Xylitol’s Health Benefits beyond Dental Health: A Comprehensive Review
Abstract
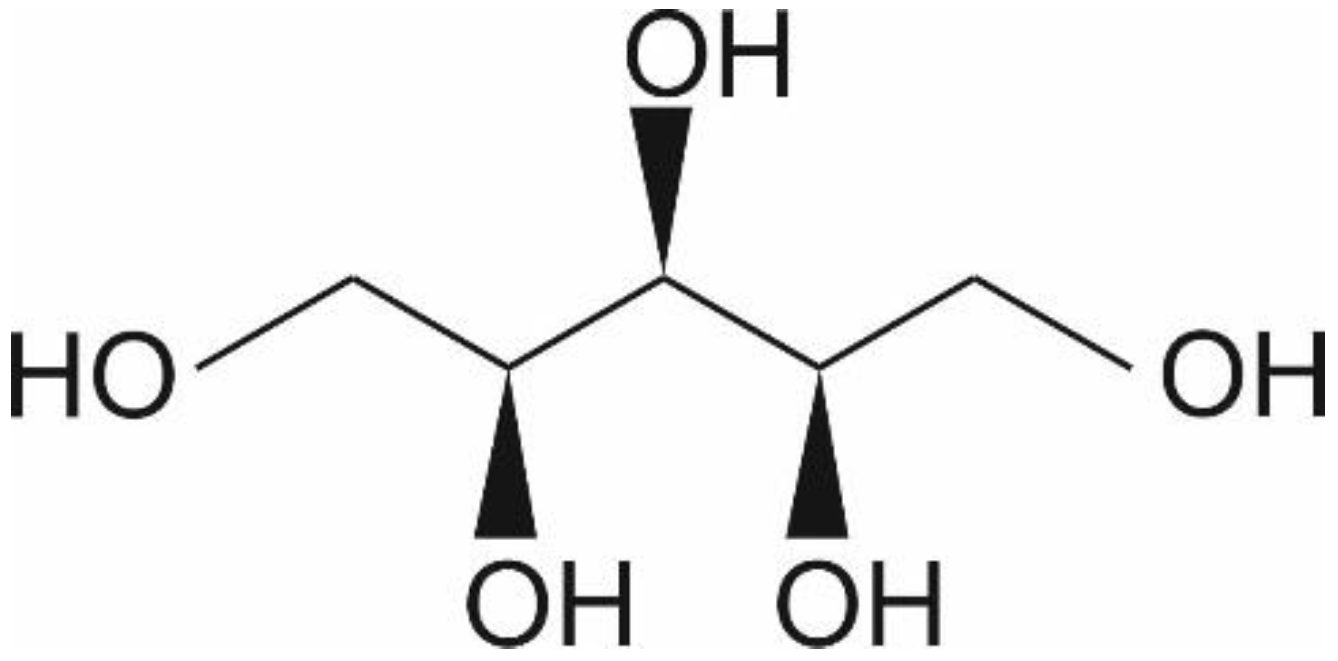
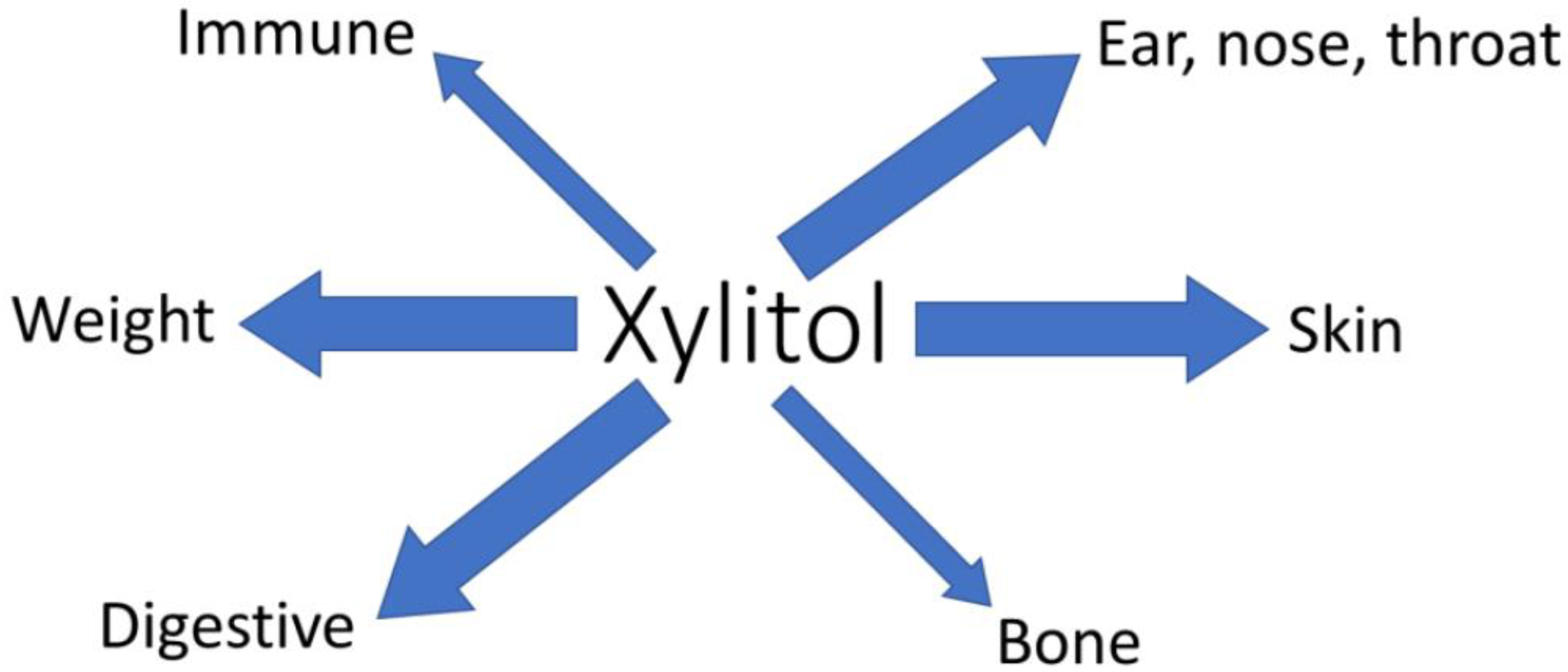
1. Introduction
2. Skin
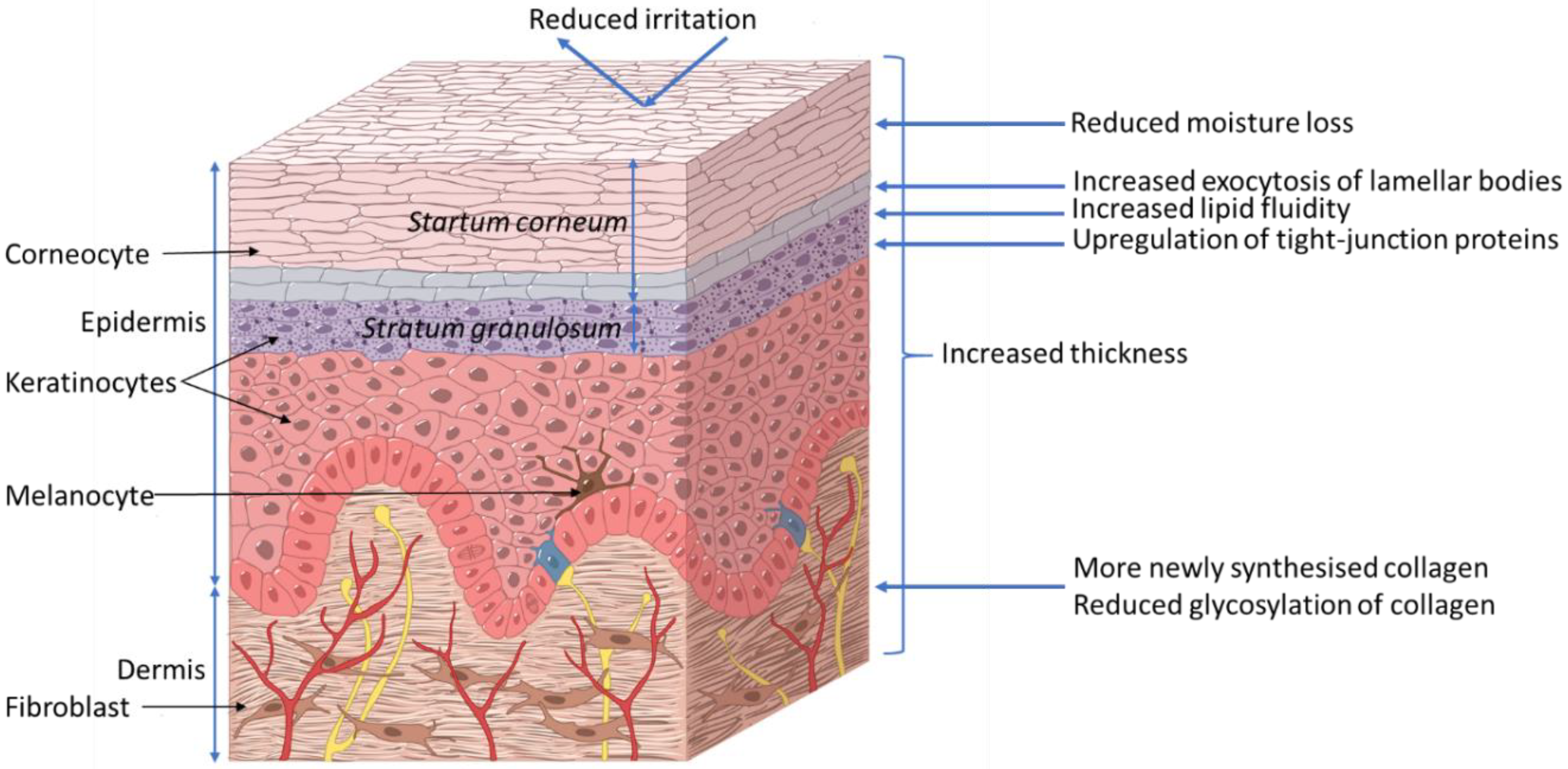
2.1. Skin Introduction
2.2. Xylitol Benefits to Skin
2.3. Conclusions
3. Digestive Tract
3.1. Introduction
3.2. Prebiotic Benefits of Xylitol
3.3. Benefits of Xylitol on Bowel Function
3.4. Conclusions
4. Nose, Throat and Ear
4.1. Introduction
4.2. Benefits of Xylitol in Respiratory Health
4.3. Benefits of Xylitol in Sinusitis
4.4. Acute Otitis Media
4.5. Conclusions
5. Bone
5.1. Introduction
5.2. Effects of Xylitol on Bone Strength
5.3. Conclusions
6. Immune Function
6.1. Introduction
6.2. Immune Modulatory Effects of Xylitol
6.3. Anti-Inflammatory Effects of Xylitol
6.4. Conclusions
7. Weight Management
7.1. Introduction
7.2. Effects of Xylitol on Weight Management
7.3. Benefits of Xylitol on Satiety
7.4. Benefits of Xylitol on Metabolic Health
7.5. Conclusions
8. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ur-Rehman, S.; Mushtaq, Z.; Zahoor, T.; Jamil, A.; Murtaza, M.A. Xylitol: A review on bioproduction, application, health benefits, and related safety issues. Crit. Rev. Food Sci. Nutr. 2015, 55, 1514–1528. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.; Dunning, N. Xylitol. In Sweeteners and Sugar Alternatives in Food Technology; Mitchell, H., Ed.; Blackwell Publishing: Oxford, UK, 2006; pp. 295–324. [Google Scholar]
- Janakiram, C.; Deepan Kumar, C.V.; Joseph, J. Xylitol in preventing dental caries: A systematic review and meta-analyses. J. Nat. Sci. Biol. Med. 2017, 8, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Mäkinen, K.K. Gastrointestinal Disturbances Associated with the Consumption of Sugar Alcohols with Special Consideration of Xylitol: Scientific Review and Instructions for Dentists and Other Health-Care Professionals. Int. J. Dent. 2016, 2016, 5967907. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Xylitol chewing gum/pastilles and reduction of the risk of tooth decay. EFSA J. 2008, 6, 852. [Google Scholar] [CrossRef]
- Livesey, G. Health potential of polyols as sugar replacers, with emphasis on low glycaemic properties. Nutr. Res. Rev. 2003, 16, 163–191. [Google Scholar] [CrossRef] [PubMed]
- Amo, K.; Arai, H.; Uebanso, T.; Fukaya, M.; Koganei, M.; Sasaki, H.; Yamamoto, H.; Taketani, Y.; Takeda, E. Effects of xylitol on metabolic parameters and visceral fat accumulation. J. Clin. Biochem. Nutr. 2011, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Storey, D.; Lee, A.; Bornet, F.; Brouns, F. Gastrointestinal tolerance of erythritol and xylitol ingested in a liquid. Eur. J. Clin. Nutr. 2007, 61, 349–354. [Google Scholar] [CrossRef]
- Abdayem, R.; Haftek, M. [The epidermal barrier]. Ann. Derm. Venereol. 2018, 145, 293–301. [Google Scholar] [CrossRef]
- Umino, Y.; Ipponjima, S.; Denda, M. Modulation of lipid fluidity likely contributes to the fructose/xylitol-induced acceleration of epidermal permeability barrier recovery. Arch. Dermatol. Res. 2019, 311, 317–324. [Google Scholar] [CrossRef]
