Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment Condition
2.2. Total RNA Extraction and Quantitative Analysis of mRNA Expression
2.3. siRNA Transfection
2.4. Western Blot Analysis
2.5. Cell Growth Assay
2.6. Amino Acid Analysis
2.7. Statistical Analyses
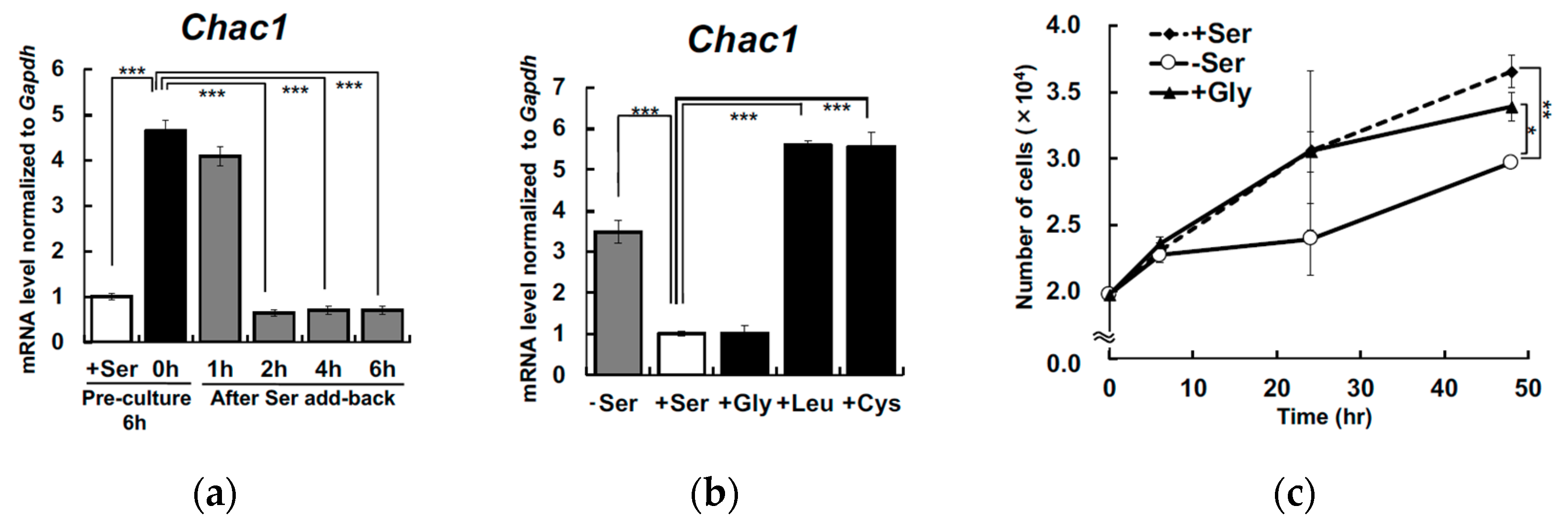
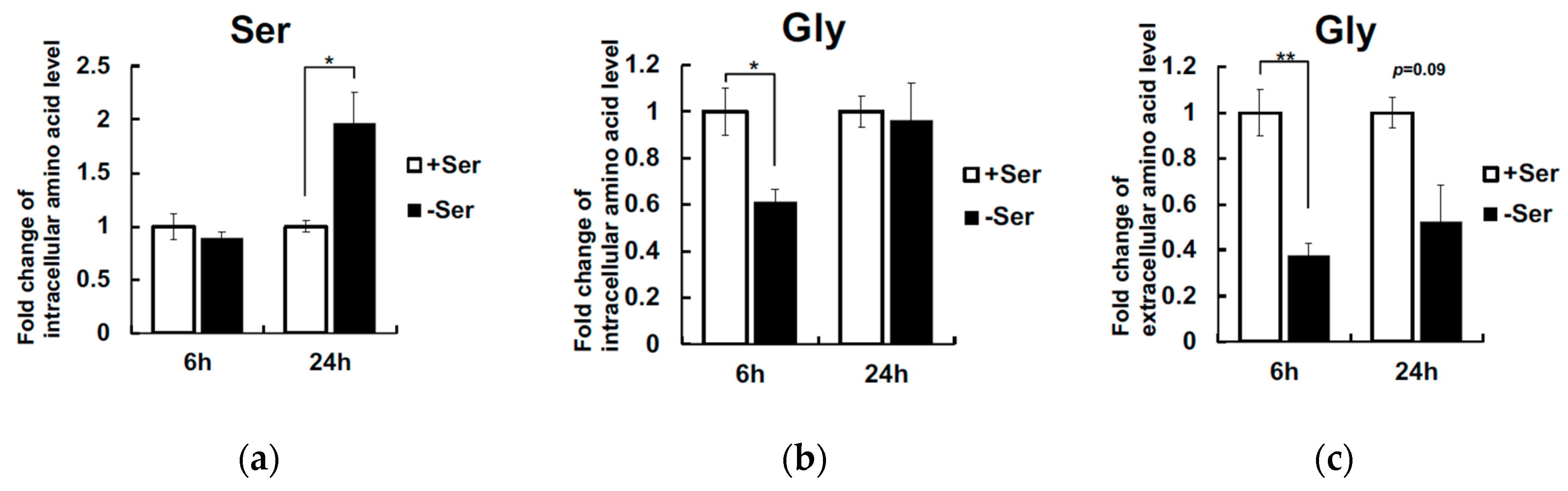
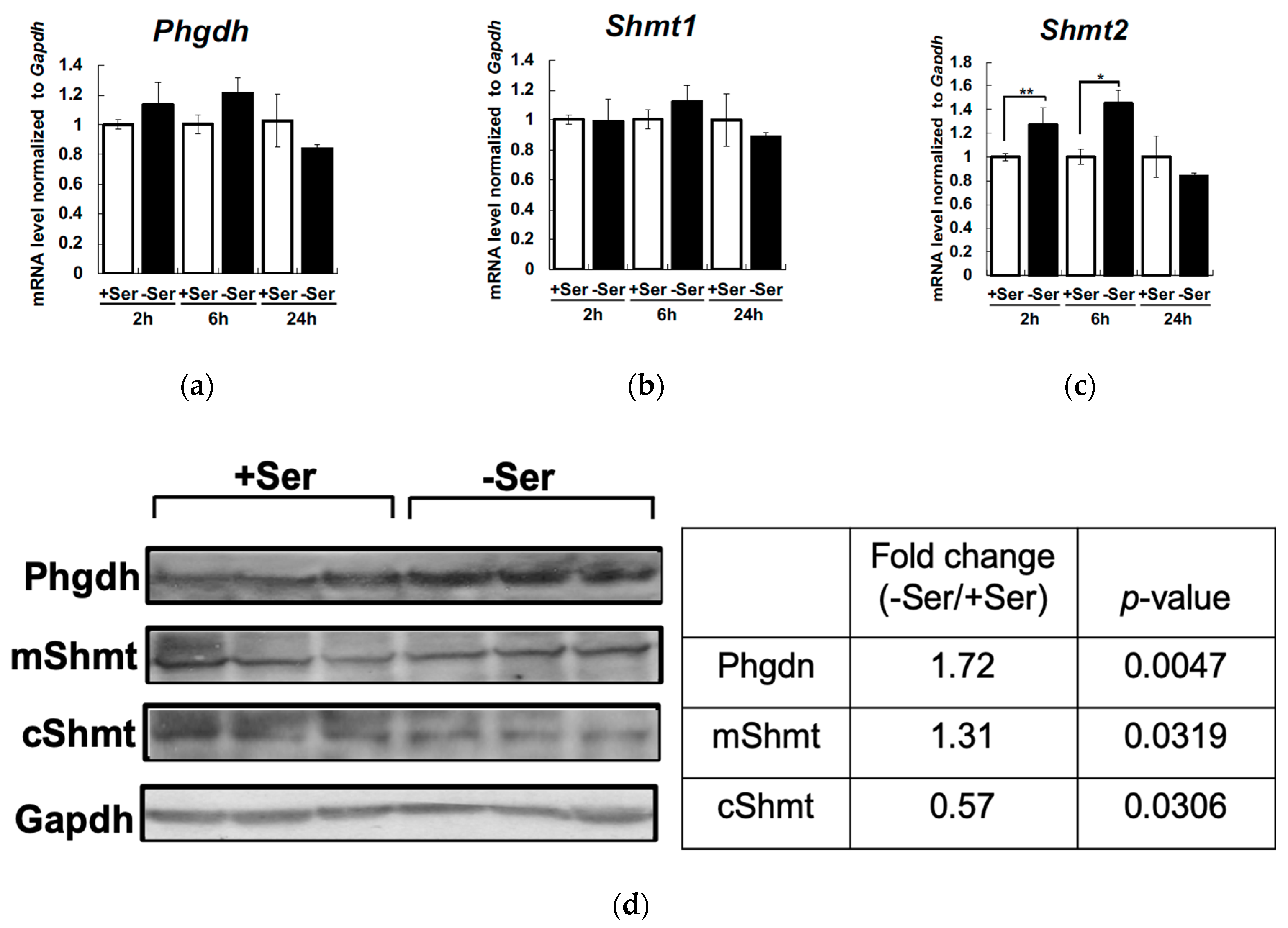
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Maddocks, O.D.K.; Berkers, C.R.; Mason, S.M.; Zheng, L.; Blyth, K.; Gottlieb, E.; Vousden, K.H. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 2013, 493, 542–546. [Google Scholar] [CrossRef]
- Samanta, D.; Park, Y.; Andrabi, S.A.; Shelton, L.M.; Gilkes, D.M.; Semenza, G.L. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016, 76, 4430–4442. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.-D.; Wu, W.K.K.; Wang, H.-U.; Li, X.-X. Serine and one-carbon metabolism, a bridge that links mTOR signaling and DNA methylation in cancer. Pharmacol. Res. 2019, 149, 104352. [Google Scholar] [CrossRef] [PubMed]
- Li, A.M.; Ye, J. Reprogramming of serine, glycine and one-carbon metabolism in cancer. Biochim. Biophys. Acta-Mol. Basis Dis. 2020, 1866, 165841. [Google Scholar] [CrossRef] [PubMed]
- Esaki, K.; Sayano, T.; Sonoda, C.; Akagi, T.; Suzuki, T.; Ogawa, T.; Okamoto, M.; Yoshikawa, T.; Hirabayashi, Y.; Furuya, S. L-serine deficiency elicits intracellular accumulation of cytotoxic deoxysphingolipids and lipid body formation. J. Biol. Chem. 2015, 290, 14595–14609. [Google Scholar] [CrossRef]
- Sayano, T.; Kawano, Y.; Kusada, W.; Arimoto, Y.; Esaki, K.; Hamano, M.; Udono, M.; Katakura, Y.; Ogawa, T.; Kato, H.; et al. Adaptive response to l-serine deficiency is mediated by p38 MAPK activation via 1-deoxysphinganine in normal fibroblasts. FEBS Open Bio 2016, 6. [Google Scholar] [CrossRef]
