Resistant Starch Is Actively Fermented by Infant Faecal Microbiota and Increases Microbial Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Participants and Intervention
2.3. Faecal Sample Collection and Processing
2.4. Carbohydrates and Chemicals
2.5. In Vitro Fermentation
2.6. SCFA and pH
2.7. Molecular and Sequence Analysis
2.7.1. DNA Extraction
2.7.2. Real-time Quantitative Polymerase Chain Reaction (qPCR)
2.7.3. Sequencing of 16S Ribosomal RNA Encoding Gene Amplicons
2.7.4. Taxonomic Assignments to 16S Reads
2.8. Statistical Analysis
3. Results
3.1. SCFA and pH Levels
3.2. Microbial Community Analysis
3.2.1. Alpha Diversity
3.2.2. Beta Diversity
3.2.3. Relative Abundance
3.2.4. Quantitative PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
Appendix A.1. Pre-Digestion Method
Appendix A.2. DNA Extraction Method
| Target | Primers | Sequence (5′–3′) | Conc (nM) | Annealing | References | |
|---|---|---|---|---|---|---|
| Temp (°C) | Time (s) | |||||
| Total Bacteria | UnivF | TCCTACGGGAGGCAGCAGT | 500 | 60 | 45 | [35] |
| UnivR | GGACTACCAGGGTATCTATCCTGTT | |||||
| Lactobacillus Spp | Lacto-F | AGCAGTAGGGAATCTTCCA | 500 | 56 | 20 | [36] |
| Lacto-R | CACCGCTACACATGGAG | |||||
| Bifidobacterium Spp | Bifi-F | TCGCGTCCGGTGTGAAAG | 500 | 56 | 20 | [37] |
| Bifi-R | CCACATCCAGCGTCCAC | |||||
Appendix A.3. qPCR
Appendix A.4. Next Generation Sequencing
- 16S Amplicon PCR Forward Primer =5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA-3′
- Amplicon PCR Reverse Primer =5′-TCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT-3′
| Taxonomy | Preweaning (n = 15) | Weaning (n = 14) | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | HAMS | p Value | Control | HAMS | p Value | mHAMS | p Value | |
| Phylum | ||||||||
| Actinobacteria | 12.50 | 15.48 | 0.35 | 10.16 | 18.55 | 0.004 | 17.88 | 0.004 |
| Bacteroidetes | 7.87 | 7.29 | 0.86 | 4.73 | 13.04 | 0.001 | 15.56 | 0.03 |
| Firmicutes | 33.33 | 37.74 | 0.09 | 33.5 | 32.51 | 0.79 | 34.37 | 0.11 |
| Proteobacteria | 46.23 | 39.39 | 0.17 | 51.27 | 35.76 | 0.001 | 32.06 | 0.004 |
| Class/subclass | ||||||||
| Actinobacteria | 12.51 | 15.48 | 0.35 | 10.17 | 18.56 | 0.004 | 17.88 | 0.004 |
| Bacilli | 13.46 | 18.74 | 0.02 | 14.48 | 16.83 | 0.32 | 19.04 | 0.08 |
| Bacteroidia | 7.87 | 7.29 | 0.86 | 4.73 | 13.04 | 0.001 | 15.56 | 0.001 |
| Clostridia | 17.71 | 16.28 | 0.58 | 16.25 | 12.31 | 0.26 | 10.79 | 0.07 |
| Gammaproteobacteria | 46.10 | 39.13 | 0.17 | 50.69 | 35.19 | <0.001 | 31.56 | <0.001 |
| Order | ||||||||
| Bacteroidales | 7.87 | 7.29 | 0.86 | 4.73 | 13.04 | 0.001 | 15.56 | 0.001 |
| Bifidobacteriales | 11.64 | 14.73 | 0.32 | 9.12 | 16.83 | 0.01 | 16.33 | 0.01 |
| Clostridiales | 17.71 | 16.27 | 0.58 | 16.24 | 12.30 | 0.26 | 10.79 | 0.07 |
| Enterobacteriales | 45.98 | 39.04 | 0.17 | 50.61 | 35.16 | <0.001 | 31.51 | <0.001 |
| Erysipelotrichales | 1.77 | 1.73 | 0.93 | 0.25 | 0.54 | 0.07 | 0.77 | 0.09 |
| Lactobacillales | 12.81 | 17.67 | 0.03 | 14.44 | 16.58 | 0.36 | 19.03 | 0.08 |
| Family | ||||||||
| Bifidobacteriaceae | 11.67 | 14.82 | 0.32 | 9.31 | 17.54 | 0.007 | 16.86 | 0.004 |
| Bacteroidaceae | 7.49 | 7.06 | 0.89 | 4.60 | 12.91 | <0.001 | 14.34 | 0.002 |
| Enterococcaceae | 12.42 | 17.04 | 0.03 | 12.79 | 13.96 | 0.61 | 16.32 | 0.16 |
| Clostridiceae | 14.58 | 13.44 | 0.67 | 7.71 | 5.22 | 0.27 | 4.98 | 0.13 |
| Erysipelotrichaceae | 1.77 | 1.73 | 0.94 | 0.26 | 0.55 | 0.07 | 0.78 | 0.08 |
| Peptostreptococcaceae | 1.99 | 1.57 | 0.45 | 3.14 | 0.94 | 0.04 | 0.54 | 0.02 |
| Ruminococcaceae | 0.42 | 0.34 | 0.34 | 1.59 | 1.46 | 0.62 | 2.02 | 0.28 |
| Enterobacteriaceae | 46.12 | 39.19 | 0.17 | 51.19 | 35.88 | <0.001 | 32.20 | <0.001 |
| Genus | ||||||||
| Bacteroides | 7.48 | 7.02 | 0.89 | 4.54 | 12.56 | 0.009 | 13.92 | 0.002 |
| Bifidobacterium | 11.64 | 14.73 | 0.32 | 9.12 | 16.83 | 0.006 | 16.33 | 0.006 |
