The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Ethics
2.3. Study Procedures
2.4. Metabolomics Analysis
2.5. Calculations
2.6. Statistical Analysis
2.7. Identification
3. Results
3.1. Baseline Characteristics
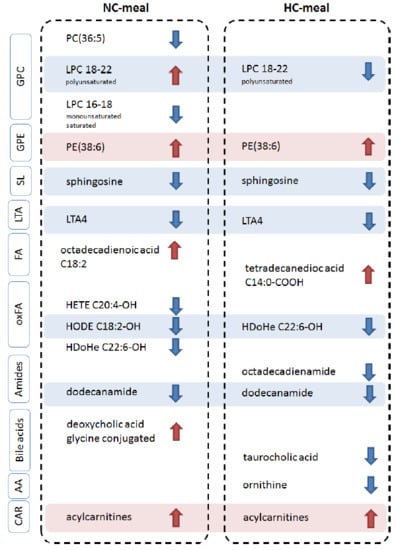
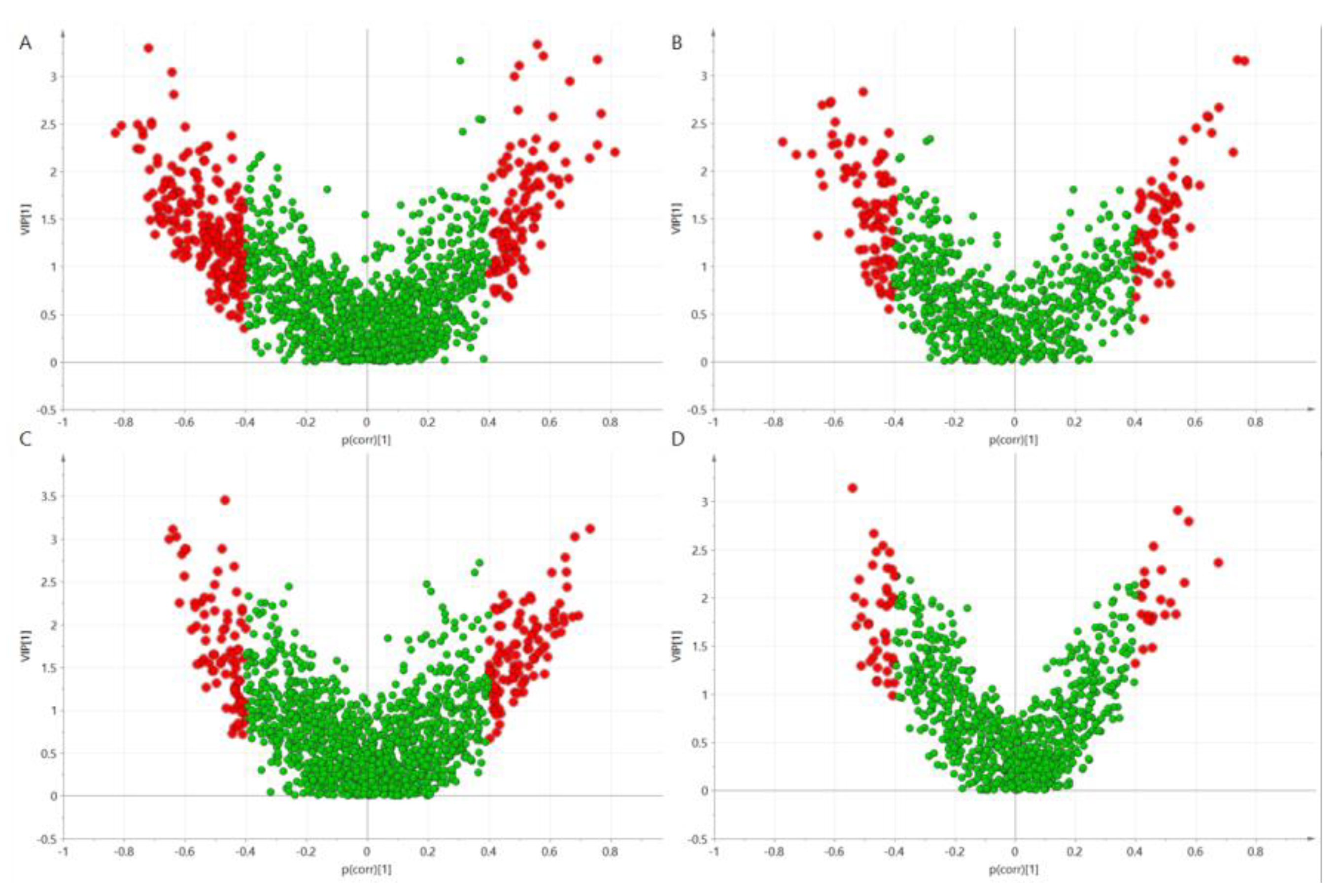
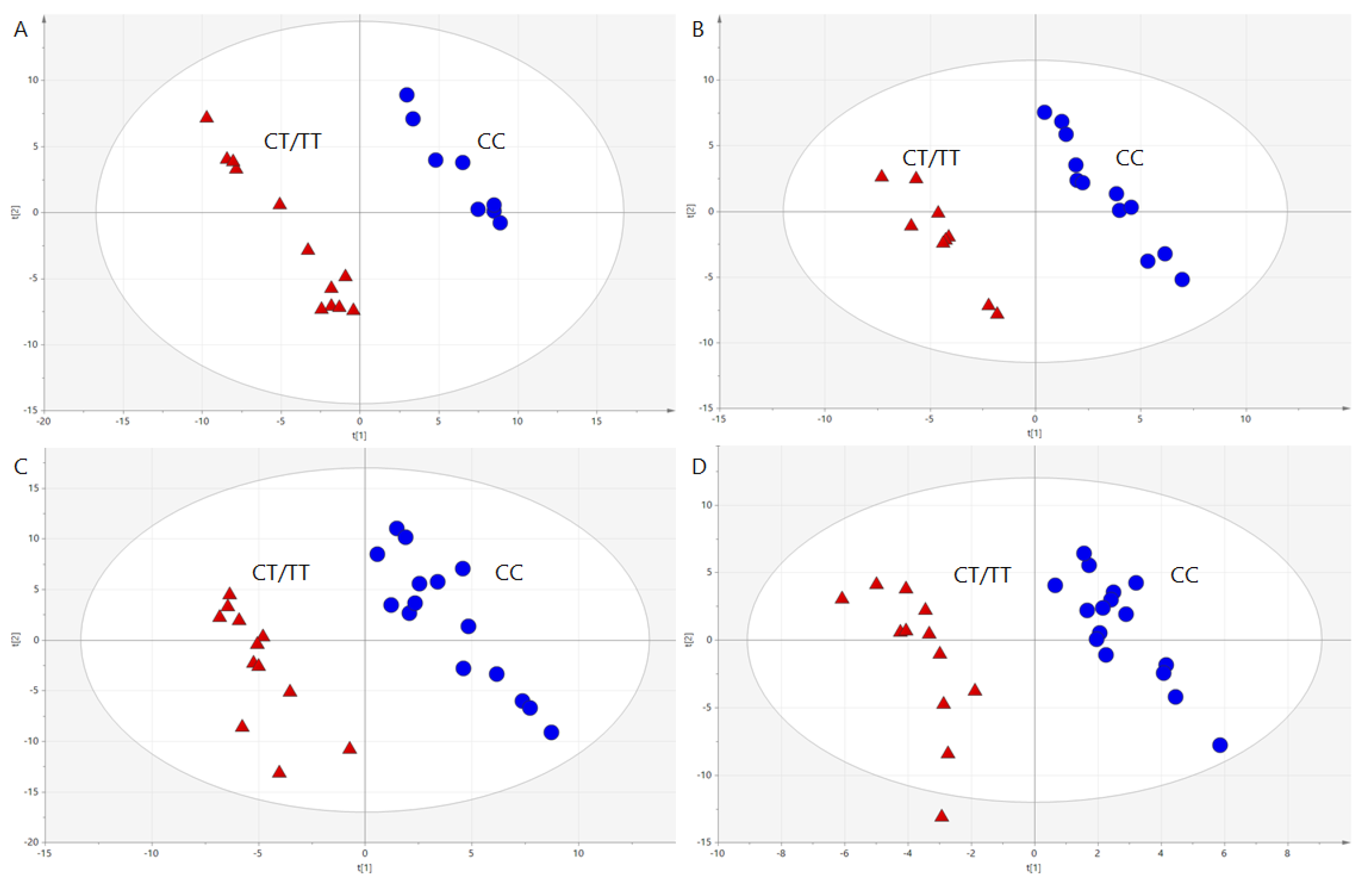
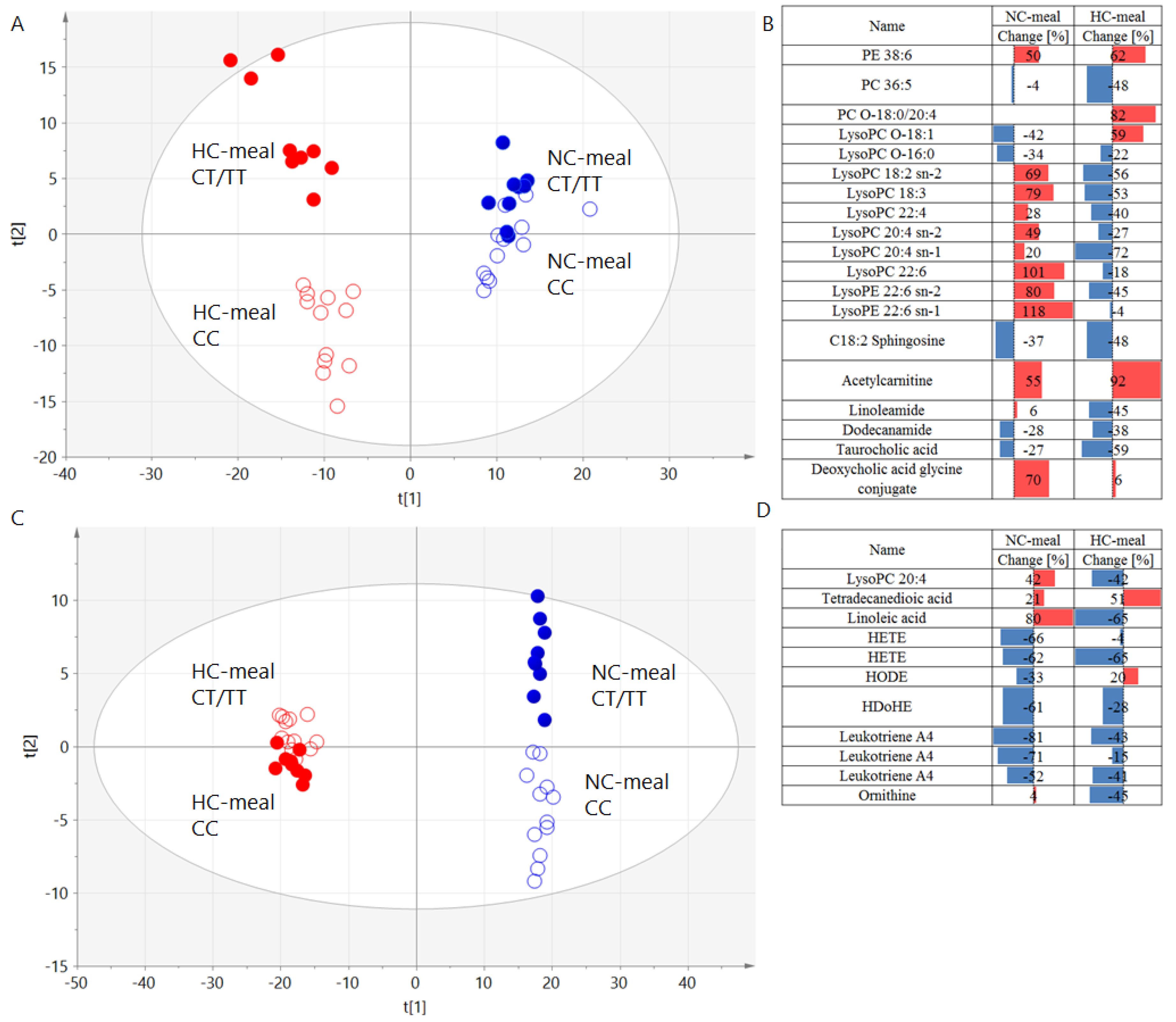
3.2. Genotype Effects on Metabolites Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- International Diabetes Federation Diabetes Atlas, 7th ed. 2015. Available online: http://www.diabetesatlas.org/ (accessed on 1 December 2018).
