Mice Microbiota Composition Changes by Inulin Feeding with a Long Fasting Period under a Two-Meals-Per-Day Schedule
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
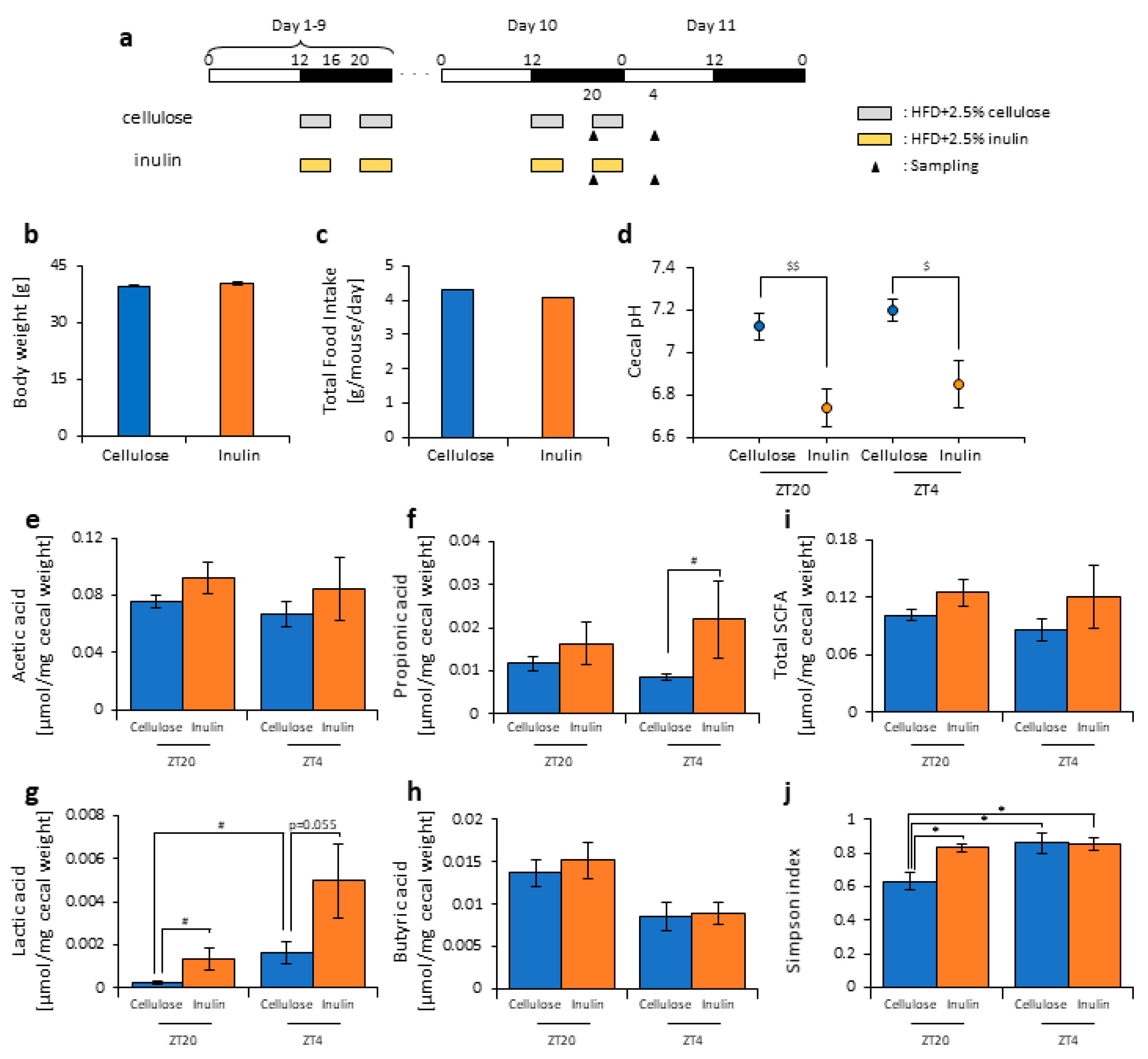
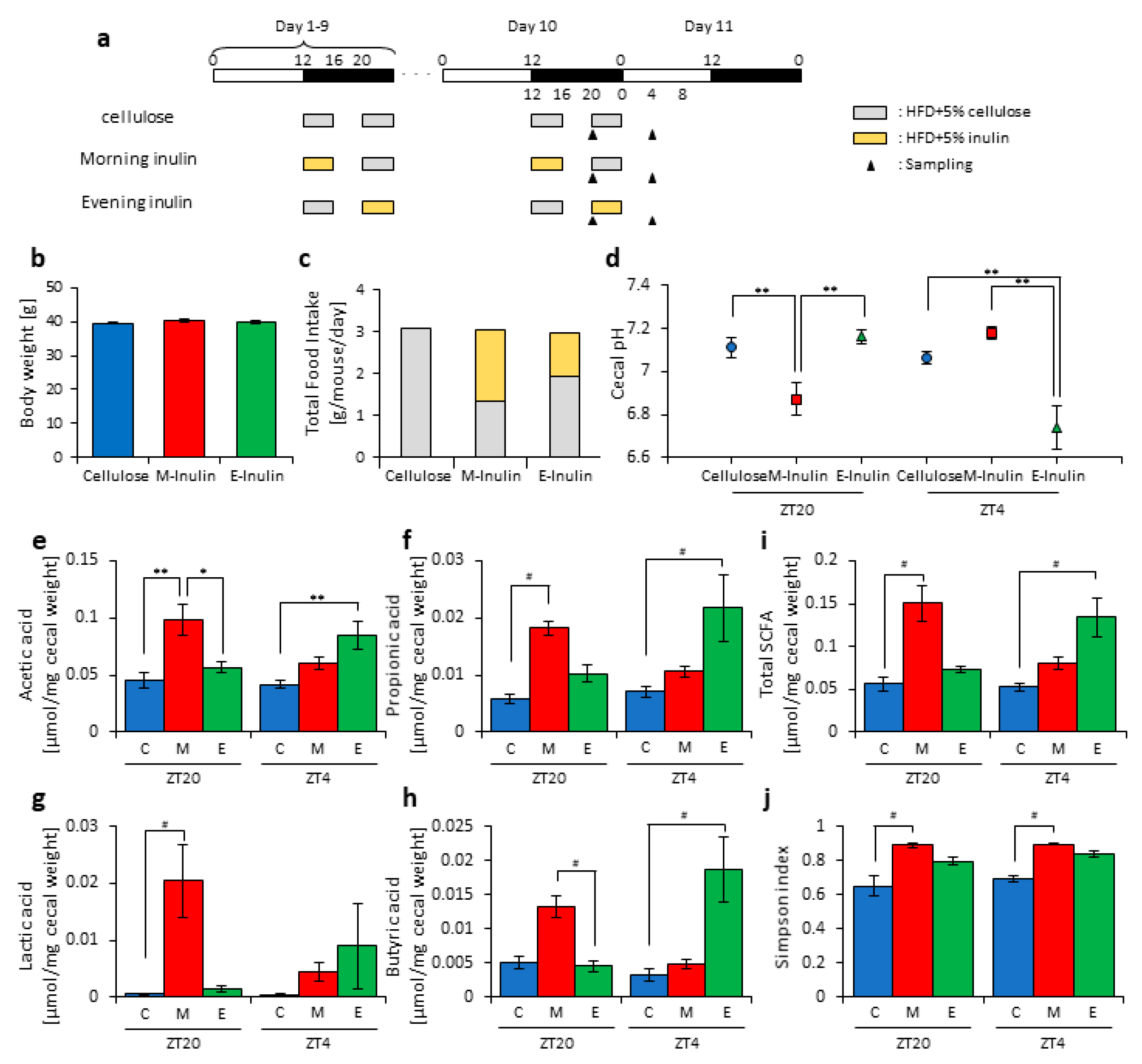
2.2. Scheduled Feeding
2.3. Cecal pH Measurement
2.4. Measurement of SCFAs
2.5. Fecal DNA Extraction
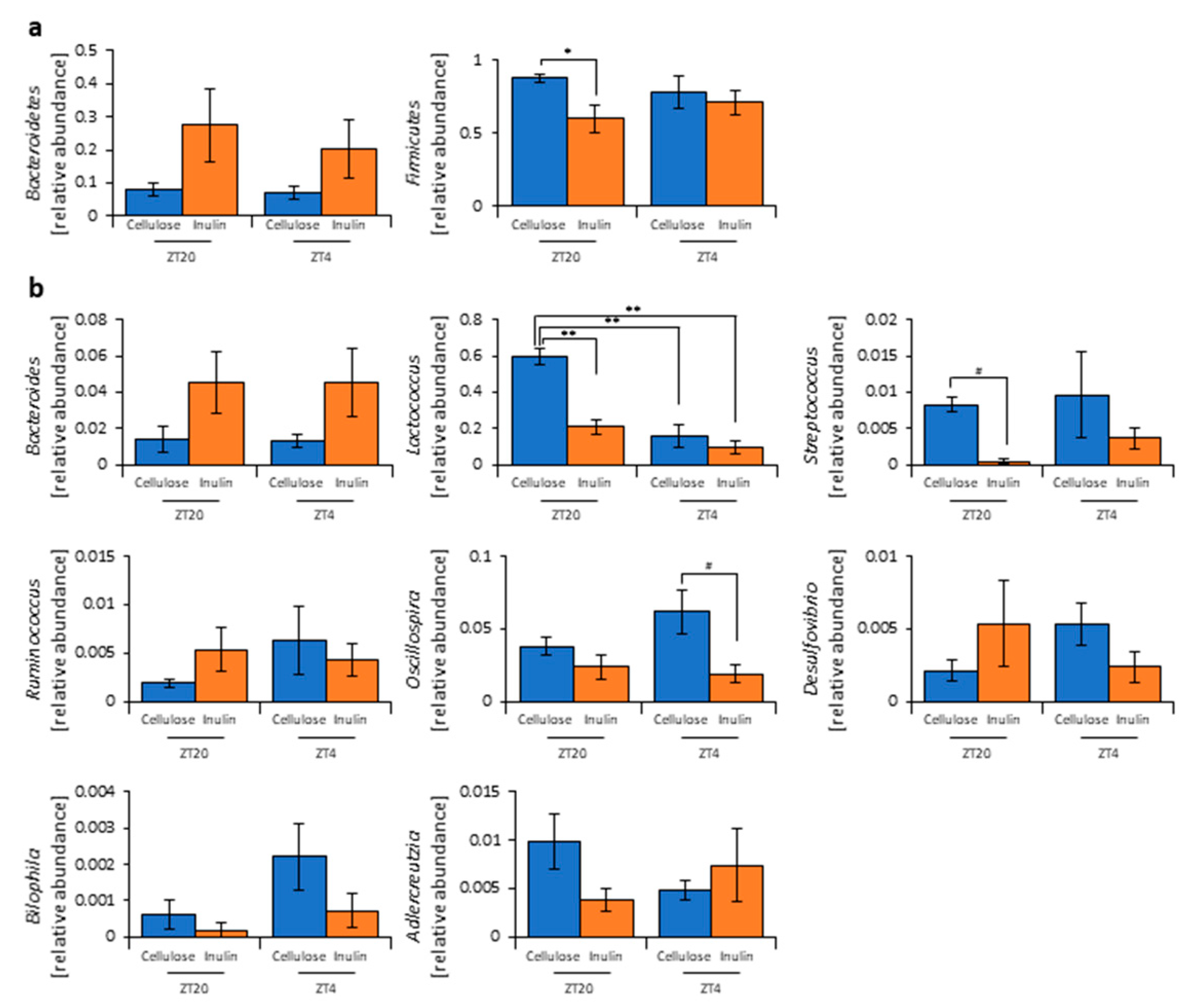
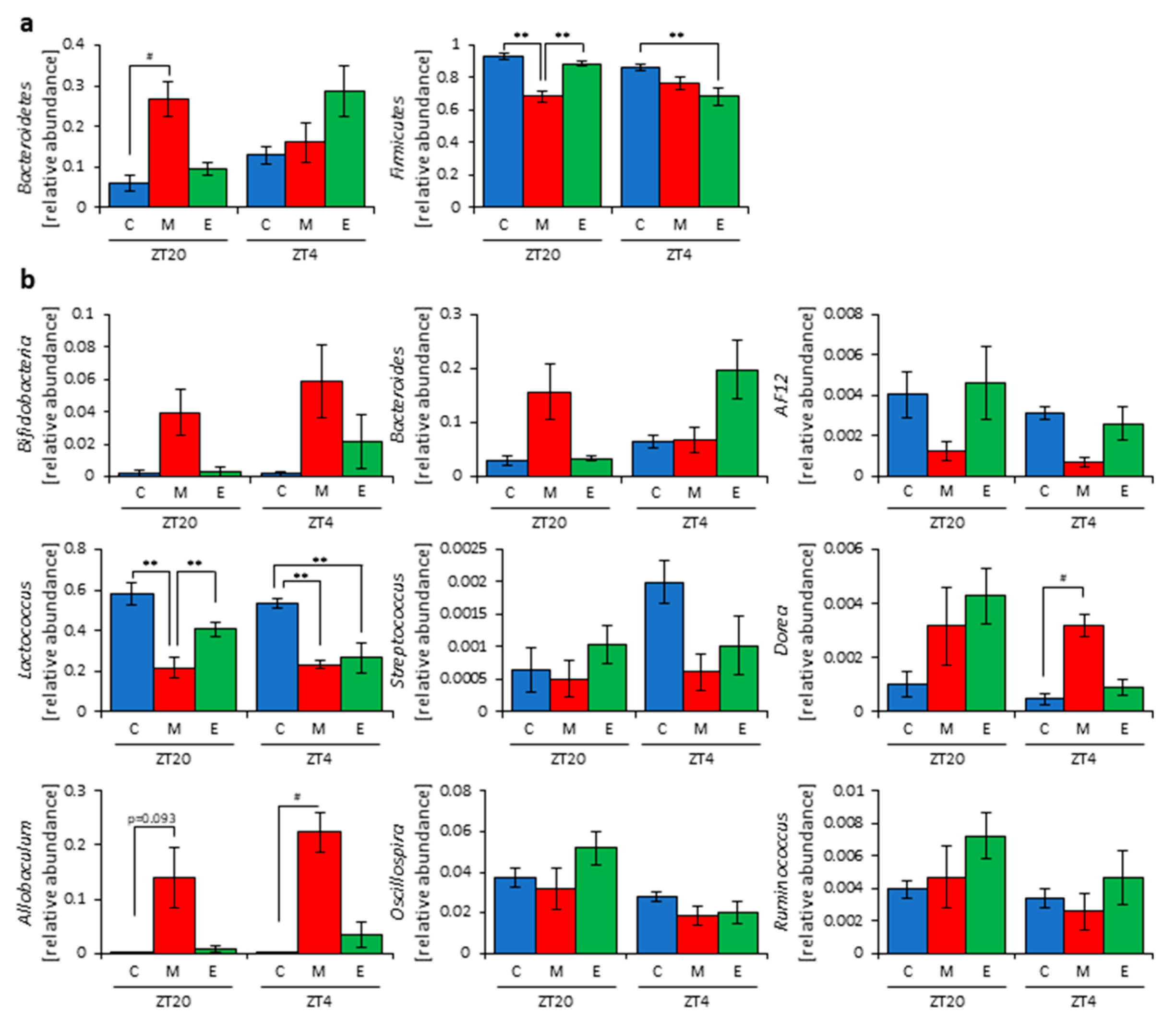
2.6. 16 S rDNA Gene Sequencing
- forward primer = 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′;
- reverse primer = 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′.
2.7. Analysis of 16S rDNA Gene Sequences
2.8. Predicted Metagenomes
2.9. Statistical Analysis
3. Results
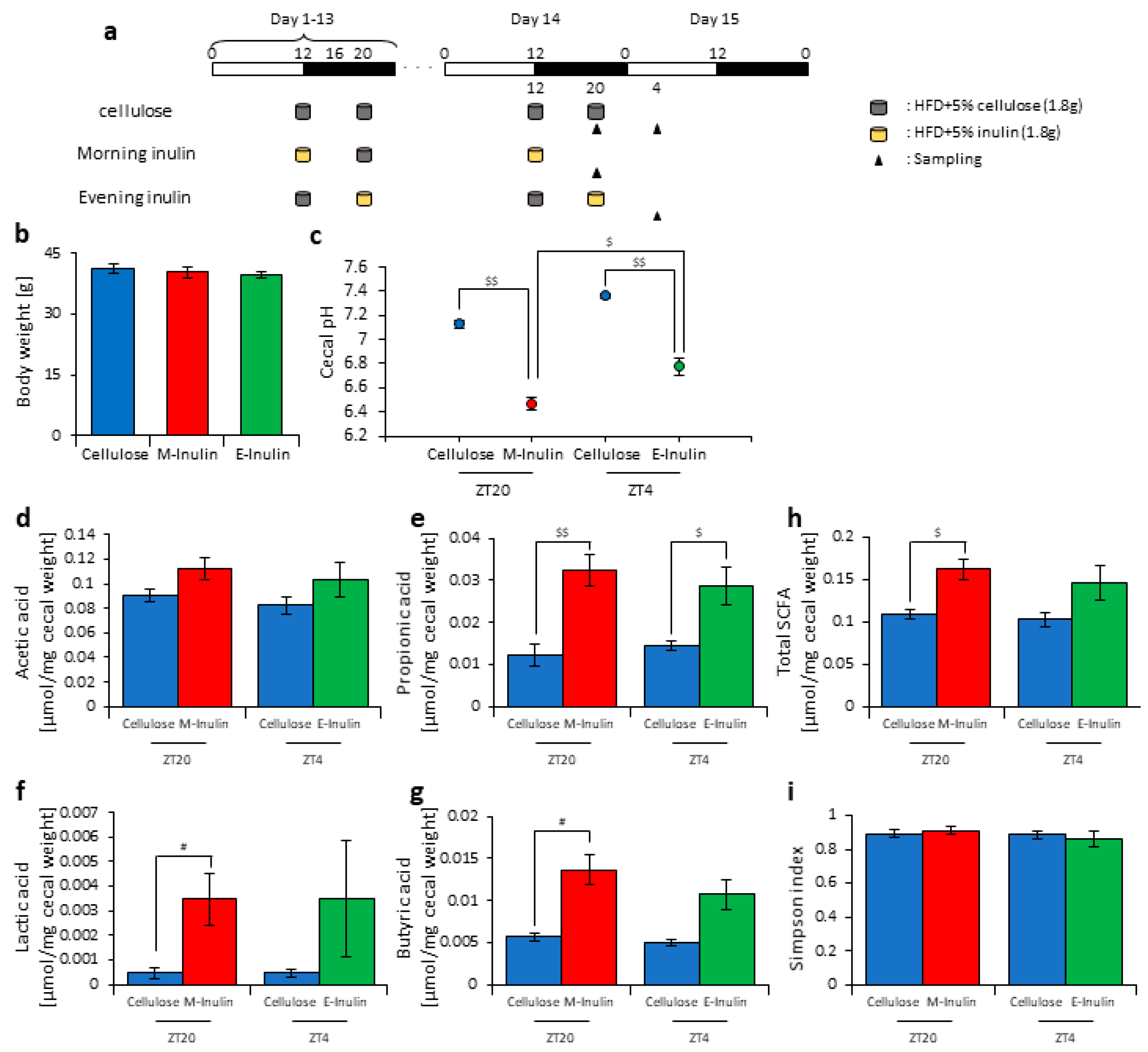
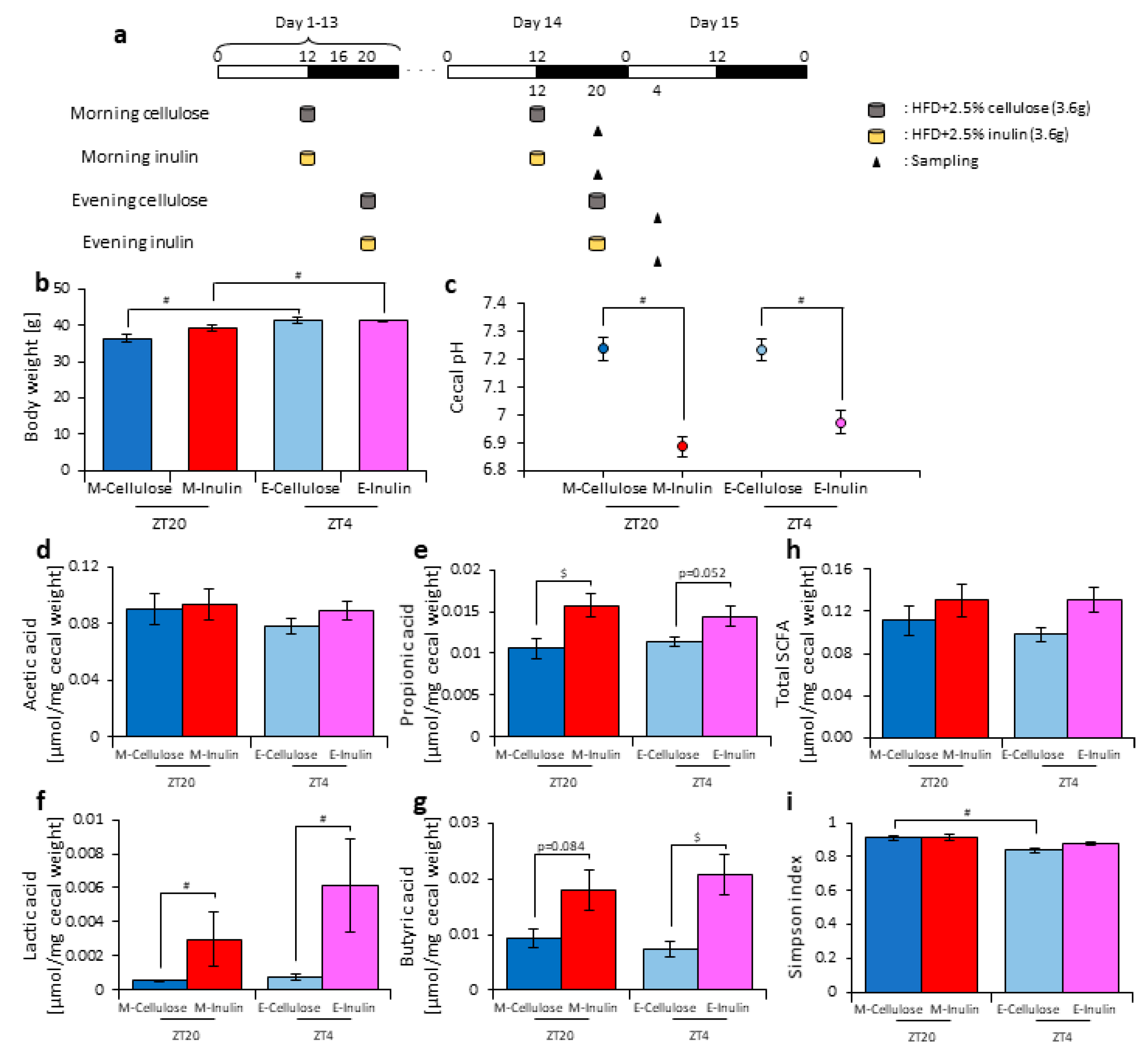
3.1. Inulin Intake Changed Microbiota Composition under Both Morning and Evening Timings
3.2. Inulin Intake in the Morning Rather than the Evening Strongly Affected the Microbiota Composition under Time-Restricted Feeding Conditions
3.3. Inulin Feeding in the Morning Affected the Microbiota Composition More than that in the Evening under Restricted Food Amount Conditions
3.4. A Relationship Was Observed between the Length of Fasting Time and Inulin Feeding Stimulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and its discontents. mBio 2017, 8, e01492–e01517. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Ahmadi Amoli, H.; Ghasemian-Safaei, H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol. Hepatol. Bed Bench 2018, 11, 101–109. [Google Scholar]
- Mai, V.; Morris, J.G., Jr. Need for prospective cohort studies to establish human gut microbiome contributions to disease risk. J. Natl. Cancer Inst. 2013, 105, 1850–1851. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef]
- Okada, T.; Fukuda, S.; Hase, K.; Nishiumi, S.; Izumi, Y.; Yoshida, M.; Hagiwara, T.; Kawashima, R.; Yamazaki, M.; Oshio, T.; et al. Microbiota-derived lactate accelerates colon epithelial cell turnover in starvation-refed mice. Nat. Commun. 2013, 4, 1654. [Google Scholar] [CrossRef]
- Kimura, I.; Ozawa, K.; Inoue, D.; Imamura, T.; Kimura, K.; Maeda, T.; Terasawa, K.; Kashihara, D.; Hirano, K.; Tani, T.; et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor gpr43. Nat. Commun. 2013, 4, 1829. [Google Scholar] [CrossRef] [PubMed]
