau, S. Fecal Metaproteomic Analysis Reveals Unique Changes of the Gut Microbiome Functions After Consumption of Sourdough Carasau Bread. Front. Microbiol. 2019, 10, 1733. [Google Scholar] [CrossRef]
- Eid, N.; Enani, S.; Walton, G.; Corona, G.; Costabile, A.; Gibson, G.; Rowland, I.; Spencer, J.P. The impact of date palm fruits and their component polyphenols, on gut microbial ecology, bacterial metabolites and colon cancer cell proliferation. J. Nutr. Sci. 2014, 3, e46. [Google Scholar] [CrossRef]
- Ali, Z.; Bhaskar, S.B. Basic statistical tools in research and data analysis. Indian J. Anaesth. 2016, 60, 662. [Google Scholar] [CrossRef]
- Fu, W.J.; Stromberg, A.J.; Viele, K.; Carroll, R.J.; Wu, G. Statistics and bioinformatics in nutritional sciences: Analysis of complex data in the era of systems biology. J. Nutr. Biochem. 2010, 21, 561–572. [Google Scholar] [CrossRef]
- Boushey, C.J.; Harris, J.; Bruemmer, B.; Archer, S.L. Publishing nutrition research: A review of sampling, sample size, statistical analysis, and other key elements of manuscript preparation, Part 2. J. Am. Diet. Assoc. 2008, 108, 679–688. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, U.; Pandey, C.M.; Mishra, P.; Pandey, G. Application of student’s t-test, analysis of variance, and covariance. Ann. Card. Anaesth. 2019, 22, 407. [Google Scholar] [CrossRef]
- Kim, T.K.; Park, J.H. More about the basic assumptions of t-test: Normality and sample size. Korean J. Anesth. 2019, 72, 331. [Google Scholar] [CrossRef]
- Gaddis, G. Advanced biostatistics: Chi-square, ANOVA, regression, and multiple regression. Doing Res. Emerg. Acute Care 2016, 213. [Google Scholar] [CrossRef]
- Khorrami-Nezhad, L.; Mirzaei, K.; Maghbooli, Z.; Keshavarz, S.A. Dietary fat intake associated with bone mineral density among visfatin genotype in obese people. Br. J. Nutr. 2018, 119, 3–11. [Google Scholar] [CrossRef]
- Pooyan, S.; Rahimi, M.H.; Mollahosseini, M.; Khorrami-Nezhad, L.; Nasir, Y.; Maghbooli, Z.; Mirzaei, K. A high-protein/low-fat diet may interact with vitamin D-binding protein gene variants to moderate the risk of depression in apparently healthy adults. Lifestyle Genom. 2018, 11, 64–72. [Google Scholar] [CrossRef]
- Cesar, L.; Suarez, S.V.; Adi, J.; Adi, N.; Vazquez-Padron, R.; Yu, H.; Ma, Q.; Goldschmidt-Clermont, P.J.; Agatston, A.; Kurlansky, P. An essential role for diet in exercise-mediated protection against dyslipidemia, inflammation and atherosclerosis in ApoE-/-mice. PLoS ONE 2011, 6, e17263. [Google Scholar] [CrossRef]
- Little, R.J.; An, H.; Johanns, J.; Giordani, B. A comparison of subset selection and analysis of covariance for the adjustment of confounders. Psychol. Methods 2000, 5, 459. [Google Scholar] [CrossRef]
- Stephens, M.A. Tests based on regression and correlation. In Goodness-of-Fit-Techniques; Routledge: Abingdon, UK, 2017; pp. 195–234. [Google Scholar]
- Smith, A.D.; Emmett, P.M.; Newby, P.; Northstone, K. Dietary patterns obtained through principal components analysis: The effect of input variable quantification. Br. J. Nutr. 2013, 109, 1881–1891. [Google Scholar] [CrossRef]
- McHugh, M.L. The chi-square test of independence. Br. J. Nutr. 2013, 23, 143–149. [Google Scholar] [CrossRef]
- Huang, S.; Chaudhary, K.; Garmire, L.X. More is better: Recent progress in multi-omics data integration methods. Front. Genet. 2017, 8, 84. [Google Scholar] [CrossRef]
