High Levels of Prebiotic Resistant Starch in Diet Modulate Gene Expression and Metabolomic Profile in Pancreatic Cancer Xenograft Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study
2.2. Preparation of RNA for RNAseq
2.3 Ingenuity Pathway Analysis
2.4. LC–MS Analysis of Serum Metabolomics
2.5. Statistical Analysis
- Reference set: ingenuity knowledge base (gene only);
- Relationship to include: direct and indirect;
- Filter: consider only the molecules and/or relationship where species = human, and confidence = experimentally observed.
3. Results
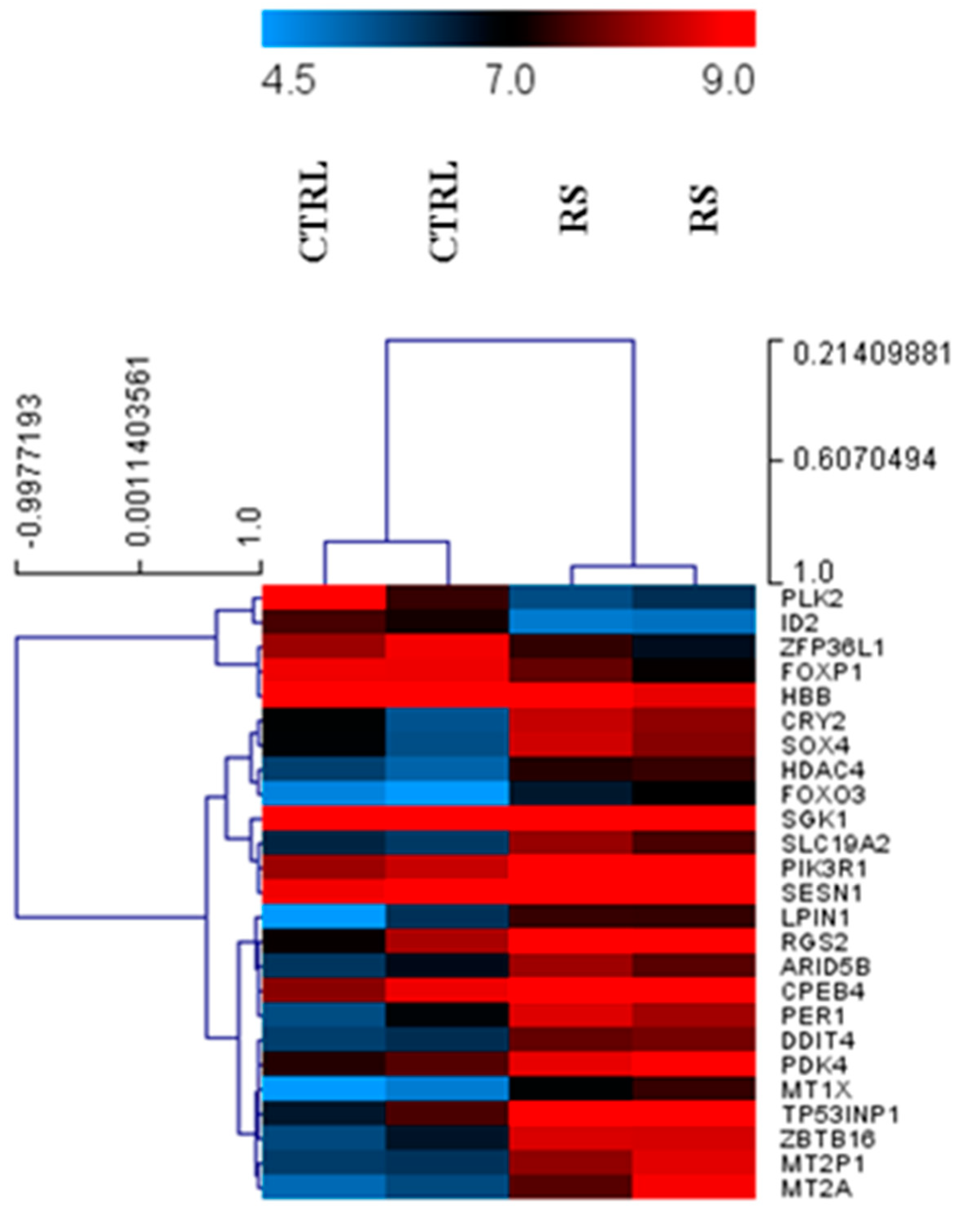
3.1. Differential Expression of Genes in PC Xenograft Mice under RS Diet
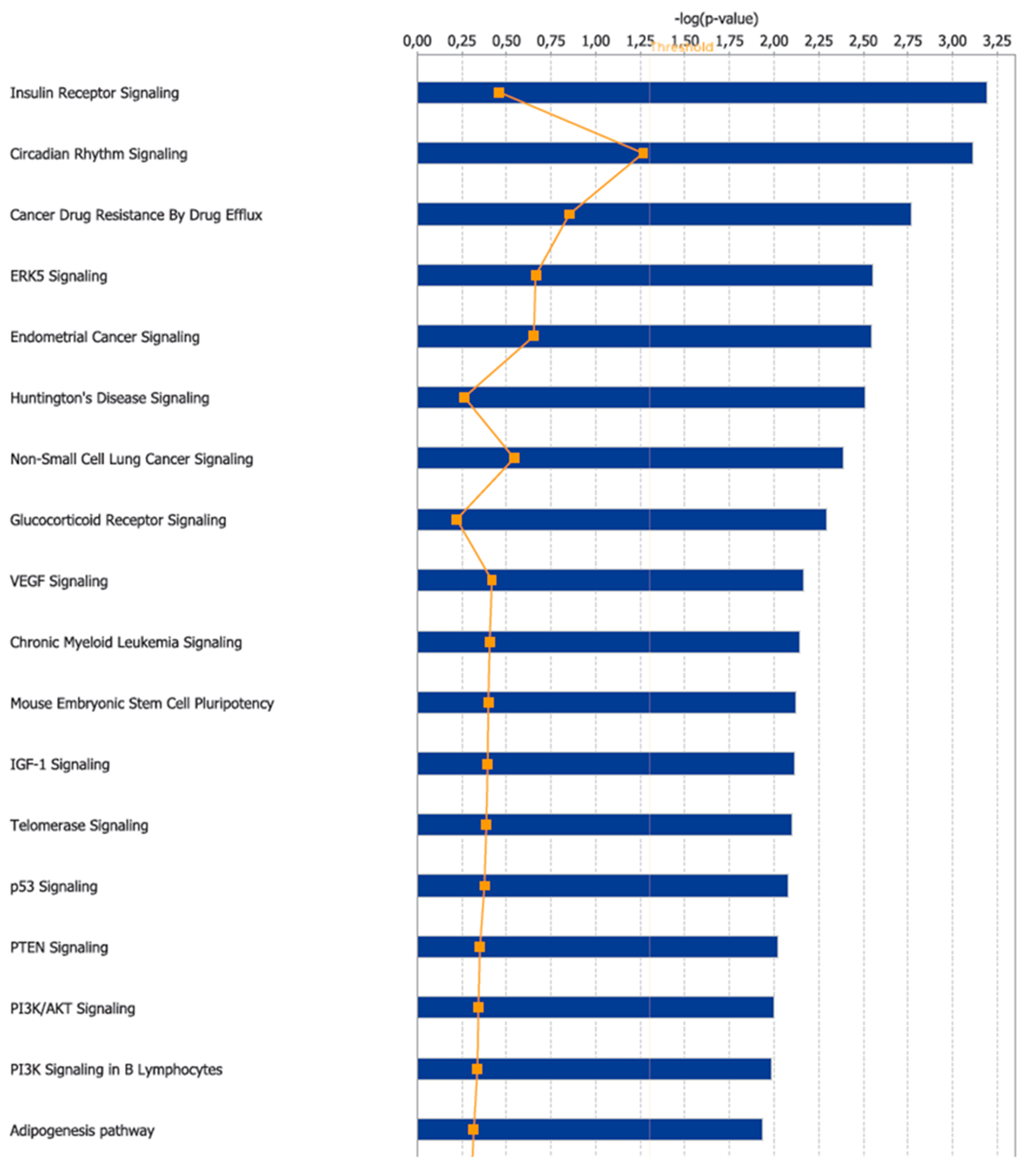
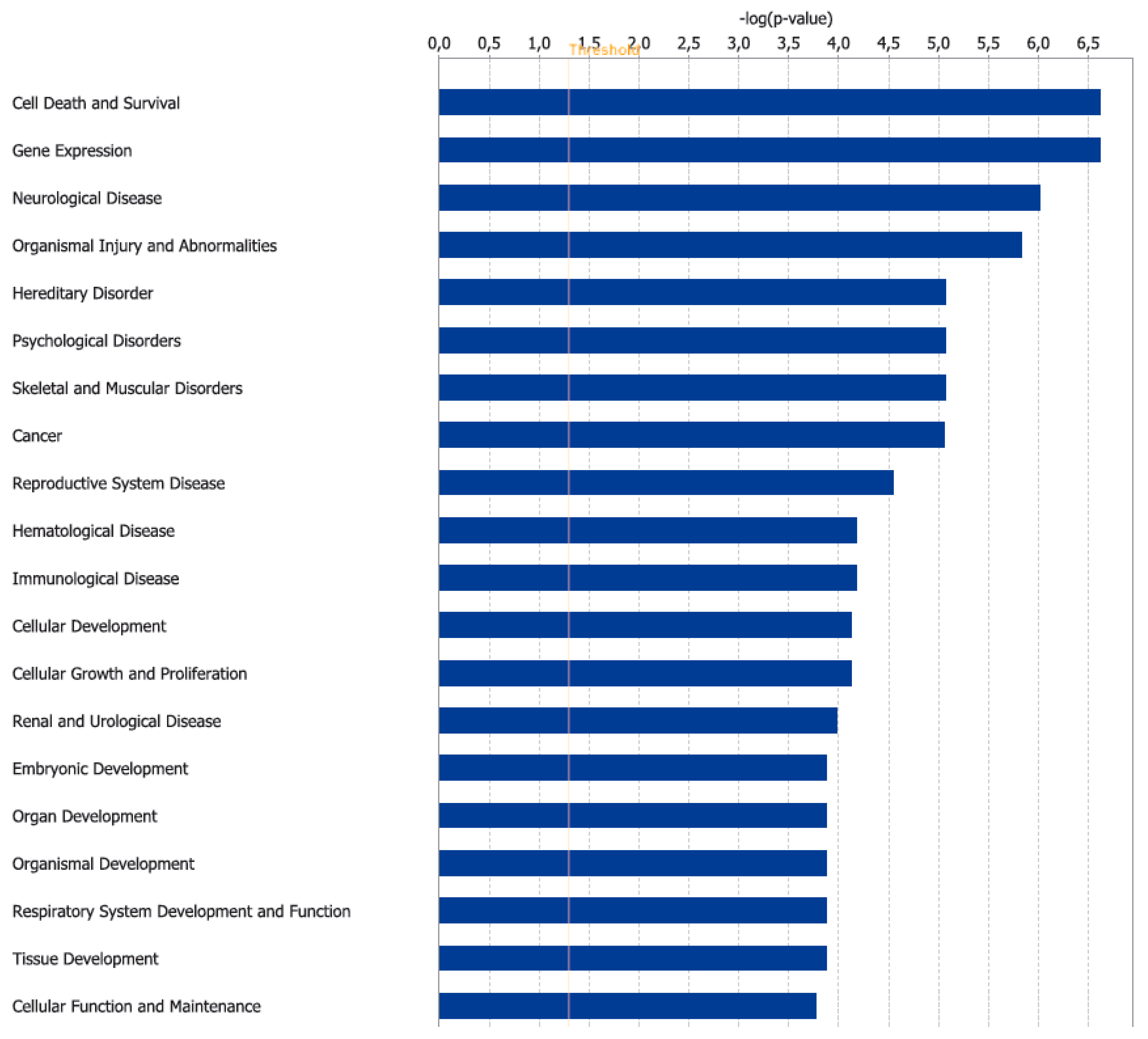
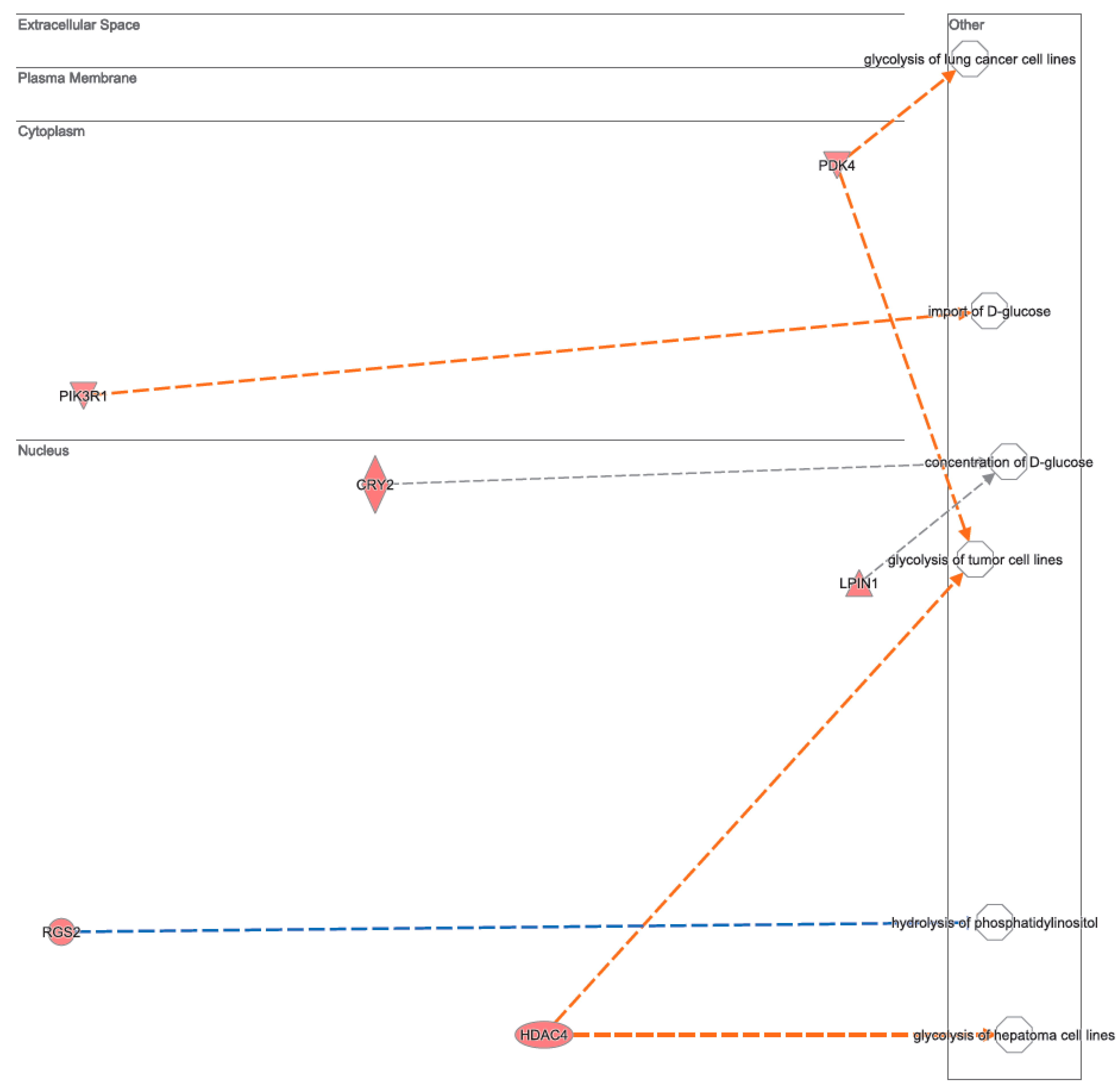
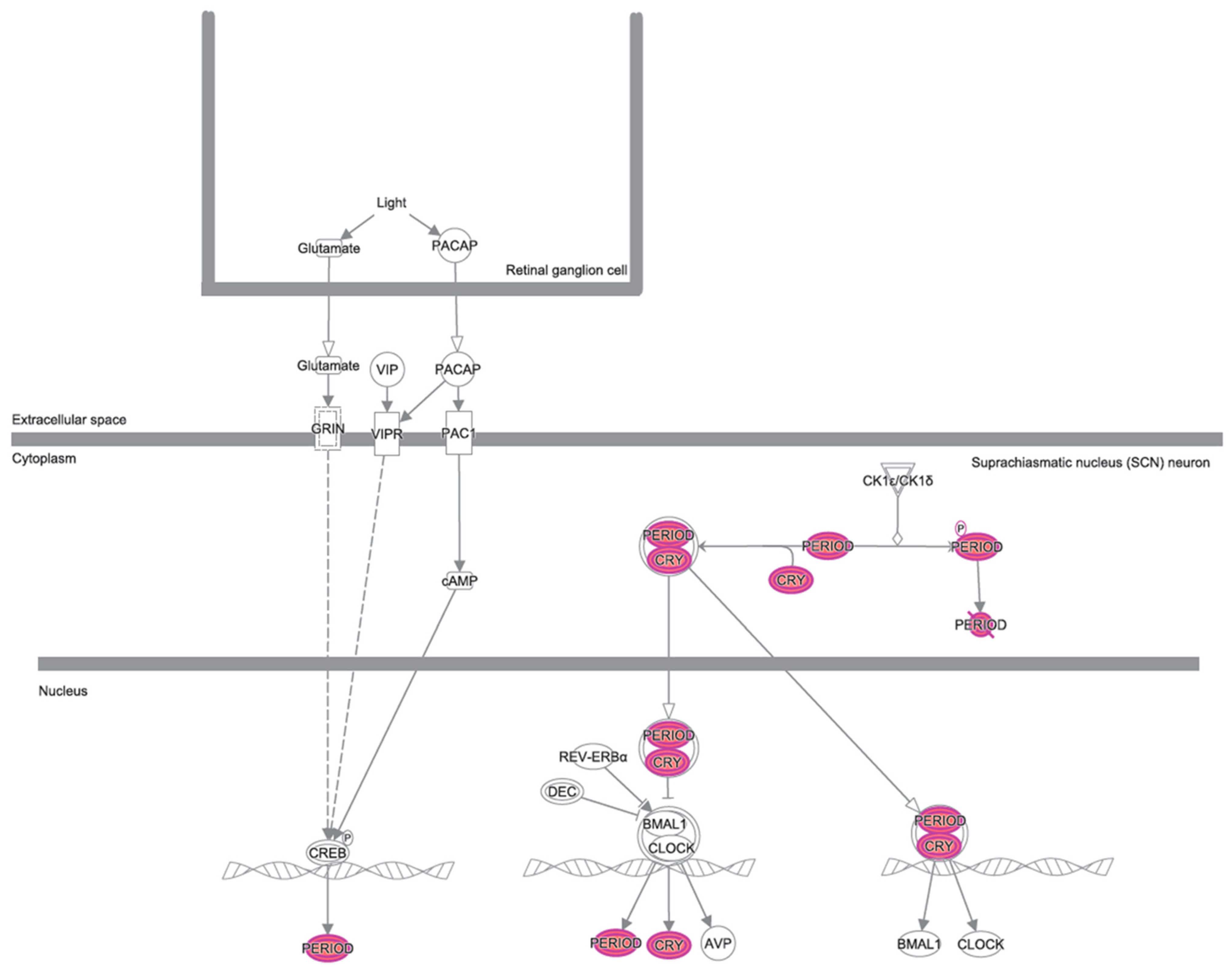
3.2. Canonical Pathways and Disease and Biological Functions Analysis
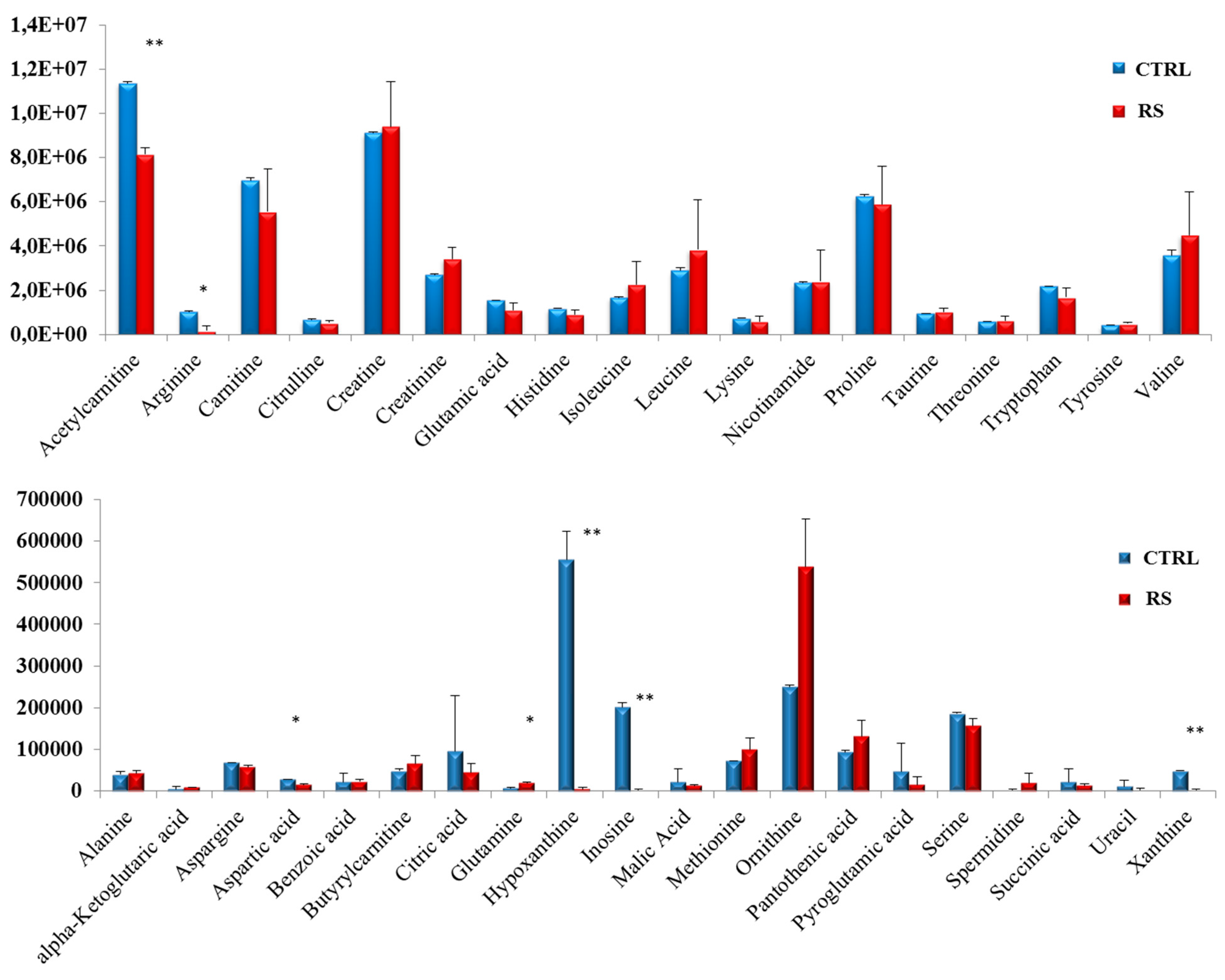
3.3. Metabolomic Profile in PC Xenograft Mice under RS Diet
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wiseman, M.J. Nutrition and cancer: Prevention and survival. Br. J. Nutr. 2018, 14, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467, 1114–1117. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a preventable disease that requires major lifestyle changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef]