- Payér, E.; Szabó-Papp, J.; Ambrus, L.; Szöllösi, A.G.; Andrási, M.; Dikstein, S.; Kemény, L.; Juhász, I.; Szegedi, A.; Bíró, T.; et al. Beyond the physico-chemical barrier: Glycerol and xylitol markedly yet differentially alter gene expression profiles and modify signalling pathways in human epidermal keratinocytes. Exp. Dermatol. 2018, 27, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, N.; Rosenthal, R.; Gunzel, D.; Moll, I.; Brandner, J.M. Tight junctions and differentiation–A chicken or the egg question? Exp. Dermatol. 2012, 21, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Korponyai, C.; Szél, E.; Behány, Z.; Varga, E.; Mohos, G.; Dura, A.; Dikstein, S.; Kemény, L.; Erös, G. Effects of Locally Applied Glycerol and Xylitol on the Hydration, Barrier Function and Morphological Parameters of the Skin. Acta Derm. Venereol. 2017, 97, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Szél, E.; Polyánka, H.; Szabó, K.; Hartmann, P.; Degovics, D.; Balázs, B.; Németh, I.B.; Korponyai, C.; Csányi, E.; Kaszaki, J.; et al. Anti-irritant and anti-inflammatory effects of glycerol and xylitol in sodium lauryl sulphate-induced acute irritation. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 2333–2341. [Google Scholar] [CrossRef] [PubMed]
- Korponyai, C.; Kovács, R.K.; Erös, G.; Dikstein, S.; Kemény, L. Antiirritant properties of polyols and amino acids. Dermatitis 2011, 22, 141–146. [Google Scholar] [PubMed]
- Odetti, P.R.; Borgoglio, A.; Rolandi, R. Age-related increase of collagen fluorescence in human subcutaneous tissue. Metabolism 1992, 41, 655–658. [Google Scholar] [CrossRef]
- Mattila, P.T.; Pelkonen, P.; Knuuttila, M.L. Effects of a long-term dietary xylitol supplementation on collagen content and fluorescence of the skin in aged rats. Gerontology 2005, 51, 166–169. [Google Scholar] [CrossRef]
- Knuuttila, M.L.; Kuoksa, T.H.; Svanberg, M.J.; Mattila, P.T.; Karjalainen, K.M.; Kolehmainen, E. Effects of dietary xylitol on collagen content and glycosylation in healthy and diabetic rats. Life Sci. 2000, 67, 283–290. [Google Scholar] [CrossRef]
- Dowd, S.E.; Sun, Y.; Smith, E.; Kennedy, J.P.; Jones, C.E.; Wolcott, R. Effects of biofilm treatments on the multi-species Lubbock chronic wound biofilm model. J. Wound Care 2009, 18, 510–512. [Google Scholar] [CrossRef]
- Ammons, M.C.; Ward, L.S.; James, G.A. Anti-biofilm efficacy of a lactoferrin/xylitol wound hydrogel used in combination with silver wound dressings. Int. Wound J. 2011, 8, 268–273. [Google Scholar] [CrossRef]
- Katsuyama, M.; Kobayashi, Y.; Ichikawa, H.; Mizuno, A.; Miyachi, Y.; Matsunaga, K.; Kawashima, M. A novel method to control the balance of skin microflora Part 2. A study to assess the effect of a cream containing farnesol and xylitol on atopic dry skin. J. Dermatol. Sci. 2005, 38, 207–213. [Google Scholar] [CrossRef]
- El Aidy, S.; Van Den Bogert, B.; Kleerebezem, M. The small intestine microbiota, nutritional modulation and relevance for health. Curr. Opin. Biotechnol. 2015, 32, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgard, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef] [PubMed]