- Hamano, M.; Sayano, T.; Kusada, W.; Kato, H.; Furuya, S. Microarray data on altered transcriptional program of Phgdh-deficient mouse embryonic fibroblasts caused by L-serine depletion. Data Br. 2016, 7, 1598–1601. [Google Scholar] [CrossRef]
- Hamano, M.; Haraguchi, Y.; Sayano, T.; Zyao, C.; Arimoto, Y.; Kawano, Y.; Moriyasu, K.; Udono, M.; Katakura, Y.; Ogawa, T.; et al. Enhanced vulnerability to oxidative stress and induction of inflammatory gene expression in 3-phosphoglycerate dehydrogenase-deficient fibroblasts. FEBS Open Bio 2018, 8, 914–922. [Google Scholar] [CrossRef]
- Sayano, T.; Kawakami, Y.; Kusada, W.; Suzuki, T.; Kawano, Y.; Watanabe, A.; Takashima, K.; Arimoto, Y.; Esaki, K.; Wada, A.; et al. L-Serine deficiency caused by genetic Phgdh deletion leads to robust induction of 4E-BP1 and subsequent repression of translation initiation in the developing central nervous system. FEBS J. 2013, 280, 1502–1517. [Google Scholar] [CrossRef]
- Yamasaki, M.; Yamada, K.; Furuya, S.; Mitoma, J.; Hirabayashi, Y.; Watanabe, M. 3-Phosphoglycerate dehydrogenase, a key enzyme for L-serine biosynthesis, is preferentially expressed in the radial glia/astrocyte lineage and olfactory ensheathing glia in the mouse brain. J. Neurosci. 2001, 21, 7691–7704. [Google Scholar] [CrossRef] [PubMed]
- Cascade, A.; Mungrue, I.N.; Pagnon, J.; Kohannim, O.; Gargalovic, P.S.; Lusis, A.J. CHAC1/MGC4504 Is a Novel Proapoptotic Component of the. J. Immunol. 2009, 210, 466–476. [Google Scholar]
- Crawford, R.R.; Prescott, E.T.; Sylvester, C.F.; Higdon, A.N.; Shan, J.; Kilberg, M.S.; Mungrue, I.N. Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a bipartite ATF/CRE regulatory element. J. Biol. Chem. 2015, 290, 15878–15891. [Google Scholar] [CrossRef] [PubMed]