- Salem, A.A.; Mackenzie, G.G. Pancreatic cancer: A critical review of dietary risk. Nutr. Res. 2018, 52, 1–13. [Google Scholar] [CrossRef]
- Gianotti, L.; Besselink, M.G.; Sandini, M.; Hackert, T.; Conlon, K.; Gerritsen, A.; Griffin, O.; Fingerhut, A.; Probst, P.; Abu Hilal, M.; et al. Nutritional support and therapy in pancreatic surgery: A position paper of the International Study Group on Pancreatic Surgery (ISGPS). Surgery 2018, 164, 1035–1048. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef]
- Panebianco, C.; Adamberg, K.; Adamberg, S.; Saracino, C.; Jaagura, M.; Kolk, K.; Di Chio, A.G.; Graziano, P.; Vilu, R.; Pazienza, V. Engineered Resistant-Starch (ERS) Diet Shapes Colon Microbiota Profile in Parallel with the Retardation of Tumor Growth in In Vitro and In Vivo Pancreatic Cancer Models. Nutrients 2017, 9, 331. [Google Scholar] [CrossRef]
- Panebianco, C.; Adamberg, K.; Jaagura, M.; Copetti, M.; Fontana, A.; Adamberg, S.; Kolk, K.; Vilu, R.; Andriulli, A.; Pazienza, V. Influence of gemcitabine chemotherapy on the microbiota of pancreatic cancer xenografted mice. Cancer Chemother. Pharmacol. 2018, 81, 773–782. [Google Scholar] [CrossRef]
- Binda, E.; Visioli, A.; Giani, F.; Trivieri, N.; Palumbo, O.; Restelli, S.; Dezi, F.; Mazza, T.; Fusilli, C.; Legnani, F.; et al. Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells. Cancer Res. 2017, 77, 996–1007. [Google Scholar] [CrossRef]