- Bass, J.; Takahashi, J.S. Circadian integration of metabolism and energetics. Science 2010, 330, 1349–1354. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Tahara, Y.; Hirao, A. The adjustment and manipulation of biological rhythms by light, nutrition, and abused drugs. Adv. Drug Deliv. Rev. 2010, 62, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Aoyama, S.; Shibata, S. The mammalian circadian clock and its entrainment by stress and exercise. J. Physiol. Sci. JPS 2017, 67, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Tahara, Y.; Shibata, S. Entrainment of the mouse circadian clock: Effects of stress, exercise, and nutrition. Free Radic. Biol. Med. 2018, 119, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Hattori, Y.; Ikeda, Y.; Kamagata, M.; Iwami, S.; Yasuda, S.; Tahara, Y.; Shibata, S. Forced rather than voluntary exercise entrains peripheral clocks via a corticosterone/noradrenaline increase in per2::Luc mice. Sci. Rep. 2016, 6, 27607. [Google Scholar] [CrossRef]
- Leone, V.; Gibbons, S.M.; Martinez, K.; Hutchison, A.L.; Huang, E.Y.; Cham, C.M.; Pierre, J.F.; Heneghan, A.F.; Nadimpalli, A.; Hubert, N.; et al. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015, 17, 681–689. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef]
- Tahara, Y.; Yamazaki, M.; Sukigara, H.; Motohashi, H.; Sasaki, H.; Miyakawa, H.; Haraguchi, A.; Ikeda, Y.; Fukuda, S.; Shibata, S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci. Rep. 2018, 8, 1395. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Materna, A.C.; Friedman, J.; Campos-Baptista, M.I.; Blackburn, M.C.; Perrotta, A.; Erdman, S.E.; Alm, E.J. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014, 15, R89. [Google Scholar] [CrossRef] [PubMed]
- Gibson, G.R.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: Introducing the concept of prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Valcheva, R.; Dieleman, L.A. Prebiotics: Definition and protective mechanisms. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Hoving, L.R.; Katiraei, S.; Pronk, A.; Heijink, M.; Vonk, K.K.D.; Amghar-El Bouazzaoui, F.; Vermeulen, R.; Drinkwaard, L.; Giera, M.; van Harmelen, V.; et al. The prebiotic inulin modulates gut microbiota but does not ameliorate atherosclerosis in hypercholesterolemic apoe*3-leiden.Cetp mice. Sci. Rep. 2018, 8, 16515. [Google Scholar] [CrossRef] [PubMed]
- Demigne, C.; Jacobs, H.; Moundras, C.; Davicco, M.J.; Horcajada, M.N.; Bernalier, A.; Coxam, V. Comparison of native or reformulated chicory fructans, or non-purified chicory, on rat cecal fermentation and mineral metabolism. Eur. J. Nutr. 2008, 47, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef]
- Wang, X.; Xue, J.; Yang, J.; Xie, M. Timed high-fat diet in the evening affects the hepatic circadian clock and pparalpha-mediated lipogenic gene expressions in mice. Genes Nutr. 2013, 8, 457–463. [Google Scholar] [CrossRef]
- Arble, D.M.; Bass, J.; Laposky, A.D.; Vitaterna, M.H.; Turek, F.W. Circadian timing of food intake contributes to weight gain. Obesity 2009, 17, 2100–2102. [Google Scholar] [CrossRef]