- Lenhart, A.; Chey, W.D. A Systematic Review of the Effects of Polyols on Gastrointestinal Health and Irritable Bowel Syndrome. Adv. Nutr. 2017, 8, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491. [Google Scholar] [CrossRef] [PubMed]
- Mäkeläinen, H.S.; Mäkivuokko, H.A.; Salminen, S.J.; Rautonen, N.E.; Ouwehand, A.C. The effects of polydextrose and xylitol on microbial community and activity in a 4-stage colon simulator. J. Food Sci. 2007, 72, M153–M159. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Kusuhara, S.; Yokoi, W.; Ito, M.; Miyazaki, K. Prebiotic potential of L-sorbose and xylitol in promoting the growth and metabolic activity of specific butyrate-producing bacteria in human fecal culture. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef] [PubMed]
- Scheppach, W.; Weiler, F. The butyrate story: Old wine in new bottles. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 563–567. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Salminen, S.; Salminen, E.; Koivistoinen, P.; Bridges, J.; Marks, V. Gut microflora interactions with xylitol in the mouse, rat and man. Food Chem. Toxicol. 1985, 23, 985–990. [Google Scholar] [CrossRef]
- Talattof, Z.; Azad, A.; Zahed, M.; Shahradnia, N. Antifungal Activity of Xylitol against Candida albicans: An in vitro Study. J. Contemp. Dent. Pract. 2018, 19, 125–129. [Google Scholar] [CrossRef]
- Uebanso, T.; Kano, S.; Yoshimoto, A.; Naito, C.; Shimohata, T.; Mawatari, K.; Takahashi, A. Effects of Consuming Xylitol on Gut Microbiota and Lipid Metabolism in Mice. Nutrients 2017, 9, 756. [Google Scholar] [CrossRef] [PubMed]
- Tamura, M.; Hoshi, C.; Hori, S. Xylitol affects the intestinal microbiota and metabolism of daidzein in adult male mice. Int. J. Mol. Sci. 2013, 14, 23993–24007. [Google Scholar] [CrossRef] [PubMed]
- Vargas, S.L.; Patrick, C.C.; Ayers, G.D.; Hughes, W.T. Modulating effect of dietary carbohydrate supplementation on Candida albicans colonization and invasion in a neutropenic mouse model. Infect. Immun. 1993, 61, 619–626. [Google Scholar] [PubMed]
- Hedberg, M.; Hasslöf, P.; Sjöström, I.; Twetman, S.; Stecksén-Blicks, C. Sugar fermentation in probiotic bacteria-an in vitro study. Oral Microbiol. Immunol. 2008, 23, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Haukioja, A.; Söderling, E.; Tenovuo, J. Acid production from sugars and sugar alcohols by probiotic lactobacilli and bifidobacteria in vitro. Caries Res. 2008, 42, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.K.; Twetman, S. Acid production in dental plaque after exposure to probiotic bacteria. BMC Oral Health 2012, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, E.; Ionescu, A.C.; Cazzaniga, G.; Ottobelli, M.; Samaranayake, L.P. Levorotatory carbohydrates and xylitol subdue Streptococcus mutans and Candida albicans adhesion and biofilm formation. J. Basic Microbiol. 2016, 56, 480–492. [Google Scholar] [CrossRef]
- Rätsep, M.; Kõljalg, S.; Sepp, E.; Smidt, I.; Truusalu, K.; Songisepp, E.; Stsepetova, J.; Naaber, P.; Mikelsaar, R.H.; Mikelsaar, M. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe 2017, 47, 94–103. [Google Scholar] [CrossRef]