- Tsunoda, S.; Avezov, E.; Zyryanova, A.; Konno, T.; Mendes-Silva, L.; Pinho Melo, E.; Harding, H.P.; Ron, D. Intact protein folding in the glutathione-depleted endoplasmic reticulum implicates alternative protein thiol reductants. Elife 2014, 3, 1–20. [Google Scholar] [CrossRef]
- Jiang, H.; Wek, S.A.; Mcgrath, B.C.; Lu, D.; Hai, T.; Harding, H.P.; Wang, X.; Ron, D.; Cavener, D.R.; Wek, R.C. Activating Transcription Factor 3 Is Integral to the Eukaryotic Initiation Factor 2 Kinase Stress Response. Mol. Cell. Biol. 2004, 24, 1365–1377. [Google Scholar] [CrossRef]
- Mazor, K.M.; Stipanuk, M.H. GCN2- and eIF2α-phosphorylation-independent, but ATF4-dependent, induction of CARE-containing genes in methionine-deficient cells. Amino Acids 2016, 48, 2831–2842. [Google Scholar] [CrossRef]
- Safory, H.; Neame, S.; Shulman, Y.; Zubedat, S.; Radzishevsky, I.; Rosenberg, D.; Sason, H.; Engelender, S.; Avital, A.; Hülsmann, S.; et al. The alanine-serine-cysteine-1 (Asc-1) transporter controls glycine levels in the brain and is required for glycinergic inhibitory transmission. EMBO Rep. 2015, 16, 590–598. [Google Scholar] [CrossRef]
- Garcia-Santos, D.; Schranzhofer, M.; Bergeron, R.; Sheftel, A.D.; Ponka, P. Extracellular glycine is necessary for optimal hemoglobinization of erythroid cells. Haematologica 2017, 102, 1314–1323. [Google Scholar] [CrossRef]
- Woo, C.C.; Chen, W.C.; Teo, X.Q.; Radda, G.K.; Lee, P.T.H. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 2016, 7, 53005–53017. [Google Scholar] [CrossRef]
- Hayashi, S.; Tanaka, T.; Naito, J.; Suda, M. Dietary and hormonal regulation of serine synthesis in the rat. J. Biochem. 1975, 77, 207–219. [Google Scholar] [CrossRef]