| Clostridium | 14.53 | 13.38 | 0.67 | 7.44 | 5.09 | 0.27 | 4.78 | 0.11 |
| Enterobacter | 30.31 | 24.89 | 0.21 | 32.32 | 18.21 | <0.001 | 15.02 | <0.001 |
| Enterococcus | 12.40 | 16.99 | 0.03 | 12.72 | 13.75 | 0.65 | 16.12 | 0.18 |
| Escherichia/Shigella | 15.05 | 13.67 | 0.73 | 16.39 | 15.88 | 0.73 | 15.59 | 0.67 |
| Ruminococcus | 0.22 | 0.17 | 0.43 | 0.50 | 0.56 | 0.58 | 1.35 | 0.01 |
References
- Albenberg, L.G.; Wu, G. Diet and the Intestinal microbiome: Associations, Functions, and Implications for Health and Disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Probert, H.M.; Loo, J.V.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Updating the concept of prebiotics. Nutr. Res. Rev. 2004, 17, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.A.; Sarbini, S.R. The potential of resistant starch as a prebiotic. Crit. Rev. Biotechnol. 2015, 36, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Englyst, H.N.; Kingman, S.M.; Cummings, J.H. Classification and measurement of nutritionally important starch fractions. Eur. J. Clin. Nutr. 1992, 46, S33–S50. [Google Scholar] [PubMed]
- Bird, A.R.; Conlon, M.A.; Christophersen, C.T.; Topping, D.L. Resistant starch, large bowel fermentation and a broader perspective of prebiotics and probiotics. Benef. Microbes 2010, 1, 423–431. [Google Scholar] [CrossRef]
- Parrett, A.M.; Edwards, C.A.; Lokerse, E. Colonic fermentation capacity in vitro: Development during weaning in breast-fed infants is slower for complex carbohydrates than for sugars. Am. J. Clin. Nutr. 1997, 65, 927–933. [Google Scholar] [CrossRef][Green Version]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef]
- Christian, M.T.; Edwards, C.A.; Preston, T.; Johnston, L.; Varley, R.; Weaver, L.T. Starch fermentation by faecal bacteria of infants, toddlers and adults: Importance for energy salvage. Eur. J. Clin. Nutr. 2003, 57, 1486–1491. [Google Scholar] [CrossRef]
- Parrett, A.M.; Edwards, C.A. In vitro fermentation of carbohydrate by breast fed and formula fed infants. Arch. Dis. Child. 1997, 76, 249–253. [Google Scholar] [CrossRef]
- Rose, D.J.; Venema, K.; Keshavarzian, A.; Hamaker, B.R. Starch-entrapped microspheres show a beneficial fermentation profile and decrease in potentially harmful bacteria during in vitro fermentation in faecal microbiota obtained from patients with inflammatory bowel disease. Br. J. Nutr. 2010, 103, 1514–1524. [Google Scholar] [CrossRef]
- Vonk, R.J.; Hagedoorn, R.E.; De Graff, R.; Elzinga, H.; Tabak, S.; Yang, Y.X.; Stellaard, F. Digestion of so-called resistant starch sources in the human small intestine. Am. J. Clin. Nutr. 2000, 72, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.R.; Usher, S.; May, B.; Topping, T.L.; Morrell, M.K. Resistant Starch: Measurements, Intakes, and Dietary Targets. In Dietary Fiber and Health 2012; CRC Press: Boca Raton, FL, USA, 2012; pp. 41–56. [Google Scholar]
- Edwards, C.A.; Gibson, G.; Champ, M.; Jensen, B.B.; Mather, J.C.; Nagengast, F.; Rumney, C.; Quehl, A. In Vitro Method for Quantification of the Fermentation of Starch by Human Faecal Bacteria. J. Sci. Food Agric. 1996, 71, 209–217. [Google Scholar] [CrossRef]
- Goñi, I.; Martin-Carrón, N. In vitro fermentation and hydration properties of commercial dietary fiber-rich supplements. Nutr. Res. 1998, 18, 1077–1089. [Google Scholar] [CrossRef]
- McOrist, A.L.; Abell, G.C.; Cooke, C.; Nyland, K. Bacterial population dynamics and faecal short-chain fatty acid (SCFA) concentrations in healthy humans. Br. J. Nutr. 2008, 100, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Payne, A.N.; Zihler, A.; Chassard, C.; Lacroix, C. Advances and perspectives in in vitro human gut fermentation modeling. Trends Biotechnol. 2012, 30, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, C.T.; Morrison, M.; Conlon, M.A. Overestimation of the Abundance of Sulfate-Reducing Bacteria in Human Feces by Quantitative PCR Targeting the Desulfovibrio 16S rRNA Gene. Appl. Environ. Microbiol. 2011, 77, 3544–3546. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Tang, M.K.; Lodge, C.J. Exmining the evidence for using synbiotics to treat or prevent atopic dermatitis. JAMA Pediatr. 2016, 170, 201–203. [Google Scholar] [CrossRef]
