Maternal Exposure to High-Fat Diet Induces Long-Term Derepressive Chromatin Marks in the Heart
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. In Vivo Experimental Studies
2.3. Heart Sampling
2.4. Protein Extraction
2.5. Western Blotting Analysis
2.6. DNA Methylation
2.7. Data Analysis
3. Results
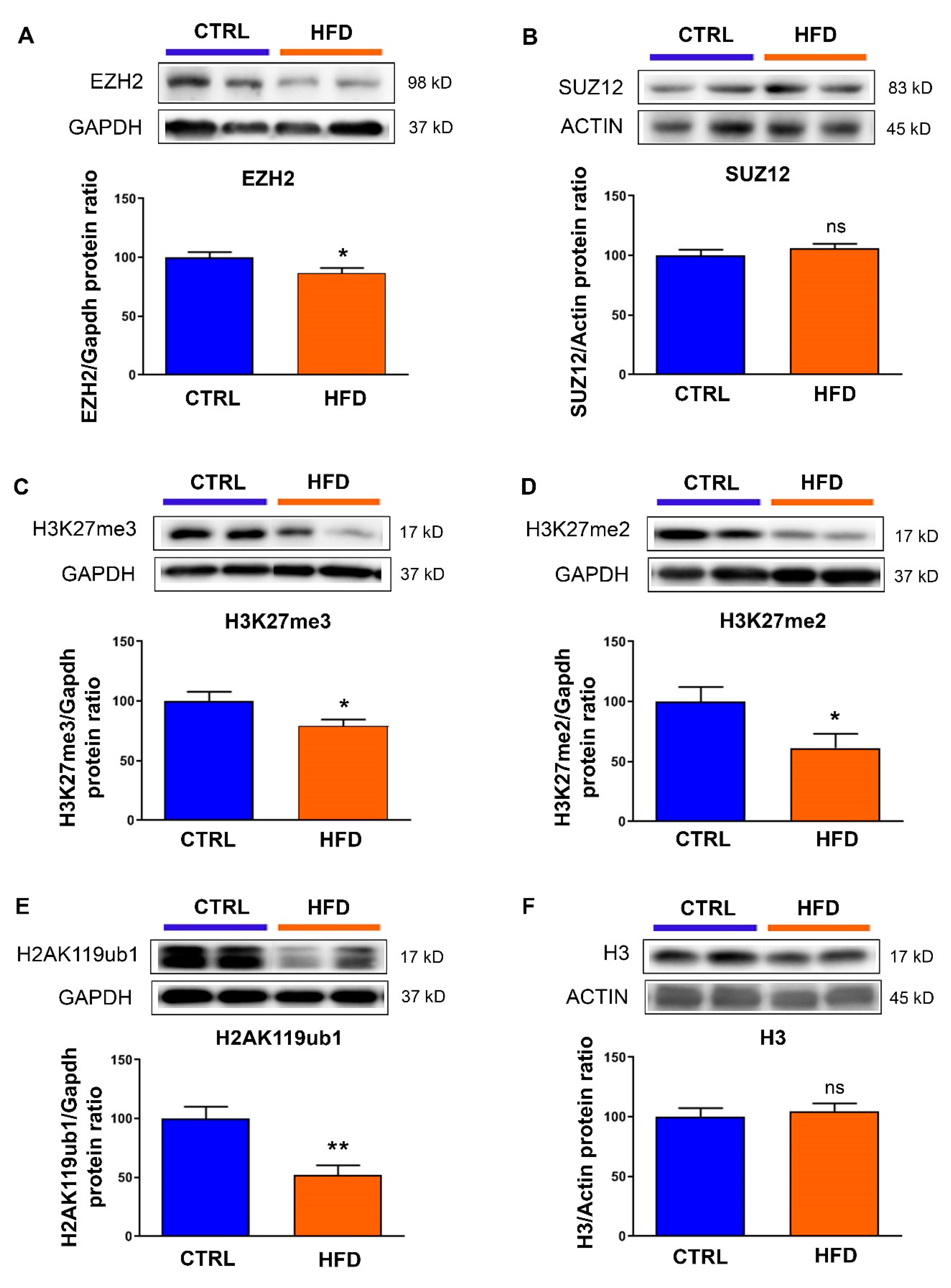
3.1. Exposure to Maternal High-Fat Diet Induces Long-Term Alterations in PRC2
3.2. Exposure to Maternal High-Fat Diet Induces Long-Term Alterations in DNA Methylation
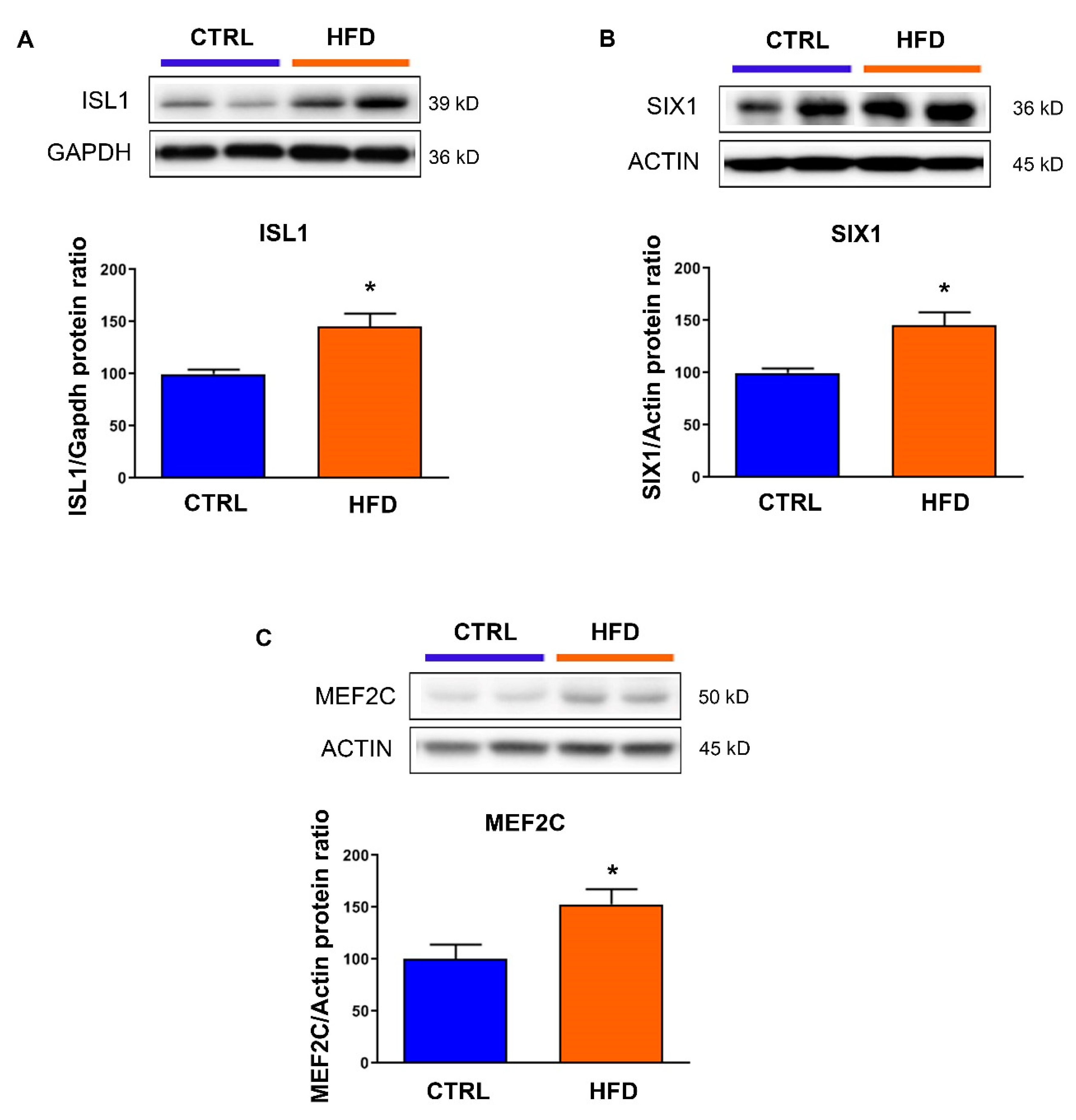
3.3. Exposure to Maternal High-Fat Diet Derepresses Genes Involved in Fibrosis and Hypertrophy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barouki, R.; Gluckman, P.D.; Grandjean, P.; Hanson, M.; Heindel, J.J. Developmental origins of non-communicable disease: Implications for research and public health. Env. Health 2012, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Collaborators, G.B.D.D. Health effects of dietary risks in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019, 393, 1958–1972. [Google Scholar] [CrossRef] [Green Version]
- Ravera, A.; Carubelli, V.; Sciatti, E.; Bonadei, I.; Gorga, E.; Cani, D.; Vizzardi, E.; Metra, M.; Lombardi, C. Nutrition and Cardiovascular Disease: Finding the Perfect Recipe for Cardiovascular Health. Nutrients 2016, 8, 363. [Google Scholar] [CrossRef]
- Siddeek, B.; Li, N.; Mauduit, C.; Chehade, H.; Rigal, E.; Tolsa, J.F.; Armengaud, J.B.; Yzydorczyk, C.; Benahmed, M.; Vergely, C.; et al. Transient postnatal over nutrition induces long-term alterations in cardiac NLRP3-inflammasome pathway. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 944–951. [Google Scholar] [CrossRef]
- Blackmore, H.L.; Niu, Y.; Fernandez-Twinn, D.S.; Tarry-Adkins, J.L.; Giussani, D.A.; Ozanne, S.E. Maternal diet-induced obesity programs cardiovascular dysfunction in adult male mouse offspring independent of current body weight. Endocrinology 2014, 155, 3970–3980. [Google Scholar] [CrossRef] [Green Version]
- Maloyan, A.; Muralimanoharan, S.; Huffman, S.; Cox, L.A.; Nathanielsz, P.W.; Myatt, L.; Nijland, M.J. Identification and comparative analyses of myocardial miRNAs involved in the fetal response to maternal obesity. Physiol. Genomics 2013, 45, 889–900. [Google Scholar] [CrossRef] [Green Version]