- Kopp, T.I.; Vogel, U.; Tjonneland, A.; Andersen, V. Meat and fiber intake and interaction with pattern recognition receptors (TLR1, TLR2, TLR4, and TLR10) in relation to colorectal cancer in a Danish prospective, case-cohort study. Am. J. Clin. Nutr. 2018, 107, 465–479. [Google Scholar] [CrossRef]
- Kohler, L.N.; Garcia, D.O.; Harris, R.B.; Oren, E.; Roe, D.J.; Jacobs, E.T. Adherence to Diet and Physical Activity Cancer Prevention Guidelines and Cancer Outcomes: A Systematic Review. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1018–1028. [Google Scholar] [CrossRef]
- Englyst, H.N.; Macfarlane, G.T. Breakdown of resistant and readily digestile starch by human gut bacteria. J. Sci. Food Agric. 1986, 37, 699–706. [Google Scholar] [CrossRef]
- Conlon, M.A.; Kerr, C.A.; McSweeney, C.S.; Dunne, R.A.; Shaw, J.M.; Kang, S.; Bird, A.R.; Morell, M.K.; Lockett, T.J.; Molloy, P.L.; et al. Resistant starches protect against colonic DNA damage and alter microbiota and gene expression in rats fed a Western diet. J. Nutr. 2012, 142, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Brito, M.; Rosch, C.; Schols, H.A.; Faas, M.M.; de Vos, P. Resistant starches differentially stimulate Toll-like receptors and attenuate proinflammatory cytokines in dendritic cells by modulation of intestinal epithelial cells. Mol. Nutr. Food Res. 2015, 59, 1814–1826. [Google Scholar] [CrossRef] [PubMed]