- Haraguchi, A.; Aoki, N.; Ohtsu, T.; Ikeda, Y.; Tahara, Y.; Shibata, S. Controlling access time to a high-fat diet during the inactive period protects against obesity in mice. Chronobiol. Int. 2014, 31, 935–944. [Google Scholar] [CrossRef]
- Hsieh, S.D.; Muto, T.; Murase, T.; Tsuji, H.; Arase, Y. Association of short sleep duration with obesity, diabetes, fatty liver and behavioral factors in japanese men. Intern. Med. 2011, 50, 2499–2502. [Google Scholar] [CrossRef] [PubMed]
- Baron, K.G.; Reid, K.J.; Kern, A.S.; Zee, P.C. Role of sleep timing in caloric intake and bmi. Obesity 2011, 19, 1374–1381. [Google Scholar] [CrossRef] [PubMed]
- Oishi, K.; Konishi, T.; Hashimoto, C.; Yamamoto, S.; Takahashi, Y.; Shiina, Y. Dietary fish oil differentially ameliorates high-fructose diet-induced hepatic steatosis and hyperlipidemia in mice depending on time of feeding. J. Nutr. Biochem. 2018, 52, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Ohguchi, M.; Iwai, Y. A novel enzyme of bacillus sp. 217c-11 that produces inulin from sucrose. Biosci. Biotechnol. Biochem. 2003, 67, 1327–1334. [Google Scholar] [CrossRef] [Green Version]
- Wada, T.; Sugatani, J.; Terada, E.; Ohguchi, M.; Miwa, M. Physicochemical characterization and biological effects of inulin enzymatically synthesized from sucrose. J. Agric. Food Chem. 2005, 53, 1246–1253. [Google Scholar] [CrossRef]
- Kowalski, T.J.; Farley, C.; Cohen-Williams, M.E.; Varty, G.; Spar, B.D. Melanin-concentrating hormone-1 receptor antagonism decreases feeding by reducing meal size. Eur. J. Pharmacol. 2004, 497, 41–47. [Google Scholar] [CrossRef]
- Oh, J.; Lee, S.R.; Hwang, K.T.; Ji, G.E. The anti-obesity effects of the dietary combination of fermented red ginseng with levan in high fat diet mouse model. Phytother. Res. PTR 2014, 28, 617–622. [Google Scholar] [CrossRef]
- Weitkunat, K.; Schumann, S.; Petzke, K.J.; Blaut, M.; Loh, G.; Klaus, S. Effects of dietary inulin on bacterial growth, short-chain fatty acid production and hepatic lipid metabolism in gnotobiotic mice. J. Nutr. Biochem. 2015, 26, 929–937. [Google Scholar] [CrossRef]
- Ponziani, F.R.; Pecere, S.; Gasbarrini, A.; Ojetti, V. Physiology and pathophysiology of liver lipid metabolism. Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1055–1067. [Google Scholar] [CrossRef]
- Federico, A.; Dallio, M.; Di Sarno, R.; Giorgio, V.; Miele, L. Gut microbiota, obesity and metabolic disorders. Minerva Gastroenterol. E Dietol. 2017, 63, 337–344. [Google Scholar]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Giustina, A.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Huazano, A.; López, M.G. Metabolism of short chain fatty acids in colon and faeces of mice after a supplementation of diets with agave fructans. Lipid Metab. 