- Lluch, A.; Maillot, M.; Gazan, R.; Vieux, F.; Delaere, F.; Vaudaine, S.; Darmon, N. Individual diet modeling shows how to balance the diet of French adults with or without excessive free sugar intakes. Nutrients 2017, 9, 162. [Google Scholar] [CrossRef]
- Nasteski, V. An overview of the supervised machine learning methods. Horizons B 2017, 4, 51–62. [Google Scholar] [CrossRef]
- Álvarez, J.D.; Matias-Guiu, J.A.; Cabrera-Martín, M.N.; Risco-Martín, J.L.; Ayala, J.L. An application of machine learning with feature selection to improve diagnosis and classification of neurodegenerative disorders. BMC Bioinform. 2019, 20, 491. [Google Scholar] [CrossRef]
- Dao, M.C.; Sokolovska, N.; Brazeilles, R.; Affeldt, S.; Pelloux, V.; Prifti, E.; Chilloux, J.; Verger, E.O.; Kayser, B.D.; Aron-Wisnewsky, J. A data integration multi-omics approach to study calorie restriction-induced changes in insulin sensitivity. Front. Physiol. 2019, 9, 1958. [Google Scholar] [CrossRef]
- Kenkel, N. On selecting an appropriate multivariate analysis. Can. J. Plant Sci. 2006, 86, 663–676. [Google Scholar] [CrossRef]
- Putri, R.A.; Sendari, S.; Widiyaningtyas, T. Classification of Toddler Nutrition Status with Anthropometry Calculation using Naïve Bayes Algorithm. In Proceedings of the 2018 International Conference on Sustainable Information Engineering and Technology (SIET), Malang, Indonesia, 10–12 November 2018; pp. 66–70. [Google Scholar]
- Kim, S.; Jhong, J.-H.; Lee, J.; Koo, J.-Y. Meta-analytic support vector machine for integrating multiple omics data. Biodata Min. 2017, 10, 2. [Google Scholar] [CrossRef]
- Ahmadi, H.; Rodehutscord, M. Application of artificial neural network and support vector machines in predicting metabolizable energy in compound feeds for pigs. Front. Nutr. 2017, 4, 27. [Google Scholar] [CrossRef]
- Panaretos, D.; Koloverou, E.; Dimopoulos, A.C.; Kouli, G.-M.; Vamvakari, M.; Tzavelas, G.; Pitsavos, C.; Panagiotakos, D.B. A comparison of statistical and machine-learning techniques in evaluating the association between dietary patterns and 10-year cardiometabolic risk (2002–2012): The ATTICA study. Br. J. Nutr. 2018, 120, 326–334. [Google Scholar] [CrossRef]
- Acharjee, A.; Kloosterman, B.; Visser, R.G.; Maliepaard, C. Integration of multi-omics data for prediction of phenotypic traits using random forest. BMC Bioinform. 2016, 17, 180. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Lin, E.; Lane, H.-Y. Machine learning and systems genomics approaches for multi-omics data. Biomark. Res. 2017, 5, 2. [Google Scholar] [CrossRef]
- Schrider, D.R.; Kern, A.D. Supervised machine learning for population genetics: A new paradigm. Trends Genet. 2018, 34, 301–312. [Google Scholar] [CrossRef]
- Huo, Z.; Tseng, G. Integrative sparse K-means with overlapping group lasso in genomic applications for disease subtype discovery. Ann. Appl. Stat. 2017, 11, 1011. [Google Scholar] [CrossRef]
- Grech, V.; Calleja, N. WASP (Write a Scientific Paper): Multivariate analysis. Early Hum. Dev. 2018, 123, 42–45. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; van Velzen, E.J.; Hoefsloot, H.C.; Smilde, A.K. Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics 2010, 6, 119–128. [Google Scholar] [CrossRef]