- Lepine, A.F.P.; de Hilster, R.H.J.; Leemhuis, H.; Oudhuis, L.; Buwalda, P.L.; de Vos, P. Higher Chain Length Distribution in Debranched Type-3 Resistant Starches (RS3) Increases TLR Signaling and Supports Dendritic Cell Cytokine Production. Mol. Nutr. Food Res. 2018, 63, e1801007. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Kettner, N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013, 119, 221–282. [Google Scholar]
- Kiessling, S.; Cermakian, N. The tumor circadian clock: A new target for cancer therapy? Future Oncol. 2017, 13, 2607–2610. [Google Scholar] [CrossRef]
- Savvidis, C.; Koutsilieris, M. Circadian rhythm disruption in cancer biology. Mol. Med. 2012, 18, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.Y.; Ward, S.M.; Murad, J.M.; Watson, N.P.; Israel, M.A.; Duffield, G.E. ID2 (inhibitor of DNA binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver. J. Biol. Chem. 2009, 284, 31735–31745. [Google Scholar] [CrossRef]
- Relles, D.; Sendecki, J.; Chipitsyna, G.; Hyslop, T.; Yeo, C.J.; Arafat, H.A. Circadian gene expression and clinicopathologic correlates in pancreatic cancer. J. Gastrointest. Surg. 2013, 17, 443–450. [Google Scholar] [CrossRef]