2013, 8, 163–182. [Google Scholar]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy japanese and its microbial and functional uniqueness. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2016, 23, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than blast. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Gibson, G.R.; Beatty, E.R.; Wang, X.; Cummings, J.H. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 1995, 108, 975–982. [Google Scholar] [CrossRef]
- Zhu, L.; Qin, S.; Zhai, S.; Gao, Y.; Li, L. Inulin with different degrees of polymerization modulates composition of intestinal microbiota in mice. FEMS Microbiol. Lett. 2017, 364, 364. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, J.; Henning, S.M.; Lee, R.; Hsu, M.; Grojean, E.; Pisegna, R.; Ly, A.; Heber, D.; Li, Z. Dietary pomegranate extract and inulin affect gut microbiome differentially in mice fed an obesogenic diet. Anaerobe 2017, 48, 184–193. [Google Scholar] [CrossRef]
- Brighenti, F.; Casiraghi, M.C.; Canzi, E.; Ferrari, A. Effect of consumption of a ready-to-eat breakfast cereal containing inulin on the intestinal milieu and blood lipids in healthy male volunteers. Eur. J. Clin. Nutr. 1999, 53, 726–733. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, M.; Ozaki, M.; Kang, M.I.; Sasaki, H.; Fukazawa, M.; Iwakami, T.; Lim, P.J.; Kim, H.K.; Aoyama, S.; Shibata, S. Effects of meal timing on postprandial glucose metabolism and blood metabolites in healthy adults. Nutrients 2018, 10, 1763. [Google Scholar] [CrossRef] [Green Version]
- Kuroda, H.; Tahara, Y.; Saito, K.; Ohnishi, N.; Kubo, Y.; Seo, Y.; Otsuka, M.; Fuse, Y.; Ohura, Y.; Hirao, A.; et al. Meal frequency patterns determine the phase of mouse peripheral circadian clocks. Sci. Rep. 2012, 2, 711. [Google Scholar] [CrossRef] [Green Version]
- Hirao, A.; Nagahama, H.; Tsuboi, T.; Hirao, M.; Tahara, Y.; Shibata, S. Combination of starvation interval and food volume determines the phase of liver circadian rhythm in per2::Luc knock-in mice under two meals per day feeding. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G1045–G1053. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, Y.; Sasaki, H.; Ohtsu, T.; Shiraishi, T.; Tahara, Y.; Shibata, S. Feeding and adrenal entrainment stimuli are both necessary for normal circadian oscillation of peripheral clocks in mice housed under different photoperiods. Chronobiol. Int. 2015, 32, 195–210. [Google Scholar] [CrossRef]
- Sasaki, H.; Hattori, Y.; Ikeda, Y.; Kamagata, M.; Iwami, S.; Yasuda, S.; Shibata, S. Phase shifts in circadian peripheral clocks caused by exercise are dependent on the feeding schedule in per2::Luc mice. Chronobiol. Int. 2016, 33, 849–862. [Google Scholar] [CrossRef]