- Benítez-Páez, A.; Kjølbæk, L.; Del Pulgar, E.M.G.; Brahe, L.K.; Astrup, A.; Matysik, S.; Schött, H.-F.; Krautbauer, S.; Liebisch, G.; Boberska, J. A multi-omics approach to unraveling the microbiome-mediated effects of arabinoxylan oligosaccharides in overweight humans. Msystems 2019, 4. [Google Scholar] [CrossRef]
- Roy, K. Advances in QSAR modeling. In Applications in Pharmaceutical, Chemical, Food, Agricultural and Environmental Sciences; Springer: Cham, Switzerland, 2017; Volume 555, p. 39. [Google Scholar]
- Zhang, F.; Tapera, T.M.; Gou, J. Application of a new dietary pattern analysis method in nutritional epidemiology. BMC Med. Res. Methodol. 2018, 18, 119. [Google Scholar] [CrossRef]
- Sundekilde, U.K.; Yde, C.C.; Honore, A.H.; Caverly Rae, J.M.; Burns, F.R.; Mukerji, P.; Mawn, M.P.; Stenman, L.; Dragan, Y.; Glover, K. An Integrated Multi-Omics Analysis Defines Key Pathway Alterations in a Diet-Induced Obesity Mouse Model. Metabolites 2020, 10, 80. [Google Scholar] [CrossRef]
- Csala, A.; Zwinderman, A.H. Multivariate Statistical Methods for High-Dimensional Multiset Omics Data Analysis. In Computational Biology [Internet]; Codon Publications: Brisbane, Australia, 2019; pp. 71–83. [Google Scholar]
- Henry, V.J.; Bandrowski, A.E.; Pepin, A.-S.; Gonzalez, B.J.; Desfeux, A. OMICtools: An informative directory for multi-omic data analysis. Database 2014, 2014, bau069. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef]
- Kato, H.; Takahashi, S.; Saito, K. Omics and integrated omics for the promotion of food and nutrition science. J. Tradit. Complement. Med. 2011, 1, 25–30. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Berry, S.E.; Valdes, A.M.; Drew, D.A.; Asnicar, F.; Mazidi, M.; Wolf, J.; Capdevila, J.; Hadjigeorgiou, G.; Davies, R.; Al Khatib, H. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 2020, 26, 964–973. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, L.; Xia, B.; Tang, S.; Liu, L.; Xie, J.; Zhang, H. Bioregional Alterations in Gut Microbiome Contribute to the Plasma Metabolomic Changes in Pigs Fed with Inulin. Microorganisms 2020, 8, 111. [Google Scholar] [CrossRef]
- Tremblay, B.L.; Guénard, F.; Lamarche, B.; Pérusse, L.; Vohl, M.-C. Integrative Network Analysis of Multi-Omics Data in the Link between Plasma Carotenoid Concentrations and Lipid Profile. Lifestyle Genom. 2020, 13, 11–19. [Google Scholar] [CrossRef]
- Wang, F.; Wan, Y.; Yin, K.; Wei, Y.; Wang, B.; Yu, X.; Ni, Y.; Zheng, J.; Huang, T.; Song, M. Lower Circulating Branched-Chain Amino Acid Concentrations Among Vegetarians are Associated with Changes in Gut Microbial Composition and Function. Mol. Nutr. Food Res. 2019, 63, 1900612. [Google Scholar] [CrossRef]
- Guirro, M.; Costa, A.; Gual-Grau, A.; Mayneris-Perxachs, J.; Torrell, H.; Herrero, P.; Canela, N.; Arola, L. Multi-omics approach to elucidate the gut microbiota activity: Metaproteomics and metagenomics connection. Electrophoresis 2018, 39, 1692–1701. [Google Scholar] [CrossRef]
- Piening, B.D.; Zhou, W.; Contrepois, K.; Röst, H.; Urban, G.J.G.; Mishra, T.; Hanson, B.M.; Bautista, E.J.; Leopold, S.; Yeh, C.Y. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018, 6, 157–170.e8. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Wu, H.; Bjornson, E.; Zhang, C.; Hakkarainen, A.; Räsänen, S.M.; Lee, S.; Mancina, R.M.; Bergentall, M.; Pietiläinen, K.H. An integrated understanding of the rapid metabolic benefits of a carbohydrate-restricted diet on hepatic steatosis in humans. Cell Metab. 2018, 27, 559–571.e5. [Google Scholar] [CrossRef]