- Locasale, J.W.; Grassian, A.R.; Melman, T.; Lyssiotis, C.A.; Mattaini, K.R.; Bass, A.J.; Heffron, G.; Metallo, C.M.; Muranen, T.; Sharfi, H.; et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 2011, 43, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Possemato, R.; Marks, K.M.; Shaul, Y.D.; Pacold, M.E.; Kim, D.; Birsoy, K.; Sethumadhavan, S.; Woo, H.; Jang, H.G.; Jha, A.K.; et al. pathway is essential in breast cancer. Nature 2011, 476, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.A.; Allen, A.E.; Liu, S.; Liberti, M.V.; Liu, P.; Liu, X.; Dai, Z.; Gao, X.; Wang, Q.; Liu, Y.; et al. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Pacold, M.E.; Brimacombe, K.R.; Chan, S.H.; Rohde, J.M.; Lewis, C.A.; Swier, L.J.Y.M.; Possemato, R.; Chen, W.W.; Sullivan, L.B.; Fiske, B.P.; et al. A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat. Chem. Biol. 2016, 12, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef]
- Gao, X.; Lee, K.; Reid, M.A.; Sanderson, S.M.; Qiu, C.; Li, S.; Liu, J.; Locasale, J.W. Serine Availability Influences Mitochondrial Dynamics and Function through Lipid Metabolism. Cell Rep. 2018, 22, 3507–3520. [Google Scholar] [CrossRef]
- Vandekeere, S.; Dubois, C.; Kalucka, J.; Sullivan, M.R.; García-Caballero, M.; Goveia, J.; Chen, R.; Diehl, F.F.; Bar-Lev, L.; Souffreau, J.; et al. Serine Synthesis via PHGDH Is Essential for Heme Production in Endothelial Cells. Cell Metab. 2018, 28, 573–587.e13. [Google Scholar] [CrossRef]
- Zhang, T.; Gillies, M.C.; Madigan, M.C.; Shen, W.; Du, J.; Grünert, U.; Zhou, F.; Yam, M.; Zhu, L. Disruption of De Novo Serine Synthesis in Müller Cells Induced Mitochondrial Dysfunction and Aggravated Oxidative Damage. Mol. Neurobiol. 2018, 55, 7025–7037. [Google Scholar] [CrossRef]