- Chamnan, P.; Simmons, R.K.; Forouhi, N.G.; Luben, R.N.; Khaw, K.T.; Wareham, N.J.; Griffin, S.J. Incidence of type 2 diabetes using proposed HbA1c diagnostic criteria in the european prospective investigation of cancer-norfolk cohort: Implications for preventive strategies. Diabetes Care 2011, 34, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Ferrannini, E.; Camastra, S. Relationship between impaired glucose tolerance, non-insulin-dependent diabetes mellitus and obesity. Eur J. Clin. Investig. 1998, 28, 3–6. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Lecompte, S.; Pasquetti, G.; Hermant, X.; Grenier-Boley, B.; Gonzalez-Gross, M.; De Henauw, S.; Molnar, D.; Stehle, P.; Béghin, L.; Moreno, L.A.; et al. Genetic and molecular insights into the role of PROX1 in glucose metabolism. Diabetes 2013, 62, 1738–1745. [Google Scholar] [CrossRef]
- Hamet, P.; Haloui, M.; Harvey, F.; Marois-Blanchet, F.C.; Sylvestre, M.P.; Tahir, M.R.; Simon, P.H.; Kanzki, B.S.; Raelson, J.; Long, C.; et al. PROX1 gene CC genotype as a major determinant of early onset of type 2 diabetes in slavic study participants from Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation study. J. Hypertens. 2017, 35, S24–S32. [Google Scholar] [CrossRef] [PubMed]
- Boesgaard, T.W.; Grarup, N.; Jørgensen, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O.; Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC). Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia 2010, 53, 1647–1655. [Google Scholar] [CrossRef]
- Wagner, R.; Dudziak, K.; Herzberg-Schäfer, S.A.; Machicao, F.; Stefan, N.; Staiger, H.; Häring, H.U.; Fritsche, A. Glucose-raising genetic variants in MADD and ADCY5 impair conversion of proinsulin to insulin. PLoS ONE 2011, 6, e23639. [Google Scholar] [CrossRef]
- Barker, A.; Sharp, S.J.; Timpson, N.J.; Bouatia-Naji, N.; Warrington, N.M.; Kanoni, S.; Beilin, L.J.; Brage, S.; Deloukas, P.; Evans, D.M.; et al. Association of genetic Loci with glucose levels in childhood and adolescence: A meta-analysis of over 6000 children. Diabetes 2011, 60, 1805–1812. [Google Scholar] [CrossRef]
- Wang, J.; Kilic, G.; Aydin, M.; Burke, Z.; Oliver, G.; Sosa-Pineda, B. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev. Biol. 2005, 286, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Kretowski, A.; Adamska, E.; Maliszewska, K.; Wawrusiewicz-Kurylonek, N.; Citko, A.; Goscik, J.; Bauer, W.; Wilk, J.; Golonko, A.; Waszczeniuk, M.; et al. The rs340874 PROX1 type 2 diabetes mellitus risk variant is associated with visceral fat accumulation and alterations in postprandial glucose and lipid metabolism. Genes Nutr. 2015, 10, 454. [Google Scholar] [CrossRef]
- Bell, D.S.; O’Keefe, J.H.; Jellinger, P. Postprandial dysmetabolism: The missing link between diabetes and cardiovascular events? Endocr. Pract. 2008, 14, 112–124. [Google Scholar] [CrossRef]
- Adamska-Patruno, E.; Ostrowska, L.; Goscik, J.; Fiedorczuk, J.; Moroz, M.; Kretowski, A.; Gorska, M. The Differences in Postprandial Serum Concentrations of Peptides That Regulate Satiety/Hunger and Metabolism after Various Meal Intake, in Men with Normal vs. Excessive BMI. Nutrients 2019, 11, 493. [Google Scholar] [CrossRef]
- Adamska, E.; Ostrowska, L.; Gościk, J.; Waszczeniuk, M.; Krętowski, A.; Górska, M. Intake of Meals Containing High Levels of Carbohydrates or High Levels of Unsaturated Fatty Acids Induces Postprandial Dysmetabolism in Young Overweight/Obese Men. Biomed. Res. Int. 2015, 2015, 147196. [Google Scholar] [CrossRef]