- Sasaki, H.; Ohtsu, T.; Ikeda, Y.; Tsubosaka, M.; Shibata, S. Combination of meal and exercise timing with a high-fat diet influences energy expenditure and obesity in mice. Chronobiol. Int. 2014, 31, 959–975. [Google Scholar] [CrossRef]
- Kilic, A.O.; Pavlova, S.I.; Ma, W.G.; Tao, L. Analysis of lactobacillus phages and bacteriocins in american dairy products and characterization of a phage isolated from yogurt. Appl. Environ. Microbiol. 1996, 62, 2111–2116. [Google Scholar]
- Gilliland, S.E. Health and nutritional benefits from lactic acid bacteria. FEMS Microbiol. Rev. 1990, 7, 175–188. [Google Scholar] [CrossRef]
- Koh, A.; Molinaro, A.; Stahlman, M.; Khan, M.T.; Schmidt, C.; Manneras-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially produced imidazole propionate impairs insulin signaling through mtorc1. Cell 2018, 175, 947–961.e917. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The human gut microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef] [Green Version]
- Bervoets, L.; Van Hoorenbeeck, K.; Kortleven, I.; Van Noten, C.; Hens, N.; Vael, C.; Goossens, H.; Desager, K.N.; Vankerckhoven, V. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Delzenne, N.M.; Cani, P.D.; Neyrinck, A.M. Modulation of glucagon-like peptide 1 and energy metabolism by inulin and oligofructose: Experimental data. J. Nutr. 2007, 137, 2547s–2551s. [Google Scholar] [CrossRef] [Green Version]
- Russo, F.; Chimienti, G.; Riezzo, G.; Pepe, G.; Petrosillo, G.; Chiloiro, M.; Marconi, E. Inulin-enriched pasta affects lipid profile and lp(a) concentrations in italian young healthy male volunteers. Eur. J. Nutr. 2008, 47, 453–459. [Google Scholar] [CrossRef]
- Letexier, D.; Diraison, F.; Beylot, M. Addition of inulin to a moderately high-carbohydrate diet reduces hepatic lipogenesis and plasma triacylglycerol concentrations in humans. Am. J. Clin. Nutr. 2003, 77, 559–564. [Google Scholar] [CrossRef] [Green Version]
- Shirouchi, B.; Kawamura, S.; Matsuoka, R.; Baba, S.; Nagata, K.; Shiratake, S.; Tomoyori, H.; Imaizumi, K.; Sato, M. Dietary guar gum reduces lymph flow and diminishes lipid transport in thoracic duct-cannulated rats. Lipids 2011, 46, 789–793. [Google Scholar] [CrossRef]
- Eastwood, M.A.; Morris, E.R. Physical properties of dietary fiber that influence physiological function: A model for polymers along the gastrointestinal tract. Am. J. Clin. Nutr. 1992, 55, 436–442. [Google Scholar] [CrossRef]
| a. Phylum Level | ||||||
| Bacterial | ZT20 | ZT4 | ||||
| Cellulose | M-Inulin | p-Value | Cellulose | E-Inulin | p-Value | |
| Actinobacteria | 0.0072 ± 0.0039 | 0.0103 ± 0.0081 | 0.7143 | 0.0690 ± 0.0647 | 0.0907 ± 0.0440 | 0.8254 |