- Boehm, G.; Moro, G. Structural and functional aspects of prebiotics used in infant nutrition. J. Nutr. 2008, 138, 1818S–1828S. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; Van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Odamaki, T.; Horiogome, A.; Sugahara, H.; Hashikura, N.; Minami, J.; Xiao, J.Z.; Abe, F. Comparative Genomics Revealed Genetic Diversity and Species/Strain-Level Differences in Carbohydrate Metabolism of Three Probiotic Bifidobacterial Species. Int. J. Genom. 2015, 2015, 12. [Google Scholar] [CrossRef] [PubMed]
- Van der Maarel, M.J.; Leemhuis, H. Starch modification with microbial alpha-glucanotransferase enzymes. Carbohydr. Polym. 2013, 93, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Storro, O.; Oien, T.; Johnsen, R.; Pope, P.; Rudi, K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol. Ecol. 2014, 87, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Lugi, G.A.; Duranti, S.; Turroni, F.; Mancabelli, L.; Ferraio, C.; Mangifesta, M.; Hevia, A.; Viappiani, A.; Scholz, M.; et al. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 2015, 5, 15782. [Google Scholar] [CrossRef] [PubMed]
- Rakoff-Nahoum, S.; Kong, Y.; Kleinstein, S.H.; Subramanian, S.; Ahern, P.P.; Gordon, J.I.; Medzhitov, R. Analysis of gene–environment interactions in postnatal development of the mammalian intestine. Proc. Natl. Acad. Sci. USA 2015, 112, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Ze, X.; Duncan, S.H.; Louis, P.; Flint, J.H. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012, 6, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Fouhse, J.M.; Ganzle, M.G.; Regmi, P.R.; Van Kempen, T.A.; Zijlstra, R.T. High Amylose Starch with Low In Vitro Digestibility Stimulates Hindgut Fermentation and Has a Bifidogenic Effect in Weaned Pigs. J. Nutr. 2015, 145, 2464–2470. [Google Scholar] [CrossRef]
- Jiang, X.; Li, B.; Su, Y.; Weiyun, Z. Shifts in bacterial community compositions during in vitro fermentation of amylopectin and resistant starch by colonic inocula of pigs. J. Food Nutr. Res. 2013, 1, 156–163. [Google Scholar]
- Turnbaugh, P.H.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Clarke, J.M.; Bird, A.R.; Topping, D.L.; Cobiac, L. Excretion of starch and esterified short-chain fatty acids by ileostomy subjects after the ingestion of acylated starches. Am. J. Clin. Nutr. 2007, 86, 1146–1151. [Google Scholar] [CrossRef] [PubMed]
- Martinez, I.; Kim, J.; Duffy, P.R.; Schlegel, V.L.; Walter, J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE 2010, 5, e15046. [Google Scholar] [CrossRef] [PubMed]
- Ashwar, B.A.; Gani, A.; Shah, A.; Wani, I.; Masoodi, F. Preparation, health benefits and applications of resistant starch—A review. Starch Stärke 2016, 68, 287–301. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E. Determination of bacterial load by real time PCR using a broad range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.C.; Hertel, C.; Tannock, G.W.; Lis, C.M.; Munro, K.; Hammes, W.P. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 2001, 67, 2578–2585. [Google Scholar] [CrossRef] [PubMed]
- Rinttila, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an extensive set of 16S rDNA targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- 16S Metagenomic Sequencing Library Preparation. Available online: http://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 1 May 2019).