- Adamska-Patruno, E.; Ostrowska, L.; Goscik, J.; Pietraszewska, B.; Kretowski, A.; Gorska, M. The relationship between the leptin/ghrelin ratio and meals with various macronutrient contents in men with different nutritional status: A randomized crossover study. Nutr. J. 2018, 17, 118. [Google Scholar] [CrossRef]
- Adamska, E.; Kretowski, A.; Goscik, J.; Citko, A.; Bauer, W.; Waszczeniuk, M.; Maliszewska, K.; Paczkowska-Abdulsalam, M.; Niemira, M.; Szczerbinski, L.; et al. The type 2 diabetes susceptibility TCF7L2 gene variants affect postprandial glucose and fat utilization in non-diabetic subjects. Diabetes Metab. 2018, 44, 379–382. [Google Scholar] [CrossRef]
- Adamska-Patruno, E.; Goscik, J.; Czajkowski, P.; Maliszewska, K.; Ciborowski, M.; Golonko, A.; Wawrusiewicz-Kurylonek, N.; Citko, A.; Waszczeniuk, M.; Kretowski, A.; et al. The MC4R genetic variants are associated with lower visceral fat accumulation and higher postprandial relative increase in carbohydrate utilization in humans. Eur. J. Nutr. 2019. [Google Scholar] [CrossRef]
- Adamska, E.; Waszczeniuk, M.; Gościk, J.; Golonko, A.; Wilk, J.; Pliszka, J.; Maliszewska, K.; Lipińska, D.; Milewski, R.; Wasilewska, A.; et al. The usefulness of glycated hemoglobin A1c (HbA1c) for identifying dysglycemic states in individuals without previously diagnosed diabetes. Adv. Med. Sci. 2012, 57, 296–301. [Google Scholar] [CrossRef]
- Ostrowska, L.; Witczak, K.; Adamska, E. Effect of nutrition and atherogenic index on the occurrence and intensity of insulin resistance. Pol. Arch. Med. Wewn. 2013, 123, 289–296. [Google Scholar] [CrossRef]
- Lu, J.; Varghese, R.T.; Zhou, L.; Vella, A.; Jensen, M.D. Glucose tolerance and free fatty acid metabolism in adults with variations in TCF7L2 rs7903146. Metabolism 2017, 68, 55–63. [Google Scholar] [CrossRef][Green Version]
- Adamska-Patruno, E.; Ostrowska, L.; Golonko, A.; Pietraszewska, B.; Goscik, J.; Kretowski, A.; Gorska, M. Evaluation of Energy Expenditure and Oxidation of Energy Substrates in Adult Males after Intake of Meals with Varying Fat and Carbohydrate Content. Nutrients 2018, 10, 627. [Google Scholar] [CrossRef]
- Samczuk, P.; Luba, M.; Godzien, J.; Mastrangelo, A.; Hady, H.R.; Dadan, J.; Barbas, C.; Gorska, M.; Kretowski, A.; Ciborowski, M. “Gear mechanism” of bariatric interventions revealed by untargeted metabolomics. J. Pharm. Biomed. Anal. 2018, 151, 219–226. [Google Scholar] [CrossRef]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef]
- Floegel, A.; Stefan, N.; Yu, Z.; Mühlenbruch, K.; Drogan, D.; Joost, H.G.; Fritsche, A.; Häring, H.U.; Hrabě de Angelis, M.; Peters, A.; et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013, 62, 639–648. [Google Scholar] [CrossRef]
- Barber, M.N.; Risis, S.; Yang, C.; Meikle, P.J.; Staples, M.; Febbraio, M.A.; Bruce, C.R. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE 2012, 7, e41456. [Google Scholar] [CrossRef]
- Tonks, K.T.; Coster, A.C.; Christopher, M.J.; Chaudhuri, R.; Xu, A.; Gagnon-Bartsch, J.; Chisholm, D.J.; James, D.E.; Meikle, P.J.; Greenfield, J.R.; et al. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity 2016, 24, 908–916. [Google Scholar] [CrossRef]
- Yea, K.; Kim, J.; Yoon, J.H.; Kwon, T.; Kim, J.H.; Lee, B.D.; Lee, H.J.; Lee, S.J.; Kim, J.I.; Lee, T.G.; et al. Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. J. Biol. Chem. 2009, 284, 33833–33840. [Google Scholar] [CrossRef]
- Tunaru, S.; Bonnavion, R.; Brandenburger, I.; Preussner, J.; Thomas, D.; Scholich, K.; Offermanns, S. 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nat. Commun. 2018, 9, 177. [Google Scholar] [CrossRef]
- Nagy, L.; Tontonoz, P.; Alvarez, J.G.; Chen, H.; Evans, R.M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998, 93, 229–240. [Google Scholar] [CrossRef]