- Kleeff, J.; Ishiwata, T.; Friess, H.; Buchler, M.W.; Israel, M.A.; Korc, M. The helix-loop-helix protein Id2 is overexpressed in human pancreatic cancer. Cancer Res. 1998, 58, 3769–3772. [Google Scholar]
- Maruyama, H.; Kleeff, J.; Wildi, S.; Friess, H.; Buchler, M.W.; Israel, M.A.; Korc, M. Id-1 and Id-2 are overexpressed in pancreatic cancer and in dysplastic lesions in chronic pancreatitis. Am. J. Pathol. 1999, 155, 815–822. [Google Scholar] [CrossRef]
- Wu, T.; Yao, C.; Huang, L.; Mao, Y.; Zhang, W.; Jiang, J.; Fu, Z. Nutrients and Circadian Rhythms in Mammals. J. Nutr. Sci. Vitaminol. (Tokyo) 2015, 61, S89–S91. [Google Scholar] [CrossRef]
- Sugden, M.C. PDK4: A factor in fatness? Obes. Res. 2003, 11, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. (Lond.) 2014, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Assaily, W.; Rubinger, D.A.; Wheaton, K.; Lin, Y.; Ma, W.; Xuan, W.; Brown-Endres, L.; Tsuchihara, K.; Mak, T.W.; Benchimol, S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell. 2011, 44, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jang, C.; Lee, K.A. Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev. Reprod. 2014, 18, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, J.; Cui, M.; Zhao, F.; Ge, C.; Chen, T.; Yao, M.; Li, J. Downregulation of FOXP1 Inhibits Cell Proliferation in Hepatocellular Carcinoma by Inducing G1/S Phase Cell Cycle Arrest. Int. J. Mol. Sci. 2016, 17, 1501. [Google Scholar] [CrossRef]