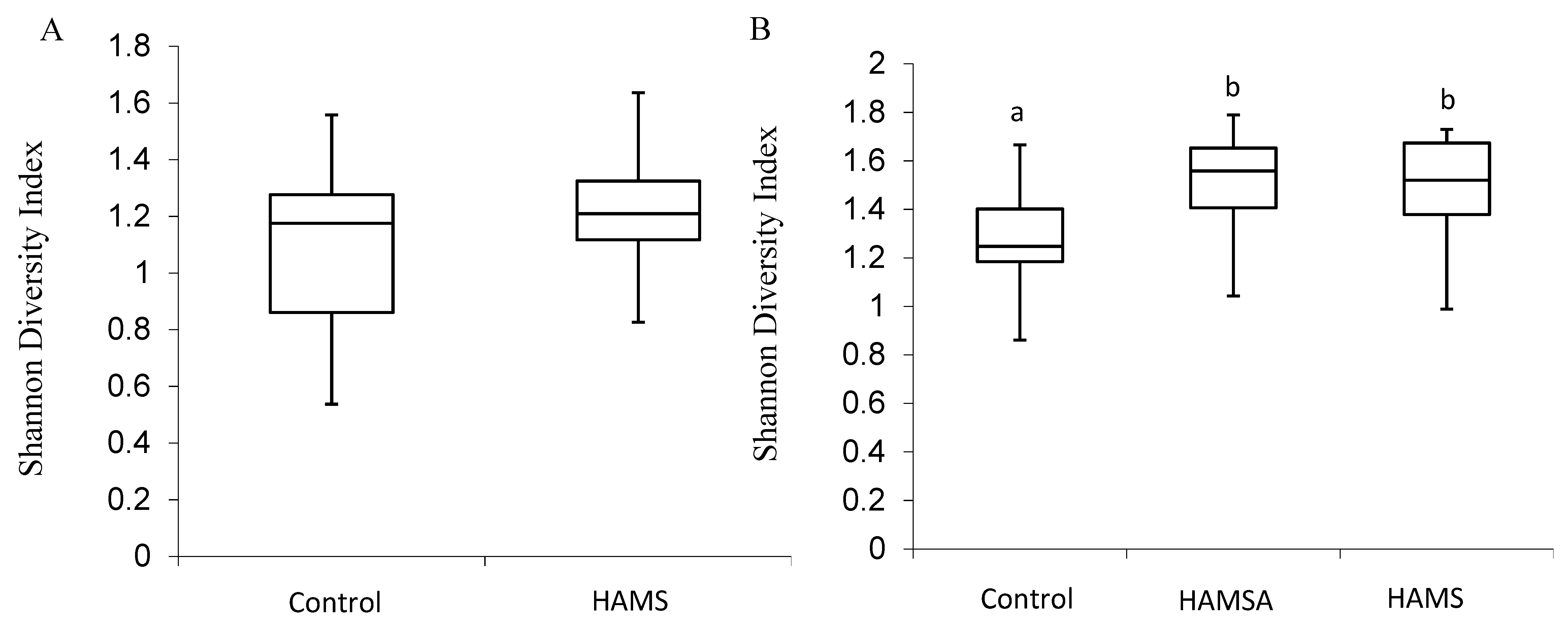
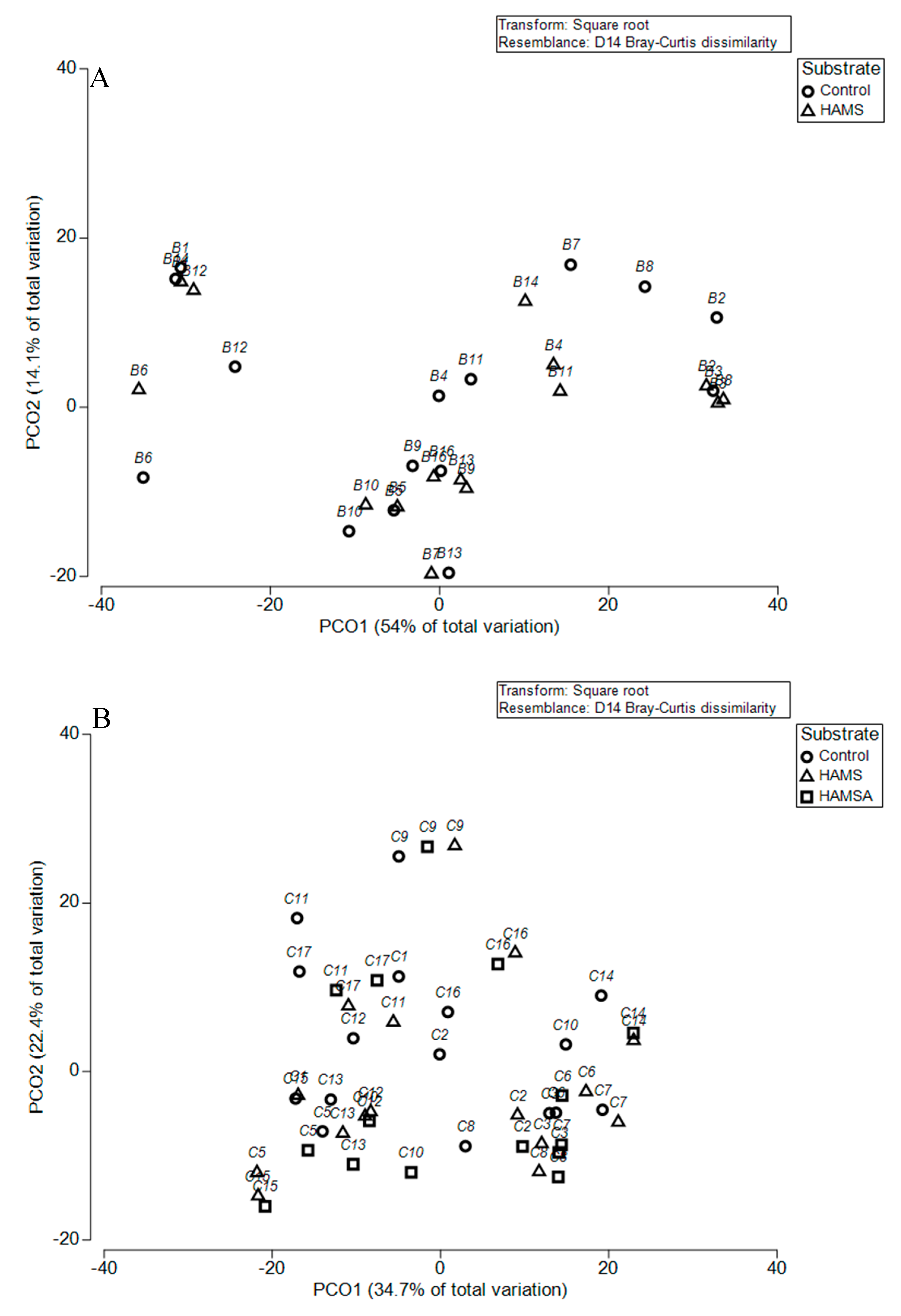
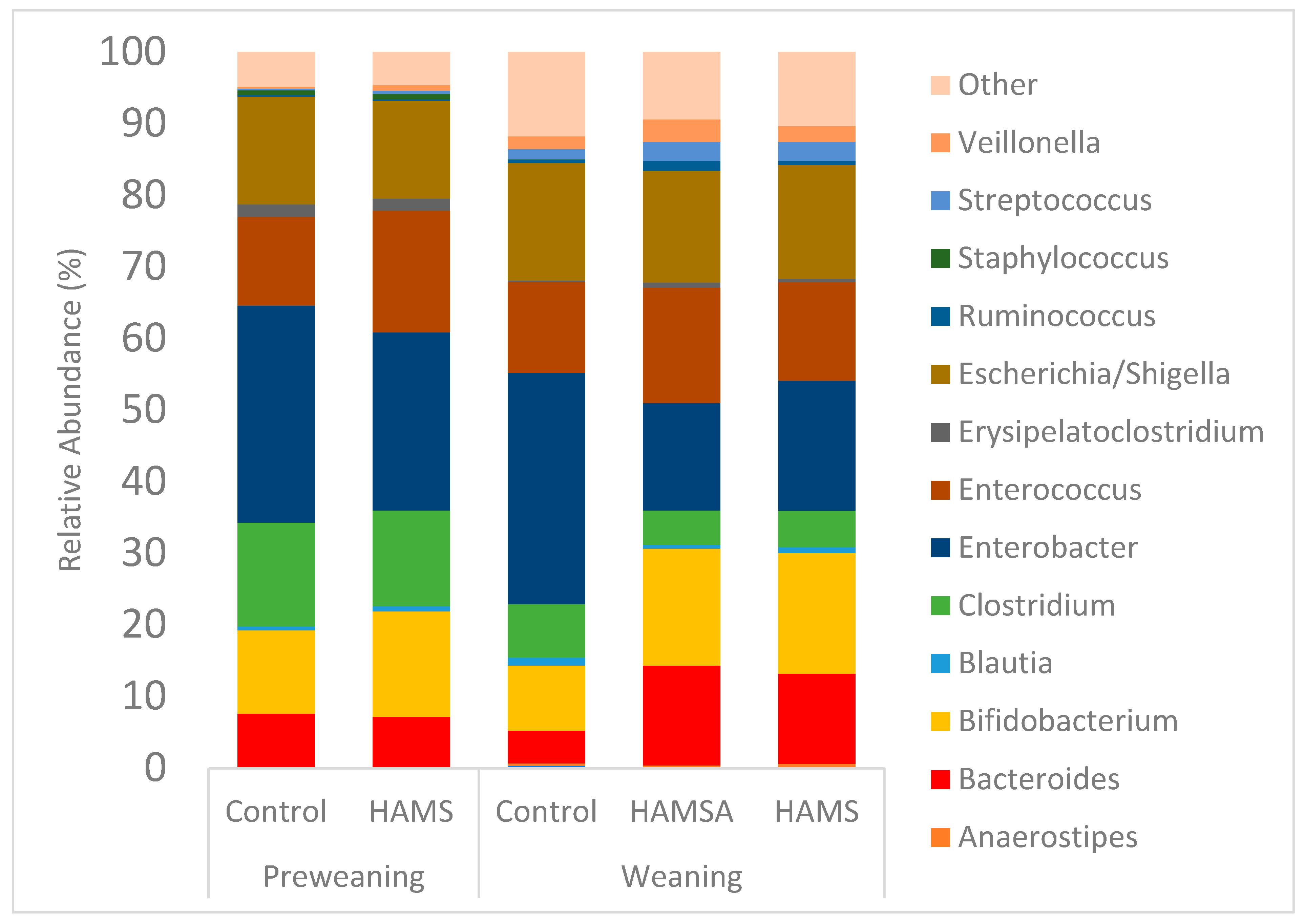
| Participants | Preweaning (n) | Weaning (n) | Age Preweaning Sample (Months) Mean ± SE | Age Weaning Sample (Months) Mean ± SE |
|---|---|---|---|---|
| Exclusively Breast-Fed | 10 | 10 | 3.32 ± 0.37 a | 7.03 ± 0.27 b |
| Mixed | 7 | 6 | 3.29 ± 0.22 a | 7.39 ± 0.21 b |
| Total | 17 | 16 | 3.31 ± 0.23 | 7.16 ± 0.18 |
| Preweaning | Weaning | ||||||
|---|---|---|---|---|---|---|---|
| Control | HAMS | Lactulose | Control | HAMS | HAMSA | Lactulose | |
| Initial pH (0 h) | 7.64 ± 0.03 a | 7.63 ± 0.03 a | 7.69 ± 0.03 a | 7.66 ± 0.03 a | 7.65 ± 0.02 a | 7.65 ± 0.03 a | 7.72 ± 0.04 a |
| Final pH (24 h) | 6.72 ± 0.08 a | 6.40 ± 0.09 b,* | 4.73 ± 0.12 c,* | 6.77 ± 0.05 a | 6.43 ± 0.05 b,* | 6.37 ± 0.04 b,* | 4.65 ± 0.08 c,* |