- Itoh, T.; Fairall, L.; Amin, K.; Inaba, Y.; Szanto, A.; Balint, B.L.; Nagy, L.; Yamamoto, K.; Schwabe, J.W. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat. Struct. Mol. Biol. 2008, 15, 924–931. [Google Scholar] [CrossRef]
- Contreras, A.V.; Torres, N.; Tovar, A.R. PPAR-α as a key nutritional and environmental sensor for metabolic adaptation. Adv. Nutr. 2013, 4, 439–452. [Google Scholar] [CrossRef]
- Semple, R.K.; Chatterjee, V.K.; O’Rahilly, S. PPAR gamma and human metabolic disease. J. Clin. Investig. 2006, 116, 581–589. [Google Scholar] [CrossRef]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef]
- Sharma, A.M.; Staels, B. Review: Peroxisome proliferator-activated receptor gamma and adipose tissue-understanding obesity-related changes in regulation of lipid and glucose metabolism. J. Clin. Endocrinol. Metab. 2007, 92, 386–395. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Mahankali, A.; Matsuda, M.; Mahankali, S.; Hardies, J.; Cusi, K.; Mandarino, L.J.; DeFronzo, R.A. Effect of pioglitazone on abdominal fat distribution and insulin sensitivity in type 2 diabetic patients. J. Clin. Endocrinol. Metab. 2002, 87, 2784–2791. [Google Scholar] [CrossRef]
- Ye, G.; Gao, H.; Wang, Z.; Lin, Y.; Liao, X.; Zhang, H.; Chi, Y.; Zhu, H.; Dong, S. PPARα and PPARγ activation attenuates total free fatty acid and triglyceride accumulation in macrophages via the inhibition of Fatp1 expression. Cell Death Dis. 2019, 10, 39. [Google Scholar] [CrossRef]
- Dirkx, R.; Meyhi, E.; Asselberghs, S.; Reddy, J.; Baes, M.; Van Veldhoven, P.P. Beta-oxidation in hepatocyte cultures from mice with peroxisomal gene knockouts. Biochem. Biophys. Res. Commun. 2007, 357, 718–723. [Google Scholar] [CrossRef]
- Filgueiras, L.R.; Serezani, C.H.; Jancar, S. Leukotriene B4 as a Potential Therapeutic Target for the Treatment of Metabolic Disorders. Front. Immunol. 2015, 6, 515. [Google Scholar] [CrossRef]
- Escobedo, N.; Oliver, G. The Lymphatic Vasculature: Its Role in Adipose Metabolism and Obesity. Cell Metab. 2017, 26, 598–609. [Google Scholar] [CrossRef]
- Tian, W.; Rockson, S.G.; Jiang, X.; Kim, J.; Begaye, A.; Shuffle, E.M.; Tu, A.B.; Cribb, M.; Nepiyushchikh, Z.; Feroze, A.H.; et al. Leukotriene B antagonism ameliorates experimental lymphedema. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef]
- Adams, S.H.; Hoppel, C.L.; Lok, K.H.; Zhao, L.; Wong, S.W.; Minkler, P.E.; Hwang, D.H.; Newman, J.W.; Garvey, W.T. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J. Nutr. 2009, 139, 1073–1081. [Google Scholar] [CrossRef]
- Yamamoto, S.; Takehara, M.; Ushimaru, M. Inhibitory action of linoleamide and oleamide toward sarco/endoplasmic reticulum Ca2+-ATPase. Biochim. Biophys. Acta 2017, 1861, 3399–3405. [Google Scholar] [CrossRef]
- Jain, M.K.; Ghomashchi, F.; Yu, B.Z.; Bayburt, T.; Murphy, D.; Houck, D.; Brownell, J.; Reid, J.C.; Solowiej, J.E.; Wong, S.M. Fatty acid amides: Scooting mode-based discovery of tight-binding competitive inhibitors of secreted phospholipases A2. J. Med. Chem. 1992, 35, 3584–3586. [Google Scholar] [CrossRef]
- Hollie, N.I.; Matlib, M.A.; Hui, D.Y. Direct inhibitory effects of phospholipase A2 enzymatic product lysophosphatidylcholine (LPC) on hepatic mitochondria. Biochemistry 2011, 25, 527. [Google Scholar] [CrossRef]
- Kitatani, K.; Idkowiak-Baldys, J.; Hannun, Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008, 20, 1010–1018. [Google Scholar] [CrossRef]
- Kowalski, G.M.; Carey, A.L.; Selathurai, A.; Kingwell, B.A.; Bruce, C.R. Plasma sphingosine-1-phosphate is elevated in obesity. PLoS ONE 2013, 8, e72449. [Google Scholar] [CrossRef]