- Ishii, C.; Nakanishi, Y.; Murakami, S.; Nozu, R.; Ueno, M.; Hioki, K.; Aw, W.; Hirayama, A.; Soga, T.; Ito, M. A metabologenomic approach reveals changes in the intestinal environment of mice fed on american diet. Int. J. Mol. Sci. 2018, 19, 4079. [Google Scholar] [CrossRef]
- Kieffer, D.A.; Piccolo, B.D.; Marco, M.L.; Kim, E.B.; Goodson, M.L.; Keenan, M.J.; Dunn, T.N.; Knudsen, K.E.B.; Martin, R.J.; Adams, S.H. Mice fed a high-fat diet supplemented with resistant starch display marked shifts in the liver metabolome concurrent with altered gut bacteria. J. Nutr. 2016, 146, 2476–2490. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M. Personalized nutrition by prediction of glycemic responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Takahashi, S.; Saito, K.; Jia, H.; Kato, H. An integrated multi-omics study revealed metabolic alterations underlying the effects of coffee consumption. PLoS ONE 2014, 9, e91134. [Google Scholar] [CrossRef]
| References | Type of Study | Population Omics | Methodology | Main Analysis Strategy | Main Finding |
|---|---|---|---|---|---|
| Berry SE. et al., 2020 [70] | Cohort study (multi-national study) | N = 1002 healthy adults (UK) N = 100 healthy adults (USA) | Nutrition assessment: Food frequency questionnaires Biochemical measurement: GCM, ADVIA chemistry triglyceride and glucose oxidase method Genomics: Illumina Infinium HumanHap610 Microbiomics: 16S rRNA and arrays | Multilinear ANOVA (Hierarchical Bayes models) Random forest regression (Unsupervised ML) PCA (Unsupervised ML) | Medium-fat and -carbohydrate lunch showed a less impact on postprandial lipemia compared to gut microbiome while genetic had a modest influence on glycemic and lipid profile. |
| Wu W. et al., 2020 [71] | Animal study | N = 12 (pigs) 6 pigs (case): a maize-soybean meal diet containing 5% a high-fermentable fiber (Inulin) 6 pigs (control): a 5% low-fermentable fiber (cellulose) control | Metabolomics: GC-TOF-MS method Microbiomics: 16S and whole metagenome | PERMANOVA (Multivariate analysis- unsupervised ML) PLS-DA (Multivariate analysis-supervised ML) Integrative analysis: mixOmics package of R software | Inulin intake has effects on the increasing the diversity of microbiota composition in the cecum along with a decrease of the circulating of metabolites including branched-chain amino acids, L-valine, L-isoleucine and an increase in the level of indole-3-propionic acid. |
| Sundekilde U.K. et al., 2020 [64] | Animal study | N = 20 mice (C57Bl/6J), males, 6-week old Group 1: 60% fat (high-fat diet; HFD) Group 2: 10% fat (low-fat diet; LFD) | Genomics: (RNA extraction, Illumina, Qiagen) Metabolomics: NMR Spectroscopy, LC-MS analysis on urine and plasma, LC-MS analysis on tissue samples, GC-MS analysis Microbiomics: 16S and whole metagenome | PCoA (Multivariate analysis-unsupervised ML) | Increase in malate, succinate and oxaloacetate levels were associated to down-regulation of gene expression of malate dehydrogenase together with gut microbiota enrichment (Lachnospiraceae, Ruminococcaceae, Streptococcaceae, Lactobacillaceae) in HFD compared to LFD mice. |