| Δ pH | −0.93 ± 0.07 a | −1.21 ± 0.06 b,* | −3.03 ± 0.06 c,* | −0.82 ± 0.07 a | −1.14 ± 0.08 b,* | −1.09 ± 0.10 b,* | −2.89 ± 0.20 c,* |
| Preweaning | Weaning | ||||||
|---|---|---|---|---|---|---|---|
| Control | HAMS | Lactulose | Control | HAMS | HAMSA | Lactulose | |
| Total SCFA # | 16.68 ± 1.7 a | 23.70 ± 1.7 b,* | 61.84 ± 5.4 c,* | 20.11 ± 2.3 a | 34.85 ± 4.0 b,* | 37.81 ± 4.2 b,* | 78.27 ± 4.5 c,* |
| Acetate | 14.70 ± 1.5 a | 20.93 ± 1.5 b,* | 56.11 ± 6.5 c,* | 16.98 ± 2.1 a | 27.73 ± 3.3 b,* | 30.27 ± 3.1 b,* | 73.42 ± 5.0 c,* |
| Propionate | 0.83 ± 0.17 a | 1.33 ± 0.33 a | 1.13 ± 0.80 a | 1.60 ± 0.32 a | 4.14 ± 0.91 b* | 5.56 ± 1.2 c* | 2.89 ± 0.74 a,b,c,* |
| Butyrate | 0.36 ± 0.13 a | 0.74 ± 0.26 b | 0.27 ± 0.12 a | 1.09 ± 0.26 a | 2.20 ± 0.59 b* | 1.23 ± 0.33 a | 1.81 ± 1.1 a |
| Total Bacteria | Lactobacillus | Bifidobacterium | |||||