| Bacteroidetes | 0.1535 ± 0.0463 | 0.2499 ± 0.0654 | 0.669 | 0.1647 ± 0.0467 | 0.3644 ± 0.1203 | 0.2085 |
| Deferribacteres | 0.0011 ± 0.0010 | 0.0005 ± 0.0004 | 0.9999 | 0.0002 ± 0.0002 | 0.0004 ± 0.0003 | 0.8413 |
| Firmicutes | 0.8221 ± 0.0467 | 0.6863 ± 0.0713 | 0.3232 | 0.7486 ± 0.0228 | 0.5294 ± 0.0865 | 0.0743 |
| Proteobacteria | 0.0111 ± 0.0033 | 0.0019 ± 0.0004 | 0.0159 # | 0.0152 ± 0.0030 | 0.0054 ± 0.0017 | 0.1905 |
| TM7 | 0.0043 ± 0.0025 | 0.0004 ± 0.0003 | 0.0159 # | 0.0017 ± 0.0002 | 0.0001 ± 0.0001 | 0.0159 # |
| Verrucomicrobia | 0.0006 ± 0.0003 | 0.0473 ± 0.0219 | 0.1905 | 0.0003 ± 0.003 | 0.0132 ± 0.0117 | 0.1032 |
| b. Genus Level | ||||||
| Bacterial | ZT20 | ZT4 | ||||
| Cellulose | M-Inulin | p-Value | Cellulose | E-Inulin | p-Value | |
| Bifidobacterium | 0.0022 ± 0.0018 | 0.0080 ± 0.0077 | 0.5556 | 0.0063 ± 0.0062 | 0.0088 ± 0.0044 | 0.6825 |
| Adlercreutzia | 0.0046 ± 0.0019 | 0.0022 ± 0.0004 | 0.2344 | 0.0047 ± 0.0014 | 0.0026 ± 0.0006 | 0.6428 |
| Bacteroides | 0.0463 ± 0.0175 | 0.1100 ± 0.0262 | 0.0993 | 0.0701 ± 0.0238 | 0.1953 ± 0.0627 | 0.1352 |
| Parabacteroides | 0.0010 ± 0.0003 | 0.0009 ± 0.0002 | 0.8247 | 0.0015 ± 0.0006 | 0.0010 ± 0.0003 | 0.6229 |
| AF12 | 0.0075 ± 0.0023 | 0.0011 ± 0.0006 | 0.0435 # | 0.0052 ± 0.0013 | 0.0009 ± 0.0002 | 0.0317 # |
| Butyricimonas | 0.0004 ± 0.0001 | 0.0008 ± 0.0004 | 0.0317 # | 0.0005 ± 0.0003 | 0.0002 ± 0.0001 | 0.7937 |
| Odoribacter | 0.0017 ± 0.0003 | 0.0006 ± 0.0002 | 0.1795 | 0.0020 ± 0.0008 | 0.0003 ± 0.0001 | 0.0308 $ |
| [Prevotella] | 0.0144 ± 0.0085 | 0.0303 ± 0.0098 | 0.2857 | 0.0118 ± 0.0064 | 0.0375 ± 0.0279 | 0.9762 |
| Staphylococcus | 0.0013 ± 0.0006 | 0.0002 ± 0.0001 | 0.0397 # | 0.0009 ± 0.0006 | 0.0005 ± 0.0002 | 0.8254 |
| Lactobacillus | 0.0294 ± 0.0230 | 0.0136 ± 0.0036 | 0.6825 | 0.1152 ± 0.0811 | 0.0631 ± 0.0323 | 0.873 |
| Lactococcus | 0.2862 ± 0.0453 | 0.0957 ± 0.0128 | 0.0159 # | 0.1691 ± 0.0446 | 0.0856 ± 0.0136 | 0.2857 |
| Streptococcus | 0.0034 ± 0.0016 | 0.0011 ± 0.0002 | 0.1545 | 0.0037 ± 0.0016 | 0.0019 ± 0.0010 | 0.357 |
| Clostridium | 0.0002 ± 0.0001 | 0.0001 ± 0.00005 | 0.3889 | 0.0014 ± 0.0012 | 0.0003 ± 0.0002 | 0.3889 |
| Dehalobacterium | 0.0018 ± 0.0002 | 0.0017 ± 0.0006 | 0.873 | 0.0010 ± 0.0001 | 0.0010 ± 0.0007 | 0.1746 |
| Coprococcus | 0.0049 ± 0.0009 | 0.0081 ± 0.0032 | 0.9999 | 0.0035 ± 0.0007 | 0.0019 ± 0.0005 | 0.1905 |
| Dorea | 0.0032 ± 0.0011 | 0.0032 ± 0.0012 | 0.9966 | 0.0041 ± 0.0031 | 0.0020 ± 0.0012 | 0.5089 |
| Roseburia | 0.0018 ± 0.0010 | 0.0053 ± 0.0038 | 0.9762 | 0.0005 ± 0.0004 | 0.0004 ± 0.0001 | 0.5635 |
| [Ruminococcus] | 0.0305 ± 0.0113 | 0.0384 ± 0.0105 | 0.6271 | 0.0327 ± 0.0129 | 0.0240 ± 0.0094 | 0.5957 |