- Velkoska, E.; Cole, T.J.; Dean, R.G.; Burrell, L.M.; Morris, M.J. Early undernutrition leads to long-lasting reductions in body weight and adiposity whereas increased intake increases cardiac fibrosis in male rats. J. Nutr. 2008, 138, 1622–1627. [Google Scholar] [CrossRef]
- Vieira, A.K.; Soares, V.M.; Bernardo, A.F.; Neves, F.A.; Mattos, A.B.; Guedes, R.M.; Cortez, E.; Andrade, D.C.; Lacerda-Miranda, G.; Garcia-Souza, E.P.; et al. Overnourishment during lactation induces metabolic and haemodynamic heart impairment during adulthood. Nutr. metab. cardiovasc. dis. 2015. [Google Scholar] [CrossRef]
- Habbout, A.; Guenancia, C.; Lorin, J.; Rigal, E.; Fassot, C.; Rochette, L.; Vergely, C. Postnatal overfeeding causes early shifts in gene expression in the heart and long-term alterations in cardiometabolic and oxidative parameters. PLoS ONE 2013, 8, e56981. [Google Scholar] [CrossRef]
- Choi, S.W.; Friso, S. Epigenetics: A New Bridge between Nutrition and Health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef]
- Martinez, S.R.; Gay, M.S.; Zhang, L. Epigenetic mechanisms in heart development and disease. Drug Discov. Today 2015, 20, 799–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddeek, B.; Mauduit, C.; Chehade, H.; Blin, G.; Liand, M.; Chindamo, M.; Benahmed, M.; Simeoni, U. Long-term impact of maternal high-fat diet on offspring cardiac health: Role of micro-RNA biogenesis. Cell Death Discov. 2019, 5, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boussios, S.; Ozturk, M.A.; Moschetta, M.; Karathanasi, A.; Zakynthinakis-Kyriakou, N.; Katsanos, K.H.; Christodoulou, D.K.; Pavlidis, N. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J. Pers. Med. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, S.; Kishino, T.; Takahashi, T.; Shimazu, T.; Charvat, H.; Kakugawa, Y.; Nakajima, T.; Lee, Y.C.; Iida, N.; Maeda, M.; et al. Genetic and epigenetic alterations in normal tissues have differential impacts on cancer risk among tissues. Proc. Natl. Acad. Sci. USA 2018, 115, 1328–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudet, F.; Hodgson, J.G.; Eden, A.; Jackson-Grusby, L.; Dausman, J.; Gray, J.W.; Leonhardt, H.; Jaenisch, R. Induction of tumors in mice by genomic hypomethylation. Science 2003, 300, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Kang, B.; Petkovich, D.A.; Bhandari, Y.R.; In, J.; Stein-O’Brien, G.; Kong, X.; Xie, W.; Zachos, N.; Maegawa, S.; et al. Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and Braf(V600E)-Induced Tumorigenesis. Cancer Cell 2019, 35, 315–328. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Luo, Q.; Sun, J.; Ju, Y.; Morita, Y.; Song, G. Chromatin organization regulated by EZH2-mediated H3K27me3 is required for OPN-induced migration of bone marrow-derived mesenchymal stem cells. Int. J. Biochem. Cell Biol. 2018, 96, 29–39. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef] [Green Version]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Delgado-Olguin, P.; Huang, Y.; Li, X.; Christodoulou, D.; Seidman, C.E.; Seidman, J.G.; Tarakhovsky, A.; Bruneau, B.G. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 2012, 44, 343–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddeek, B.; Lakhdari, N.; Inoubli, L.; Paul-Bellon, R.; Isnard, V.; Thibault, E.; Bongain, A.; Chevallier, D.; Repetto, E.; Trabucchi, M.; et al. Developmental epigenetic programming of adult germ cell death disease: Polycomb protein EZH2-miR-101 pathway. Epigenomics 2016, 8, 1459–1479. [Google Scholar] [CrossRef] [PubMed]
- Siddeek, B.; Mauduit, C.; Simeoni, U.; Benahmed, M. Sperm epigenome as a marker of environmental exposure and lifestyle, at the origin of diseases inheritance. Mutat. Res. 2018, 778, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Funtikova, A.N.; Fito, M.; Schroder, H. Prenatal nutrition and the risk of adult obesity: Long-term effects of nutrition on epigenetic mechanisms regulating gene expression. J. Nutr. Biochem. 2017, 39, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.T.; Heyne, S.; Dror, E.; Casas, E.; Leonhardt, L.; Boenke, T.; Yang, C.H.; Sagar; Arrigoni, L.; Dalgaard, K.; et al. The Polycomb-Dependent Epigenome Controls beta Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab. 2018, 27, 1294–1308.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guida, M.C.; Birse, R.T.; Dall’Agnese, A.; Toto, P.C.; Diop, S.B.; Mai, A.; Adams, P.D.; Puri, P.L.; Bodmer, R. Intergenerational inheritance of high fat diet-induced cardiac lipotoxicity in Drosophila. Nat. Commun. 2019, 10, 193. [Google Scholar] [CrossRef] [Green Version]
- Xia, B.; Gerstin, E.; Schones, D.E.; Huang, W.; Steven de Belle, J. Transgenerational programming of longevity through E(z)-mediated histone H3K27 trimethylation in Drosophila. Aging (Albany NY) 2016, 8, 2988–3008. [Google Scholar] [CrossRef] [Green Version]
- Lui, J.C.; Garrison, P.; Nguyen, Q.; Ad, M.; Keembiyehetty, C.; Chen, W.; Jee, Y.H.; Landman, E.; Nilsson, O.; Barnes, K.M.; et al. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat. Commun. 2016, 7, 13685. [Google Scholar] [CrossRef] [Green Version]