- Yu, T.; Zheng, Y.P.; Tan, J.C.; Xiong, W.J.; Wang, Y.; Lin, L. Effects of Prebiotics and Synbiotics on Functional Constipation. Am. J. Med Sci. 2017, 353, 282–292. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, Q.; Qiao, L.; Lv, D.; Ruan, J.; Chen, H.; Gong, J.; Shi, G. Xylitol Gum Chewing to Achieve Early Postoperative Restoration of Bowel Motility After Laparoscopic Surgery. Surg. Laparosc. Endosc. Percutan. Tech. 2015, 25, 303–306. [Google Scholar] [CrossRef]
- Lee, J.T.; Hsieh, M.H.; Cheng, P.J.; Lin, J.R. The Role of Xylitol Gum Chewing in Restoring Postoperative Bowel Activity After Cesarean Section. Biol. Res. Nurs. 2016, 18, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Long, W.J.; Wei, L. Chewing Xylitol Gum could Accelerate Bowel motility Recovery after Elective Open Proctectomy for Rectal Cancer. Rev. Investig. Clin. 2018, 70, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Ottman, N.; Smidt, H.; de Vos, W.M.; Belzer, C. The function of our microbiota: Who is out there and what do they do? Front. Cell. Infect. Microbiol. 2012, 2, 104. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, M.J.; Hibberd, A.A.; Mannikko, S.; Yeung, N.; Kauko, T.; Forssten, S.; Lehtoranta, L.; Lahtinen, S.J.; Stahl, B.; Lyra, A.; et al. Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci. Rep. 2018, 8, 11411. [Google Scholar] [CrossRef] [PubMed]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Brook, I. Microbiology of chronic rhinosinusitis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Azarpazhooh, A.; Lawrence, H.P.; Shah, P.S. Xylitol for preventing acute otitis media in children up to 12 years of age. Cochrane Database Syst. Rev. 2016, 8, CD007095. [Google Scholar] [CrossRef] [PubMed]
- Kontiokari, T.; Uhari, M.; Koskela, M. Effect of xylitol on growth of nasopharyngeal bacteria in vitro. Antimicrob. Agents Chemother. 1995, 39, 1820–1823. [Google Scholar] [CrossRef]
- Kontiokari, T.; Svanberg, M.; Mattila, P.; Leinonen, M.; Uhari, M. Quantitative analysis of the effect of xylitol on pneumococcal nasal colonisation in rats. FEMS Microbiol. Lett. 1999, 178, 313–317. [Google Scholar] [CrossRef]
- Zabner, J.; Seiler, M.P.; Launspach, J.L.; Karp, P.H.; Kearney, W.R.; Look, D.C.; Smith, J.J.; Welsh, M.J. The osmolyte xylitol reduces the salt concentration of airway surface liquid and may enhance bacterial killing. Proc. Natl. Acad. Sci. USA 2000, 97, 11614–11619. [Google Scholar] [CrossRef] [PubMed]
- Cingi, C.; Birdane, L.; Ural, A.; Oghan, F.; Bal, C. Comparison of nasal hyperosmolar xylitol and xylometazoline solutions on quality of life in patients with inferior turbinate hypertrophy secondary to nonallergic rhinitis. Int. Forum Allergy Rhinol. 2014, 4, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Little, P.; Stuart, B.; Wingrove, Z.; Mullee, M.; Thomas, T.; Johnson, S.; Leydon, G.; Richards-Hall, S.; Williamson, I.; Yao, L.; et al. Probiotic capsules and xylitol chewing gum to manage symptoms of pharyngitis: A randomized controlled factorial trial. CMAJ: Can. Med Assoc. J. 2017, 189, E1543–E1550. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Lee, T.; Hardcastle, T.; Biswas, K.; Radcliff, F.; Douglas, R. The in vitro effect of xylitol on chronic rhinosinusitis biofilms. Rhinology 2016, 54, 323–328. [Google Scholar] [CrossRef]