| Oscillospira | 0.0803 ± 0.0131 | 0.0290 ± 0.0088 | 0.0148 $ | 0.0600 ± 0.0173 | 0.0172 ± 0.0074 | 0.0404 $ |
| Ruminococcus | 0.0096 ± 0.0019 | 0.0067 ± 0.0032 | 0.4961 | 0.0056 ± 0.0017 | 0.0025 ± 0.0010 | 0.161 |
| Allobaculum | 0.0017 ± 0.0007 | 0.1764 ± 0.0544 | 0.1905 | 0.0662 ± 0.0649 | 0.1669 ± 0.1094 | 0.5238 |
| Bilophila | 0.0013 ± 0.0003 | 0.0003 ± 0.0002 | 0.0159 # | 0.0011 ± 0.0005 | 0.0002 ± 0.0001 | 0.1111 |
| Desulfovibrio | 0.0037 ± 0.0011 | 0.0006 ± 0.0004 | 0.047 # | 0.0021 ± 0.0007 | 0.0019 ± 0.0002 | 0.371 |
| Akkermansia | 0.0006 ± 0.0002 | 0.0473 ± 0.0219 | 0.1905 | 0.0003 ± 0.0003 | 0.0132 ± 0.0117 | 0.1032 |
| a. Phylum Level | ||||||
| Bacterial | ZT20 | ZT4 | ||||
| Cellulose | M-Inulin | p-Value | Cellulose | E-Inulin | p-Value | |
| Actinobacteria | 0.0041 ± 0.0017 | 0.0364 ± 0.0199 | 0.0174 # | 0.0321 ± 0.0154 | 0.0871 ± 0.0380 | 0.1797 |
| Bacteroidetes | 0.0759 ± 0.0296 | 0.1372 ± 0.0337 | 0.3748 | 0.1024 ± 0.0275 | 0.1609 ± 0.0411 | 0.4069 |
| Deferribacteres | 0.0037 ± 0.0016 | 0.0018 ± 0.0006 | 0.5714 | 0.0007 ± 0.0004 | 0.0002 ± 0.00006 | 0.4459 |
| Firmicutes | 0.80427 ± 0.0240 | 0.7513 ± 0.0306 | 0.1797 | 0.7832 ± 0.0207 | 0.6906 ± 0.0373 | 0.1298 |
| Proteobacteria | 0.1119 ± 0.0161 | 0.0706 ± 0.0120 | 0.0799 | 0.0790 ± 0.0141 | 0.0568 ± 0.0102 | 0.4449 |
| Verrucomicrobia | 0.0001 ± 0.0001 | 0.0025 ± 0.0011 | 0.0606 | 0.0025 ± 0.0022 | 0.0042 ± 0.0036 | 0.5455 |
| b. Genus Level | ||||||
| Bacterial | ZT20 | ZT4 | ||||
| Cellulose | M-Inulin | p-Value | Cellulose | E-Inulin | p-Value | |
| Bifidobacterium | 0.0004 ± 0.0003 | 0.0305 ± 0.0192 | 0.0043 ## | 0.0266 ± 0.0151 | 0.0785 ± 0.0367 | 0.1775 |
| Adlercreutzia | 0.0036 ± 0.0017 | 0.0037 ± 0.0010 | 0.5714 | 0.0051 ± 0.0008 | 0.0041 ± 0.0008 | 0.3874 |
| Bacteroides | 0.0222 ± 0.0103 | 0.0327 ± 0.0078 | 0.4395 | 0.0149 ± 0.0051 | 0.0313 ± 0.0101 | 0.1801 |
| Parabacteroides | 0.0059 ± 0.0027 | 0.0032 ± 0.0010 | 0.8983 | 0.0048 ± 0.0012 | 0.0025 ± 0.0005 | 0.3874 |
| Butyricimonas | 0.0002 ± 0.00008 | 0.0001 ± 0.00004 | 0.6623 | 0.0003 ± 0.0001 | 0.0002 ± 0.0001 | 0.6591 |
| Odoribacter | 0.0015 ± 0.0003 | 0.0022 ± 0.0012 | 0.8983 | 0.0023 ± 0.0004 | 0.0031 ± 0.0007 | 0.3874 |
| [Prevotella] | 0.0066 ± 0.0031 | 0.0151 ± 0.0102 | 0.5541 | 0.0033 ± 0.0018 | 0.0056 ± 0.0045 | 0.9805 |
| Mucispirillum | 0.0037 ± 0.0016 | 0.0018 ± 0.0006 | 0.5714 | 0.0007 ± 0.0004 | 0.0001 ± 0.00006 | 0.4459 |
| Staphylococcus | 0.0005 ± 0.0002 | 0.00005 ± 0.00003 | 0.145 | 0.0009 ± 0.0004 | 0.00007 ± 0.00002 | 0.0022 ## |
| Lactobacillus | 0.0080 ± 0.0031 | 0.0088 ± 0.0027 | 0.8983 | 0.1050 ± 0.0530 | 0.01225 ± 0.0046 | 0.1797 |
| Lactococcus | 0.2490 ± 0.0332 | 0.1468 ± 0.0205 | 0.094 | 0.3408 ± 0.0271 | 0.1824 ± 0.0494 | 0.0078 $$ |