- Hallmann, D.; Trümper, K.; Trusheim, H.; Ueki, K.; Kahn, C.R.; Cantley, L.C.; Fruman, D.A.; Hörsch, D. Altered signaling and cell cycle regulation in embryonal stem cells with a disruption of the gene for phosphoinositide 3-kinase regulatory subunit p85alpha. J. Biol. Chem. 2003, 278, 5099–5108. [Google Scholar] [CrossRef]
- Ben Sahra, I.; Regazzetti, C.; Robert, G.; Laurent, K.; Le Marchand-Brustel, Y.; Auberger, P.; Tanti, J.F.; Giorgetti-Peraldi, S.; Bost, F. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. 2011, 71, 4366–4372. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Yeyati, P.L.; Shaknovich, R.; Boterashvili, S.; Li, J.; Ball, H.J.; Waxman, S.; Nason-Burchenal, K.; Dmitrovsky, E.; Zelent, A.; Licht, J.D. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene 1999, 18, 925–934. [Google Scholar] [CrossRef]
- Hur, W.; Rhim, H.; Jung, C.K.; Kim, J.D.; Bae, S.H.; Jang, J.W.; Yang, J.M.; Oh, S.T.; Kim, D.G.; Wang, H.J.; et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: Clinical implication and functional analysis in vitro. Carcinogenesis 2010, 31, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Tindall, D.J. FOXOs, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312–2319. [Google Scholar] [CrossRef]