- Brown, C.L.; Graham, S.M.; Cable, B.B.; Ozer, E.A.; Taft, P.J.; Zabner, J. Xylitol enhances bacterial killing in the rabbit maxillary sinus. Laryngoscope 2004, 114, 2021–2024. [Google Scholar] [CrossRef] [PubMed]
- Weissman, J.D.; Fernandez, F.; Hwang, P.H. Xylitol nasal irrigation in the management of chronic rhinosinusitis: A pilot study. Laryngoscope 2011, 121, 2468–2472. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Tang, X.; Wei, J.; Dai, F.; Sun, G. Xylitol nasal irrigation in the treatment of chronic rhinosinusitis. Am. J. Otolaryngol. 2017, 38, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Hardcastle, T.; Jain, R.; Radcliff, F.; Waldvogel-Thurlow, S.; Zoing, M.; Biswas, K.; Douglas, R. The in vitro mucolytic effect of xylitol and dornase alfa on chronic rhinosinusitis mucus. Int. Forum Allergy Rhinol. 2017, 7, 889–896. [Google Scholar] [CrossRef]
- Tapiainen, T.; Sormunen, R.; Kaijalainen, T.; Kontiokari, T.; Ikäheimo, I.; Uhari, M. Ultrastructure of Streptococcus pneumoniae after exposure to xylitol. J. Antimicrob. Chemother. 2004, 54, 225–228. [Google Scholar] [CrossRef]
- Kurola, P.; Tapiainen, T.; Kaijalainen, T.; Uhari, M.; Saukkoriipi, A. Xylitol and capsular gene expression in Streptococcus pneumoniae. J. Med. Microbiol. 2009, 58, 1470–1473. [Google Scholar] [CrossRef]
- Kurola, P.; Tapiainen, T.; Sevander, J.; Kaijalainen, T.; Leinonen, M.; Uhari, M.; Saukkoriipi, A. Effect of xylitol and other carbon sources on Streptococcus pneumoniae biofilm formation and gene expression in vitro. Apmis 2011, 119, 135–142. [Google Scholar] [CrossRef]
- Persaud, N.; Laupacis, A.; Azarpazhooh, A.; Birken, C.; Hoch, J.S.; Isaranuwatchai, W.; Maguire, J.L.; Mamdani, M.M.; Thorpe, K.; Allen, C.; et al. Xylitol for the prevention of acute otitis media episodes in children aged 2–4 years: Protocol for a pragmatic randomised controlled trial. BMJ Open 2018, 8, e020941. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Ide, Y.; Nasu, M.; Numabe, Y. The effects of oral xylitol administration on bone density in rat femur. Odontology 2011, 99, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.T.; Svanberg, M.J.; Mäkinen, K.K.; Knuuttila, M.L. Dietary xylitol, sorbitol and D-mannitol but not erythritol retard bone resorption in rats. J. Nutr. 1996, 126, 1865–1870. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.; Svanberg, M.; Knuuttila, M. Diminished bone resorption in rats after oral xylitol administration: A dose-response study. Calcif. Tissue Int. 1995, 56, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.T.; Svanberg, M.J.; Pökkä, P.; Knuuttila, M.L. Dietary xylitol protects against weakening of bone biomechanical properties in ovariectomized rats. J. Nutr. 1998, 128, 1811–1814. [Google Scholar] [CrossRef] [PubMed]