|---|---|---|---|---|---|---|---|
| Substrate | 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | |
| Preweaning | control (n = 17) | 6.6 ± 0.15 | 7.18 ± 0.24 a,* | 4.26 ± 0.4 | 4.19 ± 0.23 a | 6.06 ± 0.09 | 6.08 ± 0.32 a |
| HAMS (n = 17) | 7.21 ± 0.1 a,* | 4.02 ± 0.28 a | 6.39 ± 0.25 a | ||||
| Weaning | Control (n = 16) | 6.65 ± 0.07 | 7.32 ± 0.06 a,* | 2.9 ± 0.27 | 3.34 ± 0.86 a | 5.09 ± 0.31 | 5.56 ± 0.21 a |
| HAMSA (n = 16) | 7.42 ± 0.06 a,* | 3.52 ± 0.91 a | 6.21 ± 0.10 b,* | ||||
| HAMS (n = 15) | 7.47 ± 0.05 a,* | 3.49 ± 0.89 a | 6.04 ± 0.19 b,* | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gopalsamy, G.; Mortimer, E.; Greenfield, P.; Bird, A.R.; Young, G.P.; Christophersen, C.T. Resistant Starch Is Actively Fermented by Infant Faecal Microbiota and Increases Microbial Diversity. Nutrients 2019, 11, 1345. https://doi.org/10.3390/nu11061345
Gopalsamy G, Mortimer E, Greenfield P, Bird AR, Young GP, Christophersen CT. Resistant Starch Is Actively Fermented by Infant Faecal Microbiota and Increases Microbial Diversity. Nutrients. 2019; 11(6):1345. https://doi.org/10.3390/nu11061345
Chicago/Turabian StyleGopalsamy, Geetha, Elissa Mortimer, Paul Greenfield, Anthony R. Bird, Graeme P. Young, and Claus T. Christophersen. 2019. "Resistant Starch Is Actively Fermented by Infant Faecal Microbiota and Increases Microbial Diversity" Nutrients 11, no. 6: 1345. https://doi.org/10.3390/nu11061345
APA StyleGopalsamy, G., Mortimer, E., Greenfield, P., Bird, A. R., Young, G. P., & Christophersen, C. T. (2019). Resistant Starch Is Actively Fermented by Infant Faecal Microbiota and Increases Microbial Diversity. Nutrients, 11(6), 1345. https://doi.org/10.3390/nu11061345