- Seux, M.; Peuget, S.; Montero, M.P.; Siret, C.; Rigot, V.; Clerc, P.; Gigoux, V.; Pellegrino, E.; Pouyet, L.; N’Guessan, P.; et al. TP53INP1 decreases pancreatic cancer cell migration by regulating SPARC expression. Oncogene 2011, 30, 3049–3061. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential Mechanisms Connecting Purine Metabolism and Cancer Therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef] [PubMed]
- Son, J.; Lyssiotis, C.A.; Ying, H.; Wang, X.; Hua, S.; Ligorio, M.; Perera, R.M.; Ferrone, C.R.; Mullarky, E.; Shyh-Chang, N.; et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 2013, 496, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Tataranni, T.; Agriesti, F.; Ruggieri, V.; Mazzoccoli, C.; Simeon, V.; Laurenzana, I.; Scrima, R.; Pazienza, V.; Capitanio, N.; Piccoli, C. Rewiring carbohydrate catabolism differentially affects survival of pancreatic cancer cell lines with diverse metabolic profiles. Oncotarget 2017, 8, 41265–41281. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.J.; Guo, L.Y.; Li, P.; Zhao, Z.; Zhou, H.; Di, L.J. Molecular link between glucose and glutamine consumption in cancer cells mediated by CtBP and SIRT4. Oncogenesis 2018, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Wellen, K.E.; Lu, C.; Mancuso, A.; Lemons, J.M.; Ryczko, M.; Dennis, J.W.; Rabinowitz, J.D.; Coller, H.A.; Thompson, C.B. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010, 24, 2784–2799. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Riganti, C.; Borgogno, S.F.; Curto, R.; Curcio, C.; Catanzaro, V.; Digilio, G.; Padovan, S.; Puccinelli, M.P.; Isabello, M.; et al. Endogenous glutamine decrease is associated with pancreatic cancer progression. Oncotarge 2017, 8, 95361–95376. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panebianco, C.; Villani, A.; Pazienza, V. High Levels of Prebiotic Resistant Starch in Diet Modulate Gene Expression and Metabolomic Profile in Pancreatic Cancer Xenograft Mice. Nutrients 2019, 11, 709. https://doi.org/10.3390/nu11040709
Panebianco C, Villani A, Pazienza V. High Levels of Prebiotic Resistant Starch in Diet Modulate Gene Expression and Metabolomic Profile in Pancreatic Cancer Xenograft Mice. Nutrients. 2019; 11(4):709. https://doi.org/10.3390/nu11040709
Chicago/Turabian StylePanebianco, Concetta, Annacandida Villani, and Valerio Pazienza. 2019. "High Levels of Prebiotic Resistant Starch in Diet Modulate Gene Expression and Metabolomic Profile in Pancreatic Cancer Xenograft Mice" Nutrients 11, no. 4: 709. https://doi.org/10.3390/nu11040709
APA StylePanebianco, C., Villani, A., & Pazienza, V. (2019). High Levels of Prebiotic Resistant Starch in Diet Modulate Gene Expression and Metabolomic Profile in Pancreatic Cancer Xenograft Mice. Nutrients, 11(4), 709. https://doi.org/10.3390/nu11040709