- Kaivosoja, S.M.; Mattila, P.T.; Knuuttila, M.L. Dietary xylitol protects against the imbalance in bone metabolism during the early phase of collagen type II-induced arthritis in dark agouti rats. Metabolism 2008, 57, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Mattila, P.T.; Knuuttila, M.L.; Svanberg, M.J. Dietary xylitol supplementation prevents osteoporotic changes in streptozotocin-diabetic rats. Metabolism 1998, 47, 578–583. [Google Scholar] [CrossRef]
- Castelo-Branco, C.; Soveral, I. Phytoestrogens and bone health at different reproductive stages. Gynecol. Endocrinol. 2013, 29, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.M.; Yu, M.; Darby, T.M.; Vaccaro, C.; Li, J.Y.; Owens, J.A.; Hsu, E.; Adams, J.; Weitzmann, M.N.; Jones, R.M.; et al. The Microbial Metabolite Butyrate Stimulates Bone Formation via T Regulatory Cell-Mediated Regulation of WNT10B Expression. Immunity 2018, 49, 1116–1131.e7. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Bosch, J. The human immune system. In Excercise Immunology; Gleeson, M., Bishop, N., Walsh, N., Eds.; Routledge: London, UK, 2013. [Google Scholar]
- Jakob, A.; Williamson, J.R.; Asakura, T. Xylitol metabolism in perfused rat liver. Interactions with gluconeogenesis and ketogenesis. J. Biol. Chem. 1971, 246, 7623–7631. [Google Scholar] [PubMed]
- Takahashi, K.; Akiba, Y. Single administration of xylitol to newly hatched chicks enhances growth, digestive enzyme activity and immune responses by 12 d of age. Br. Poult. Sci. 2005, 46, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Renko, M.; Valkonen, P.; Tapiainen, T.; Kontiokari, T.; Mattila, P.; Knuuttila, M.; Svanberg, M.; Leinonen, M.; Karttunen, R.; Uhari, M. Xylitol-supplemented nutrition enhances bacterial killing and prolongs survival of rats in experimental pneumococcal sepsis. BMC Microbiol. 2008, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Salli, K.M.; Gürsoy, U.K.; Söderling, E.M.; Ouwehand, A.C. Effects of Xylitol and Sucrose Mint Products on Streptococcus mutans Colonization in a Dental Simulator Model. Curr. Microbiol. 2017, 74, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.L.; Wi, G.R.; Kim, H.J.; Kim, H.J. Ameliorating Effect of Dietary Xylitol on Human Respiratory Syncytial Virus (hRSV) Infection. Biol. Pharm. Bull. 2016, 39, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.Y.; Kim, H.J.; Kim, H.J. Protective effect of dietary xylitol on influenza A virus infection. PloS ONE 2014, 9, e84633. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Mashiko, T.; Akiba, Y. Effect of dietary concentration of xylitol on growth in male broiler chicks during immunological stress. Poult. Sci. 2000, 79, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Grundy, S.M. Metabolic syndrome update. Trends Cardiovasc. Med. 2016, 26, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Islam, S. Xylitol improves pancreatic islets morphology to ameliorate type 2 diabetes in rats: A dose response study. J. Food Sci. 2014, 79, H1436–H1442. [Google Scholar] [CrossRef] [PubMed]
- Geidenstam, N.; Al-Majdoub, M.; Ekman, M.; Spegel, P.; Ridderstrale, M. Metabolite profiling of obese individuals before and after a one year weight loss program. Int. J. Obes. (Lond.) 2017, 41, 1369–1378. [Google Scholar] [CrossRef]