| Tremblay B.L. et al., 2020 [72] | Observational study | N = 48 healthy Parents = 22, age, 42.3 year Children = 26, age, 11.3 year | Nutrition assessment: carotenoid measurements High-performance liquid chromatograph (HPLC), ChemStation software Biochemical measurement: Enzymatic assays, Friedewald formula, the rocket immunoelectrophoretic method Genomics: DNA and RNA extraction (microarray platform, Illumina) Epigenomics: Methylation; Infinium Human Methylation 450 array | One-Sample Wilcoxon Signed Rank Test Linear regression Clustering method (Unsupervised ML) Correlation based analyses WGCNA (Data mining method) | Genes expression in lipid metabolism and inflammatory pathways together with DNA methylation have a mediatory role in the association between total carotenoids and lipid profile in plasma. |
| Benitez-Paez A. et al., 2019 [61] | Randomized crossover study | N = 15 overweight subjects Duration: 4-weeks for each phase 10.4 g/day AXOS (Arabinoxylan-oligosaccharides) | Biochemical measurement: plasma and fecal bile acids (LC-MS/MS) fecal lipid species (LC-MS/HRMS) Microbiomics: 16S and whole metagenome Metabolomics: NMR Spectroscopy | Paired and one-sided t-test or Wilcox signed-rank test Logistic regression model PCA (Unsupervised ML) OPLS-DA (supervised ML) | Increase in the abundance of Actinobacteria, Bifidobacteriaceae, Bifidobacterium and change the host metabolism including glucose homeostasis (reduction in fasting insulin and HOMA-IR) after consumption of AXOS. |
| Wang F. et al., 2019 [73] | Preliminary study | N = 36, age = 28.1 Duration of study = 6 months Vegan = 12 Lacto-ovo vegetarian = 12 Omnivorous = 12 | Nutrition assessment: 3-day food records Metabolomics: Gas chromatography coupled to time-of-flight mass spectrometry system Metatranscriptomic: Illumina HiSeq 4000, KEGG, using BLASTP Microbiomics: 16S and whole metagenome | Chi-square and t-test for PCoA (Multivariate analysis-unsupervised ML) Clustering method (Unsupervised ML) | Decrease concentrations of BCAAs, the abundance of Prevotella and Bacteroides were increased and decreased, respectively, among vegetarians compared with omnivores. |
| Tang Z.Z. et al., 2019 [4] | Cross-sectional study | N = 150 healthy (55 M, 95 F) Age 18–50 year | Nutrition assessment: 3-day food records and food frequency questionnaires Metabolomics: (untargeted LC-MS) Microbiomics: 16S and whole metagenome sequencing from stool | Correlation based analyses Sparse Linear Log-Contrast Model (Supervised ML) Network analysis-WGCNA) (Data mining method) | Mediatory role of Ruminococcaceae in the association of plant-derived food and artificial sweeteners with bile acids in stool. |
| Guirro M. et al., 2018 [74] | Animal study | N = 24 male Sprague-Dawley rats (8-week old) Duration: 9-week Two groups: N = 12, cafeteria (CAF) N = 12 standard chow (STD) After intervention (8-week): Each diet group supplemented: 1. Low-fat condensed milk (n = 6) 2. Hesperidin dissolved with low-fat condensed milk (n = 6) | Metaproteomics: NanoLC-(Orbitrap) MS/MS analysis Microbiomics: 16S and whole metagenome sequencing from stool | Univariate statistical analysis (Student’s t-test) PCA (Multivariate analysis-unsupervised ML) | Increase the abundance of Bacteroidetes and Firmicutes, which are related to down-regulation of proteins in energy metabolism pathways such as the tricarboxylic acid cycle or ATP-binding pathways after CAF diet. |