- He, A.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Xie, H.; Zhang, B.; Hsing, M.; Christodoulou, D.C.; Cahan, P.; et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ. Res. 2012, 110, 406–415. [Google Scholar] [CrossRef] [Green Version]
- Bai, J.; Xi, Q. Crosstalk between TGF-beta signaling and epigenome. Acta biochim. biophys. Sin. 2018, 50, 322. [Google Scholar] [CrossRef] [Green Version]
- He, A.; Shen, X.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Wang, G.; Marquez, V.E.; Orkin, S.H.; Pu, W.T. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012, 26, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Rajagopal, S.K.; Ma, Q.; Obler, D.; Shen, J.; Manichaikul, A.; Tomita-Mitchell, A.; Boardman, K.; Briggs, C.; Garg, V.; Srivastava, D.; et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J. Mol. Cell Cardiol. 2007, 43, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathison, M.; Singh, V.P.; Sanagasetti, D.; Yang, L.; Pinnamaneni, J.P.; Yang, J.; Rosengart, T.K. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J. Thorac. Cardiovasc. Surg. 2017, 154, 1601–1610. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Li, J.; Green, C.D.; Yu, X.; Tang, X.; Han, D.; Xian, B.; Wang, D.; Huang, X.; Cao, X.; et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011, 14, 161–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bracken, A.P.; Kleine-Kohlbrecher, D.; Dietrich, N.; Pasini, D.; Gargiulo, G.; Beekman, C.; Theilgaard-Monch, K.; Minucci, S.; Porse, B.T.; Marine, J.C.; et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007, 21, 525–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Meunier, L.; Siddeek, B.; Vega, A.; Lakhdari, N.; Inoubli, L.; Bellon, R.P.; Lemaire, G.; Mauduit, C.; Benahmed, M. Perinatal programming of adult rat germ cell death after exposure to xenoestrogens: Role of microRNA miR-29 family in the down-regulation of DNA methyltransferases and Mcl-1. Endocrinology 2012, 153, 1936–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Chen, O.; Zheng, M.; Wang, L.; Zhou, Y.; Yin, C.; Liu, J.; Qian, L. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem. Cell Res. 2016, 16, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Pepin, M.E.; Drakos, S.; Ha, C.M.; Tristani-Firouzi, M.; Selzman, C.H.; Fang, J.C.; Wende, A.R.; Wever-Pinzon, O. DNA methylation reprograms cardiac metabolic gene expression in end-stage human heart failure. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H674–H684. [Google Scholar] [CrossRef]