- Wölnerhanssen, B.K.; Cajacob, L.; Keller, N.; Doody, A.; Rehfeld, J.F.; Drewe, J.; Peterli, R.; Beglinger, C.; Meyer-Gerspach, A.C. Gut hormone secretion, gastric emptying, and glycemic responses to erythritol and xylitol in lean and obese subjects. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E1053–E1061. [Google Scholar] [CrossRef]
- King, N.A.; Craig, S.A.S.; Pepper, T.; Blundell, J.E. Evaluation of the independent and combined effects of xylitol and polydextrose consumed as a snack on hunger and energy intake over 10 d. Br. J. Nutr. 2005, 93, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Salminen, E.K.; Salminen, S.J.; Porkka, L.; Kwasowski, P.; Marks, V.; Koivistoinen, P.E. Xylitol vs glucose: Effect on the rate of gastric emptying and motilin, insulin, and gastric inhibitory polypeptide release. Am. J. Clin. Nutr. 1989, 49, 1228–1232. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Mochiki, E.; Kuwano, H. The roles of motilin and ghrelin in gastrointestinal motility. Int. J. Pept. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Shafer, R.B.; Levine, A.S.; Marlette, J.M.; Morley, J.E. Effects of xylitol on gastric emptying and food intake. Am. J. Clin. Nutr. 1987, 45, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Natah, S.S.; Hussien, K.R.; Tuominen, J.A.; Koivisto, V.A. Metabolic response to lactitol and xylitol in healthy men. Am. J. Clin. Nutr. 1997, 65, 947–950. [Google Scholar] [CrossRef]
- Islam, S.; Indrajit, M. Effects of xylitol on blood glucose, glucose tolerance, serum insulin and lipid profile in a type 2 diabetes model of rats. Ann. Nutr. Metab. 2012, 61, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S. Effects of xylitol as a sugar substitute on diabetes-related parameters in nondiabetic rats. J. Med. Food 2011, 14, 505–511. [Google Scholar] [CrossRef]
- Förster, H.; Quadbeck, R.; Gottstein, U. Metabolic tolerance to high doses of oral xylitol in human volunteers not previously adapted to xylitol. Int. J. Vitam. Nutr. Res. Suppl. 1982, 22, 67–88. [Google Scholar]
- Islam, M.S. Xylitol increases serum triglyceride in normal but not in a type 2 diabetes model of rats. J. Med. Food 2012, 15, 677. [Google Scholar] [CrossRef]
- Nevitt, S.J.; Thornton, J.; Murray, C.S.; Dwyer, T. Inhaled mannitol for cystic fibrosis. Cochrane Database Syst. Rev. 2018, 2, CD008649. [Google Scholar] [CrossRef]
- Drakoularakou, A.; Hasselwander, O.; Edinburgh, M.; Ouwehand, A.C. Lactitol, an emerging prebiotic: Functional properties with a focus on digestive health. Food Sci. Technol. Bull. 2007, 3, 71–80. [Google Scholar] [CrossRef]
- Sarmiento-Rubiano, L.A.; Zuniga, M.; Perez-Martinez, G.; Yebra, M.J. Dietary supplementation with sorbitol results in selective enrichment of lactobacilli in rat intestine. Res. Microbiol. 2007, 158, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Lissner, S.A.; Wald, A. Constipation in adults. BMJ Clin. Evid. 2010, 7, 413. [Google Scholar]