- Haus, J.M.; Kashyap, S.R.; Kasumov, T.; Zhang, R.; Kelly, K.R.; Defronzo, R.A.; Kirwan, J.P. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes 2009, 58, 337–343. [Google Scholar] [CrossRef]
- Qin, J.; Gao, D.M.; Jiang, Q.F.; Zhou, Q.; Kong, Y.Y.; Wang, Y.; Xie, Y.H. Prospero-related homeobox (Prox1) is a corepressor of human liver receptor homolog-1 and suppresses the transcription of the cholesterol 7-alpha-hydroxylase gene. Mol. Endocrinol. 2004, 18, 2424–2439. [Google Scholar] [CrossRef][Green Version]
- Prawitt, J.; Caron, S.; Staels, B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr. Diab. Rep. 2011, 11, 160–166. [Google Scholar] [CrossRef]
- Tomkin, G.H.; Owens, D. Obesity diabetes and the role of bile acids in metabolism. J. Transl. Int. Med. 2016, 4, 73–80. [Google Scholar] [CrossRef]
- Kuipers, F.; Bloks, V.W.; Groen, A.K. Beyond intestinal soap--bile acids in metabolic control. Nat. Rev. Endocrinol. 2014, 10, 488–498. [Google Scholar] [CrossRef]
- Zaborska, K.E.; Lee, S.A.; Garribay, D.; Cha, E.; Cummings, B.P. Deoxycholic acid supplementation impairs glucose homeostasis in mice. PLoS ONE 2018, 13, e0200908. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Zamora, S.; Méndez-Rodríguez, M.L.; Olguín-Martínez, M.; Sánchez-Sevilla, L.; Quintana-Quintana, M.; García-García, N.; Hernández-Muñoz, R. Increased erythrocytes by-products of arginine catabolism are associated with hyperglycemia and could be involved in the pathogenesis of type 2 diabetes mellitus. PLoS ONE 2013, 8, e66823. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.R.; Lara, A.; Zhang, R.; Park, Y.M.; DeFronzo, R.A. Insulin reduces plasma arginase activity in type 2 diabetic patients. Diabetes Care 2008, 31, 134–139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, C.; Zhang, R.; Wang, C.; Wang, J.; Ma, X.; Hou, X.; Lu, J.; Yu, W.; Jiang, F.; Bao, Y.; et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS ONE 2010, 5, e15542. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Are oxidative stress-activated signaling pathways mediators of insulin resistance and beta-cell dysfunction? Diabetes 2003, 52, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, S.; Samocha-Bonet, D.; Heilbronn, L.K.; Campbell, L.V. Inflammatory and oxidative stress responses to high-carbohydrate and high-fat meals in healthy humans. J. Nutr. Metab. 2012, 2012, 238056. [Google Scholar] [CrossRef] [PubMed]
| CC Genotype | CT/TT | p-Value * | |
|---|---|---|---|
| Age (years) | 35.3 ± 9.5 | 36.3 ± 7.0 | 0.75 |
| Weight (kg) | 93.6 ± 24.5 | 89.1 ± 16.1 | 0.95 |
| Body mass index (BMI) (kg/m2) | 29.1 ± 8.1 | 27.3 ± 4.2 | 0.74 |
| Body fat content (%) | 23.8 ± 10.1 | 23.2 ± 7.8 | 0.87 |
| Fat free mass (%) | 69.6 ± 11.0 | 67.6 ± 8.3 | 0.60 |
| Waist (cm) | 99.6 ± 21.1 | 95.7 ± 13.6 | 0.77 |
| Hip (cm) | 104.3 ± 14.8 | 99.6 ± 8.7 | 0.76 |
| WHR | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.81 |
| Fasting glucose concentration (mg/dl) | 86.2 ± 8.0 | 86.7 ± 6.4 | 0.85 |
| Fasting insulin concentration (IU/mL) | 10.4 ± 9.1 | 8.9 ± 5.4 | 0.84 |
| HOMA-IR | 2.2 ± 2.0 | 1.9 ± 1.3 | 0.81 |
| HOMA-B | 188.2 ± 163.3 | 143.7 ± 88.9 | 0.78 |
| HbA1c | 5.2 ± 0.5 | 5.2 ± 0.2 | 0.90 |
| Name | Molecular Weight, Da | RT, Min | NC-Meal | HC-Meal | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Change, % | p-Value | p (corr) | VIP | Change, % | p-Value | p (corr) | VIP | |||
| PE 38:6 | 763.5152 | 9.40 | 50 | 0.25 | 0.43 | 1.38 | 62 | 0.18 | −0.56 | 1.98 |
| PC 36:5 | 779.5465 | 7.95 | −4 | 0.93 | −0.51 | 1.35 | −48 | 0.19 | 0.30 | 1.43 |
| PC O-18:0/20:4 | 795.6141 | 10.20 | −0.45 | 1.52 | −0.16 | 1.34 | ||||