- Yang, X.J.; Seto, E. HATs and HDACs: From structure, function and regulation to novel strategies for therapy and prevention. Oncogene 2007, 26, 5310–5318. [Google Scholar] [CrossRef]
- De Jong, K.A.; Barrand, S.; Wood-Bradley, R.J.; de Almeida, D.L.; Czeczor, J.K.; Lopaschuk, G.D.; Armitage, J.A.; McGee, S.L. Maternal high fat diet induces early cardiac hypertrophy and alters cardiac metabolism in Sprague Dawley rat offspring. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 600–609. [Google Scholar] [CrossRef]
- Sun, H.; Yang, X.; Zhu, J.; Lv, T.; Chen, Y.; Chen, G.; Zhong, L.; Li, Y.; Huang, X.; Huang, G.; et al. Inhibition of p300-HAT results in a reduced histone acetylation and down-regulation of gene expression in cardiac myocytes. Life sciences 2010, 87, 707–714. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Olson, E.N. Cardiac histone acetylation--therapeutic opportunities abound. Trends Genet. 2004, 20, 206–213. [Google Scholar] [CrossRef]
- Upadhyaya, B.; Larsen, T.; Barwari, S.; Louwagie, E.J.; Baack, M.L.; Dey, M. Prenatal Exposure to a Maternal High-Fat Diet Affects Histone Modification of Cardiometabolic Genes in Newborn Rats. Nutrients 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.K.; Ramchandani, S.; Cervoni, N.; Szyf, M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature 1999, 397, 579–583. [Google Scholar] [CrossRef]
- Hill, P.W.; Amouroux, R.; Hajkova, P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: An emerging complex story. Genomics 2014, 104, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Saavedra, D.; Moody, L.; Xu, G.B.; Chen, H.; Pan, Y.X. Epigenetic Regulation of Metabolism and Inflammation by Calorie Restriction. Adv. Nutr. 2019, 10, 520–536. [Google Scholar] [CrossRef]
- Lopez-Camarillo, C.; Gallardo-Rincon, D.; Alvarez-Sanchez, M.E.; Marchat, L.A. Pharmaco-epigenomics: On the Road of Translation Medicine. Adv. Exp. Med. Biol. 2019, 1168, 31–42. [Google Scholar] [CrossRef]
- Ideraabdullah, F.Y.; Zeisel, S.H. Dietary Modulation of the Epigenome. Physiol. Rev. 2018, 98, 667–695. [Google Scholar] [CrossRef] [Green Version]
- Keyes, M.K.; Jang, H.; Mason, J.B.; Liu, Z.; Crott, J.W.; Smith, D.E.; Friso, S.; Choi, S.W. Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. J. Nutr. 2007, 137, 1713–1717. [Google Scholar] [CrossRef]