- Andre, P.; Villain, F. Free radical scavenging properties of mannitol and its role as a constituent of hyaluronic acid fillers: A literature review. Int. J. Cosmet. Sci. 2017, 39, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kubota, N.; Takahashi, T.; To, M.; Hayashi, T.; Shimizu, T.; Kamata, Y.; Saruta, J.; Tsukinoki, K. Continuous combined intake of polydextrose and lactitol stimulates cecal fermentation and salivary IgA secretion in rats. J. Oral. Sci. 2017, 59, 603–610. [Google Scholar] [CrossRef]
- Chukwuma, C.I.; Ibrahim, M.A.; Islam, M.S. Maltitol inhibits small intestinal glucose absorption and increases insulin mediated muscle glucose uptake ex vivo but not in normal and type 2 diabetic rats. Int. J. Food Sci. Nutr. 2017, 68, 73–81. [Google Scholar] [CrossRef]
- Chukwuma, C.I.; Mopuri, R.; Nagiah, S.; Chuturgoon, A.A.; Islam, M.S. Erythritol reduces small intestinal glucose absorption, increases muscle glucose uptake, improves glucose metabolic enzymes activities and increases expression of Glut-4 and IRS-1 in type 2 diabetic rats. Eur. J. Nutr. 2018, 57, 2431–2444. [Google Scholar] [CrossRef]
- Chukwuma, C.I.; Islam, M.S. Sorbitol increases muscle glucose uptake ex vivo and inhibits intestinal glucose absorption ex vivo and in normal and type 2 diabetic rats. Appl. Physiol. Nutr. Metab. 2017, 42, 377–383. [Google Scholar] [CrossRef]
| Organisms Reported to Grow on Xylitol | Reference | Organisms Reported Not to Grow on Xylitol | Reference |
|---|---|---|---|
| Anaerostipes hadrus (strain dependent), A. caccae | [27] | Lactobacillus plantarum 299v, L. plantarum 931, L. rhamnosus GG, L. rhamnosus LB21, L. paracasei F19, L. reuteri PTA5289 | [35] |
| Bifidobacterium lactis 1100, B. lactis Bb-12, B. longum 913, B. lactis 420, L. acidophilus NCFM, L. casei 921, L. casei Shirota, L. bulgaricus 365, L. johnsonii LA1, L. paracasei F19, L. plantarum 299v, L. reuteri SD2112, L. rhamnosus GG, L. rhamnosus, Lc-705, Streptococcus mutans Ingbritt | [36] | ||
| L. plantarum 299v, L. reuteri DSM17938 | [37] | ||
| Coprococcus catus, Eubacterium halli, E. limosum, E. rectale, Faecalibacterium prausnitzii, Megasphera elsedenii, Ruminococcus faecis, R. hominis, R. intestinalis, R. inulinivoruans | [27] | ||
| S. pneumoniae | |||
| S. mutans, S. salivarius, S. sanguis | |||
| Candida albicans, S. mutans | [38] | ||
| Staphylococcus epidermidis, Staphylococcus aureus, Pseudomonas aeruginosa |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salli, K.; Lehtinen, M.J.; Tiihonen, K.; Ouwehand, A.C. Xylitol’s Health Benefits beyond Dental Health: A Comprehensive Review. Nutrients 2019, 11, 1813. https://doi.org/10.3390/nu11081813
Salli K, Lehtinen MJ, Tiihonen K, Ouwehand AC. Xylitol’s Health Benefits beyond Dental Health: A Comprehensive Review. Nutrients. 2019; 11(8):1813. https://doi.org/10.3390/nu11081813
Chicago/Turabian StyleSalli, Krista, Markus J. Lehtinen, Kirsti Tiihonen, and Arthur C. Ouwehand. 2019. "Xylitol’s Health Benefits beyond Dental Health: A Comprehensive Review" Nutrients 11, no. 8: 1813. https://doi.org/10.3390/nu11081813
APA StyleSalli, K., Lehtinen, M. J., Tiihonen, K., & Ouwehand, A. C. (2019). Xylitol’s Health Benefits beyond Dental Health: A Comprehensive Review. Nutrients, 11(8), 1813. https://doi.org/10.3390/nu11081813