| Streptococcus | 0.0048 ± 0.0011 | 0.0013 ± 0.0002 | 0.0087 ## | 0.0037 ± 0.0005 | 0.0023 ± 0.0006 | 0.0931 |
| SMB53 | 0.0102 ± 0.0079 | 0.0130 ± 0.0078 | 0.9394 | 0.0467 ± 0.0224 | 0.0132 ± 0.0076 | 0.1688 |
| Dehalobacterium | 0.0017 ± 0.0004 | 0.0024 ± 0.0006 | 0.5628 | 0.0007 ± 0.0002 | 0.0021 ± 0.0008 | 0.132 |
| Blautia | 0.0004 ± 0.0001 | 0.0003 ± 0.00008 | 0.7381 | 0.0002 ± 0.0001 | 0.0003 ± 0.0001 | 0.1991 |
| Coprococcus | 0.0093 ± 0.0009 | 0.0116 ± 0.0026 | 0.7879 | 0.0028 ± 0.0005 | 0.0048 ± 0.0016 | 0.3874 |
| Dorea | 0.0017 ± 0.0004 | 0.0034 ± 0.0007 | 0.077 | 0.0011 ± 0.0003 | 0.0042 ± 0.0006 | 0.0016 $$ |
| Roseburia | 0.00006 ± 0.00002 | 0.00005 ± 0.00003 | 0.9242 | 0.00007 ± 0.00002 | 0.0001 ± 0.00007 | 0.3398 |
| [Ruminococcus] | 0.0652 ± 0.0085 | 0.0509 ± 0.0082 | 0.3544 | 0.0299 ± 0.0059 | 0.0347 ± 0.0073 | 0.882 |
| Anaerotruncus | 0.0004 ± 0.0001 | 0.0001 ± 0.00005 | 0.1797 | 0.0001 ± 0.00004 | 0.0003 ± 0.0001 | 0.8312 |
| Oscillospira | 0.0473 ± 0.0076 | 0.0213 ± 0.0052 | 0.0043 $$ | 0.0217 ± 0.0042 | 0.0158 ± 0.0022 | 0.6797 |
| Ruminococcus | 0.0082 ± 0.0009 | 0.0034 ± 0.0007 | 0.0012 $$ | 0.0035 ± 0.0008 | 0.0020 ± 0.0003 | 0.7285 |
| Allobaculum | 0.0011 ± 0.0006 | 0.1122 ± 0.0562 | 0.0022 ## | 0.0327 ± 0.0157 | 0.1703 ± 0.0513 | 0.0449 # |
| Catenibacterium | 0.0005 ± 0.0001 | 0.0003 ± 0.0001 | 0.3874 | 0.0004 ± 0.00009 | 0.0003 ± 0.0001 | 0.474 |
| Desulfovibrio | 0.0010 ± 0.0005 | 0.0011 ± 0.0007 | 0.9073 | 0.0006 ± 0.0002 | 0.0023 ± 0.0009 | 0.1001 |
| Citrobacter | 0.0030 ± 0.0011 | 0.0026 ± 0.0004 | 0.7879 | 0.0026 ± 0.0004 | 0.0015 ± 0.0003 | 0.0931 |
| Klebsiella | 0.0417 ± 0.0147 | 0.0328 ± 0.0138 | 0.5714 | 0.0412 ± 0.0105 | 0.0291 ± 0.0124 | 0.1797 |
| Akkermansia | 0.0001 ± 0.0001 | 0.0025 ± 0.0011 | 0.4545 | 0.0024 ± 0.0022 | 0.0041 ± 0.0036 | 0.5455 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasaki, H.; Miyakawa, H.; Watanabe, A.; Nakayama, Y.; Lyu, Y.; Hama, K.; Shibata, S. Mice Microbiota Composition Changes by Inulin Feeding with a Long Fasting Period under a Two-Meals-Per-Day Schedule. Nutrients 2019, 11, 2802. https://doi.org/10.3390/nu11112802
Sasaki H, Miyakawa H, Watanabe A, Nakayama Y, Lyu Y, Hama K, Shibata S. Mice Microbiota Composition Changes by Inulin Feeding with a Long Fasting Period under a Two-Meals-Per-Day Schedule. Nutrients. 2019; 11(11):2802. https://doi.org/10.3390/nu11112802
Chicago/Turabian StyleSasaki, Hiroyuki, Hiroki Miyakawa, Aya Watanabe, Yuki Nakayama, Yijin Lyu, Koki Hama, and Shigenobu Shibata. 2019. "Mice Microbiota Composition Changes by Inulin Feeding with a Long Fasting Period under a Two-Meals-Per-Day Schedule" Nutrients 11, no. 11: 2802. https://doi.org/10.3390/nu11112802