- Park, L.K.; Friso, S.; Choi, S.W. Nutritional influences on epigenetics and age-related disease. Proc. Nutr.Soc. 2012, 71, 75–83. [Google Scholar] [CrossRef]
- Urvalek, A.M.; Gudas, L.J. Retinoic acid and histone deacetylases regulate epigenetic changes in embryonic stem cells. J. Biol. Chem. 2014, 289, 19519–19530. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, G.F.S.; Silva, G.D.B.; Pavan, A.R.; Chiba, D.E.; Chin, C.M.; Dos Santos, J.L. Epigenetic Regulatory Mechanisms Induced by Resveratrol. Nutrients 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Hassan, F.U.; Rehman, M.S.; Khan, M.S.; Ali, M.A.; Javed, A.; Nawaz, A.; Yang, C. Curcumin as an Alternative Epigenetic Modulator: Mechanism of Action and Potential Effects. Front. Genet. 2019, 10, 514. [Google Scholar] [CrossRef] [Green Version]
- Kaufman-Szymczyk, A.; Majewski, G.; Lubecka-Pietruszewska, K.; Fabianowska-Majewska, K. The Role of Sulforaphane in Epigenetic Mechanisms, Including Interdependence between Histone Modification and DNA Methylation. Int. J. Mol. Sci. 2015, 16, 29732–29743. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.H.; Lai, C.S.; Wu, J.C.; Ho, C.T. Epigenetic and disease targets by polyphenols. Curr. Pharm. Des. 2013, 19, 6156–6185. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blin, G.; Liand, M.; Mauduit, C.; Chehade, H.; Benahmed, M.; Simeoni, U.; Siddeek, B. Maternal Exposure to High-Fat Diet Induces Long-Term Derepressive Chromatin Marks in the Heart. Nutrients 2020, 12, 181. https://doi.org/10.3390/nu12010181
Blin G, Liand M, Mauduit C, Chehade H, Benahmed M, Simeoni U, Siddeek B. Maternal Exposure to High-Fat Diet Induces Long-Term Derepressive Chromatin Marks in the Heart. Nutrients. 2020; 12(1):181. https://doi.org/10.3390/nu12010181
Chicago/Turabian StyleBlin, Guillaume, Marjorie Liand, Claire Mauduit, Hassib Chehade, Mohamed Benahmed, Umberto Simeoni, and Benazir Siddeek. 2020. "Maternal Exposure to High-Fat Diet Induces Long-Term Derepressive Chromatin Marks in the Heart" Nutrients 12, no. 1: 181. https://doi.org/10.3390/nu12010181
APA StyleBlin, G., Liand, M., Mauduit, C., Chehade, H., Benahmed, M., Simeoni, U., & Siddeek, B. (2020). Maternal Exposure to High-Fat Diet Induces Long-Term Derepressive Chromatin Marks in the Heart. Nutrients, 12(1), 181. https://doi.org/10.3390/nu12010181