| Dao M.C. et al., 2018 [48] | Cohort study | 27 F (24), M (3) overweight or obese adults 6-week calorie restriction (CR): 1200 kcal/day (F) 1500 kcal/day (M) | Nutrition assessment: 7-day food records Genomics: DNA (microarray platform, Illumina) Transcriptomics: Microarray platform (Illumina) Metabolomics: Gas chromatography system (GC–MS) and H-NMR Spectroscopy | Nutrition analysis (Profile Dossier v3 & Profile Dossier x029) PLSR (Supervised ML) | Increase in insulin sensitivity and BCAA after CR associated with gut microbiota, metabolomics and adipose tissue genes in both genders. |
| Piening B.D. et al., 2018 [75] | Case-control study | 13 Insulin resistance (IR) participants F (7), M (6)-age: 58, BMI: 30.5 10 Insulin sensitivity (IS) participants BMI 25–35 kg/m2, F (7), M (3)-age: 56, BMI: 28.5 First, hypercaloric diet for a period of 30-day (increase 880 kcal per day) Second, iso-caloric diet for 7 days Third, a caloric-restricted (CR) diet 60-day period | Proteomics: Untargeted liquid chromatography (LC-MS) Metabolomics: Untargeted LC-MS Microbiomics: 16S and whole metagenome sequencing from stool) | Correlation and regression-based analyses Random forest classification (Supervised ML) Interaction model: generalized linear models (GLMs) | Dysregulation of antimicrobial response (CAMP, LFT, and defensins) was reflected in proteome and circulating cytokines in IR compared to IS participants. |
| Mardinoglu A. et al., 2018 [76] | Short term intervention study | N = 10 (NAFLD), F (2)–M (8) BMI: 34.1, age: 47 Intervention: Isocaloric, low-carbohydrate diet (23–30 g/day) with increased protein (24% of total energy)-14 days | Transcriptomics: Microarray platform (Illumina) Metabolomics: Untargeted Analyses UPLC/MS/MS-GC/MS | Correlation based analyses Linear mixed effect model The multivariate analysis based on mix DIABLO (Supervised and unsupervised ML) | Increase in serum concentration of β-hydroxybutyrate concentrations, mitochondrial β-oxidation, and folate producing Streptococcus and serum folate after intervention. |
| Ishii C. et al., 2018 [77] | Case-control study | Mice (C57BL/6J) 52 weeks: Normal diet (n = 6) American diet (n = 5) | Metabolomics: (Ultrafree MC) Metagenomics: (microarray platform, Affymetrix) | Correlation based analysis PICRUST software analyses (Supervised and unsupervised ML) | Abundance of genes associated with butyrate metabolism is positively correlated with butyrate producing bacteria (Oscillospira and Ruminococcus). |
| Kieffer D. et al., 2016 [78] | Animal study | 45% kcal from fat + high-amylose-maize resistant starch type 2 (HAMRS2), Case, N = 14 5-week old male (C57BL/6J mice) Digestible starch, Control, N = 15 5-week old male C57BL/6J mice Duration: 5 weeks | Metabolomics: Untargeted GC-MS Microbiomics: 16S and whole metagenome sequencing from stool, analyzed by QIIME | Correlation and t-test-based analyses PLS-DA (Multivariate analysis- supervised ML) | Changes in hepatic metabolism and gene expression related to fatty acids metabolism together with increases in Tenericutes, Bacteroidetes, Verrucomicrobia and decrease in Proteobacteria and Firmicutes phyla after HAMRS2 diet. |