| LysoPC O-18:1 | 507.3689 | 5.95 | −42 | 0.17 | −0.59 | 2.21 | 59 | 0.45 | 0.27 | 0.46 |
| LysoPC O-16:0 | 481.3532 | 5.80 | −34 | 0.50 | −0.64 | 2.81 | −22 | 0.61 | 0.34 | 1.30 |
| LysoPC 18:2 sn-2 | 519.3325 | 5.40 | 69 | 0.30 | 0.30 | 1.07 | −56 | 0.049 | 0.55 | 1.90 |
| LysoPC 18:3 | 517.3168 | 5.05 | 79 | 0.28 | 0.16 | 0.71 | −53 | 0.26 | 0.53 | 1.72 |
| LysoPC 22:4 | 571.3638 | 5.85 | 28 | 0.47 | 0.55 | 1.51 | −40 | 0.018 | 0.56 | 1.96 |
| LysoPC 20:4 | 543.3325 | 5.40 | 42 | 0.46 | 0.21 | 1.01 | −42 | 0.01 | 0.62 | 2.16 |
| LysoPC 20:4 sn-2 | 543.3325 | 5.35 | 49 | 0.07 | −0.50 | 1.43 | −27 | 0.11 | 0.45 | 1.76 |
| LysoPC 20:4 sn-1 | 543.3325 | 5.35 | 20 | 0.74 | −0.01 | 0.07 | −72 | 0.02 | 0.54 | 2.91 |
| LysoPC 22:6 | 567.3325 | 5.40 | 101 | 0.04 | 0.81 | 2.21 | −18 | 0.40 | 0.26 | 1.11 |
| LysoPE 22:6 sn-2 | 525.2855 | 5.35 | 80 | 0.23 | −0.54 | 2.35 | −45 | 0.29 | −0.34 | 1.26 |
| LysoPE 22:6 sn-1 | 525.2855 | 5.35 | 118 | 0.09 | −0.56 | 2.03 | −4 | 0.86 | −0.27 | 1.10 |
| Tetradecanedioic acid | 258.1831 | 4.35 | 21 | 0.44 | −0.20 | 0.78 | 51 | 0.005 | −0.53 | 2.01 |
| Linoleic acid | 280.2402 | 7.05 | 80 | 0.01 | −0.59 | 2.29 | −65 | 0.13 | −0.12 | 0.67 |
| HETE | 320.2351 | 5.70 | −66 | 0.10 | 0.64 | 2.56 | −4 | 0.93 | 0.38 | 1.69 |
| HETE | 320.2351 | 5.70 | −62 | 0.10 | 0.57 | 1.89 | −65 | 0.13 | 0.43 | 2.15 |
| HODE | 298.2508 | 5.85 | −33 | 0.09 | 0.51 | 1.43 | 20 | 0.43 | −0.26 | 1.06 |
| HDoHE | 344.2351 | 5.70 | −61 | 0.11 | 0.57 | 1.90 | −28 | 0.53 | 0.43 | 2.15 |
| C18:2 Sphingosine | 297.2668 | 5.85 | −37 | 0.25 | −0.41 | 1.60 | −48 | 0.01 | 0.65 | 2.78 |
| Leukotriene A4 | 318.2195 | 5.45 | −81 | 0.02 | 0.74 | 3.16 | −43 | 0.21 | 0.16 | 0.79 |
| Leukotriene A4 | 318.2195 | 5.45 | −71 | 0.01 | 0.64 | 2.58 | −15 | 0.71 | 0.24 | 1.14 |
| Leukotriene A4 | 318.2195 | 5.45 | −52 | 0.10 | 0.58 | 1.40 | −41 | 0.14 | 0.41 | 2.03 |
| Acetylcarnitine | 203.1158 | 0.25 | 55 | 0.06 | 0.54 | 2.00 | 92 | 0.002 | −0.60 | 2.88 |
| Linoleamide | 279.2562 | 5.30 | 6 | 0.93 | −0.01 | 0.05 | −45 | 0.09 | 0.53 | 1.51 |
| Dodecanamide | 199.1936 | 5.20 | −28 | 0.49 | −0.81 | 2.48 | −38 | 0.16 | 0.48 | 1.10 |
| Taurocholic acid | 515.2917 | 2.30 | −27 | 0.35 | −0.39 | 0.39 | −59 | 0.07 | 0.66 | 2.43 |
| Deoxycholic acid glycine conjugate | 449.3141 | 4.30 | 70 | 0.14 | 0.55 | 1.42 | 6 | 0.75 | −0.29 | 0.91 |
| Ornithine | 132.0899 | 0.25 | 4 | 0.83 | 0.12 | 0.05 | −45 | 0.047 | 0.53 | 1.83 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adamska-Patruno, E.; Godzien, J.; Ciborowski, M.; Samczuk, P.; Bauer, W.; Siewko, K.; Gorska, M.; Barbas, C.; Kretowski, A. The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men. Nutrients 2019, 11, 882. https://doi.org/10.3390/nu11040882
Adamska-Patruno E, Godzien J, Ciborowski M, Samczuk P, Bauer W, Siewko K, Gorska M, Barbas C, Kretowski A. The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men. Nutrients. 2019; 11(4):882. https://doi.org/10.3390/nu11040882
Chicago/Turabian StyleAdamska-Patruno, Edyta, Joanna Godzien, Michal Ciborowski, Paulina Samczuk, Witold Bauer, Katarzyna Siewko, Maria Gorska, Coral Barbas, and Adam Kretowski. 2019. "The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men" Nutrients 11, no. 4: 882. https://doi.org/10.3390/nu11040882
APA StyleAdamska-Patruno, E., Godzien, J., Ciborowski, M., Samczuk, P., Bauer, W., Siewko, K., Gorska, M., Barbas, C., & Kretowski, A. (2019). The Type 2 Diabetes Susceptibility PROX1 Gene Variants Are Associated with Postprandial Plasma Metabolites Profile in Non-Diabetic Men. Nutrients, 11(4), 882. https://doi.org/10.3390/nu11040882