- Chen, J.; Chung, F.; Yang, G.G.; Pu, M.; Gao, H.; Jiang, W.; Yin, H.; Capka, V.; Kasibhatla, S.; Laffitte, B.; et al. Phosphoglycerate dehydrogenase is dispensable for breast tumor maintenance and growth. Oncotarget 2013, 4, 2502–2511. [Google Scholar] [CrossRef]
- Zhang, B.; Zheng, A.; Hydbring, P.; Ambroise, G.; Ouchida, A.T.; Goiny, M.; Vakifahmetoglu-Norberg, H.; Norberg, E. PHGDH Defines a Metabolic Subtype in Lung Adenocarcinomas with Poor Prognosis. Cell Rep. 2017, 19, 2289–2303. [Google Scholar] [CrossRef]
- Sullivan, M.R.; Mattaini, K.R.; Dennstedt, E.A.; Nguyen, A.A.; Sivanand, S.; Reilly, M.F.; Meeth, K.; Muir, A.; Darnell, A.M.; Bosenberg, M.W.; et al. Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell Metab. 2019, 29, 1410–1421.e4. [Google Scholar] [CrossRef] [PubMed]
- Razak, M.A.; Begum, P.S.; Viswanath, B.; Rajagopal, S. Multifarious Beneficial Effect of Nonessential Amino Acid, Glycine: A Review. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamano, M.; Tomonaga, S.; Osaki, Y.; Oda, H.; Kato, H.; Furuya, S. Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells. Nutrients 2020, 12, 3018. https://doi.org/10.3390/nu12103018
Hamano M, Tomonaga S, Osaki Y, Oda H, Kato H, Furuya S. Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells. Nutrients. 2020; 12(10):3018. https://doi.org/10.3390/nu12103018
Chicago/Turabian StyleHamano, Momoko, Shozo Tomonaga, Yusuke Osaki, Hiroaki Oda, Hisanori Kato, and Shigeki Furuya. 2020. "Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells" Nutrients 12, no. 10: 3018. https://doi.org/10.3390/nu12103018
APA StyleHamano, M., Tomonaga, S., Osaki, Y., Oda, H., Kato, H., & Furuya, S. (2020). Transcriptional Activation of Chac1 and Other Atf4-Target Genes Induced by Extracellular l-Serine Depletion is negated with Glycine Consumption in Hepa1-6 Hepatocarcinoma Cells. Nutrients, 12(10), 3018. https://doi.org/10.3390/nu12103018