| Zhang C. et al., 2015 [79] | Case-control study | N = 17, Prader–Willi syndrome (PWS), duration 90 day N = 21, heathy obese, duration 30 days Intervention: WTP diet (whole grains, traditional Chinese medicinal foods, and prebiotics) | Nutrition assessment: 24-h dietary record Biochemical analysis: Serum glucose, CRP, total cholesterol, triglycerides, free fatty acids, ALT and AST (automatic biochemical analyzer (ADVIA® 1800 Clinical Chemistry System), Insulin (immunochemiluminometric assays), HbAlc (HPLC) Metabolomics: NMR Spectroscopy Microbiomics: 16S and whole metagenome sequencing from stool | Wilcoxon matched-pairs signed rank test (two-tailed) Independent Mann–Whitney U test (two-tailed) OPLS-DA | Balance of gut microbiota composition which contributes to the alleviation of metabolic deterioration in obesity among children with Prader–Willi syndrome after consumption of a diet rich in fermentable carbohydrates. Children genetically obese with Prader–Willi syndrome shared a similar dysbiosis in their gut microbiota with those having diet-related obesity. |
| Zeevi D. et al., 2015 [80] | Cohort study | N = 800 healthy, F (60%)–M (40%) 54% Overweight 22% Obese | Nutrition assessment: Dietary habits (www.personalnutrition.org) Biochemical analysis: Glucose was measured for 7 days using the iPro2 CGM with Enlite sensors Genomics: DNA extraction (microarray platform, Illumina) Microbiomics: 16S and whole metagenome sequencing from stool | Correlation and regression-based analyses PCoA) (Multivariate analysis-unsupervised ML) | Lower postprandial responses are related to alterations Proteobacteria and Enterobacteriaceae based on the ML algorithm. |
| Takahashi S et al., 2014 [81] | Case-control Study | Mice (C57BL/6J) 5 groups (n = 8–9), (9 weeks) Caffeine (2 g coffee powder/140 mL) -Normal diet group = 10% fat -High-fat diet group = 60% fat -Caffeinated coffee group = A high-fat diet + 2% caffeine -Decaffeinated coffee group = A high-fat diet + 2% decaffeinated coffee. -Green unroasted coffee group = A high-fat diet + 2% unroasted caffeinated coffee | Biochemical measurement: Hepatic lipid composition (the Folch method (Folch, Lees, & Sloane, 1957)), plasma liver enzymes (The transaminase C II-test WAKO kit) Genomics: (DNA microarray platform, Affymetrix) Proteomics: two-dimensional electrophoresis combined with MALDI-TOF mass spectrometry Metabolomics: Millipore Ultrafree-MC PLHCC HMT/CE-TOF-MS) analysis | PCA (Unsupervised ML) | Up-regulation of the iso-citrate dehydrogenase, lipid metabolism and ATP turnover were related anti-obesity effects of different types of coffee. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khorraminezhad, L.; Leclercq, M.; Droit, A.; Bilodeau, J.-F.; Rudkowska, I. Statistical and Machine-Learning Analyses in Nutritional Genomics Studies. Nutrients 2020, 12, 3140. https://doi.org/10.3390/nu12103140
Khorraminezhad L, Leclercq M, Droit A, Bilodeau J-F, Rudkowska I. Statistical and Machine-Learning Analyses in Nutritional Genomics Studies. Nutrients. 2020; 12(10):3140. https://doi.org/10.3390/nu12103140
Chicago/Turabian StyleKhorraminezhad, Leila, Mickael Leclercq, Arnaud Droit, Jean-François Bilodeau, and Iwona Rudkowska. 2020. "Statistical and Machine-Learning Analyses in Nutritional Genomics Studies" Nutrients 12, no. 10: 3140. https://doi.org/10.3390/nu12103140
APA StyleKhorraminezhad, L., Leclercq, M., Droit, A., Bilodeau, J.-F., & Rudkowska, I. (2020). Statistical and Machine-Learning Analyses in Nutritional Genomics Studies. Nutrients, 12(10), 3140. https://doi.org/10.3390/nu12103140






