Impact of Donor Human Milk in the Preterm Very Low Birth Weight Gut Transcriptome Profile by Use of Exfoliated Intestinal Cells
Abstract
:1. Introduction
2. Material and Methods
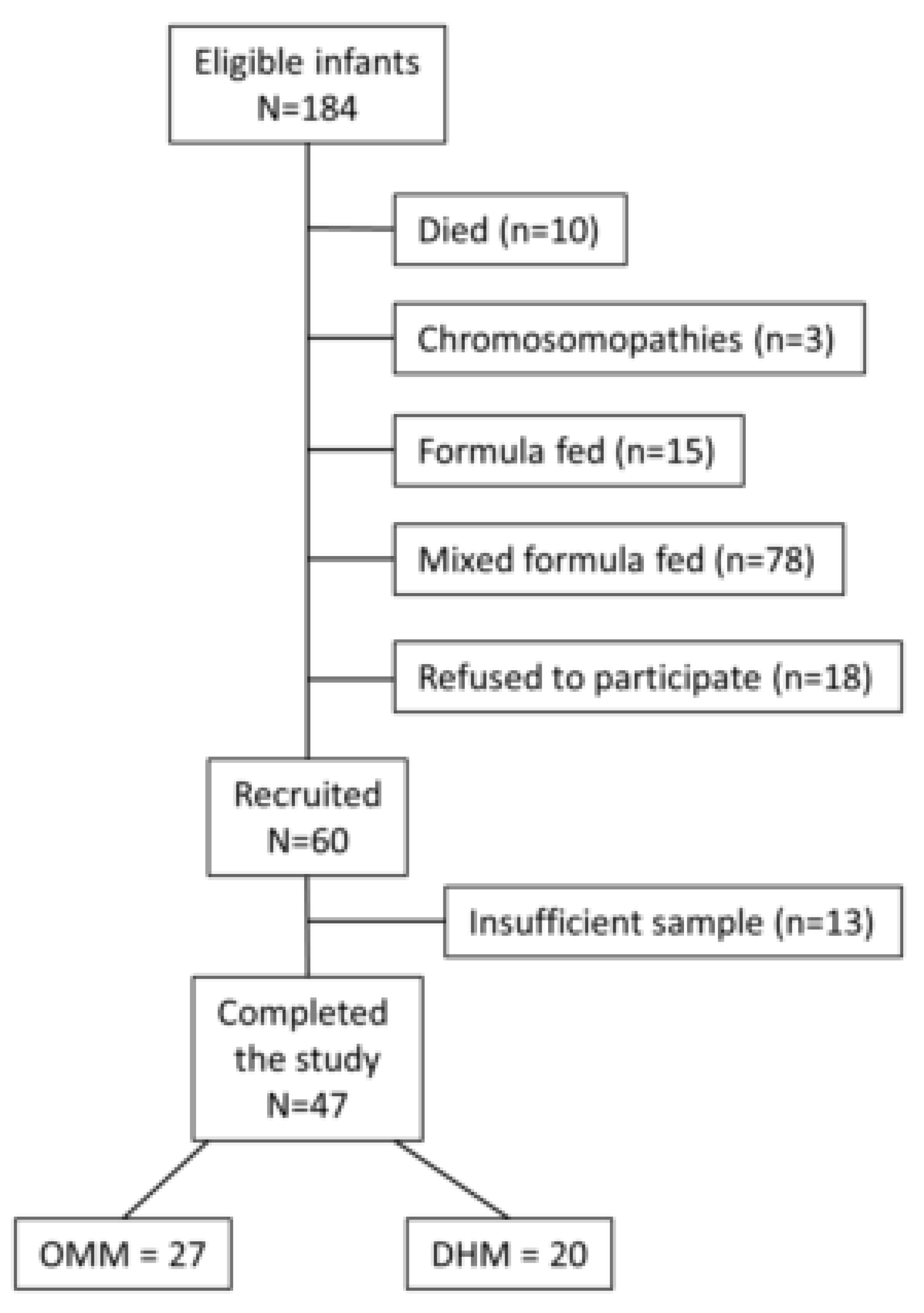
2.1. Study Design and Patients’ Characteristics
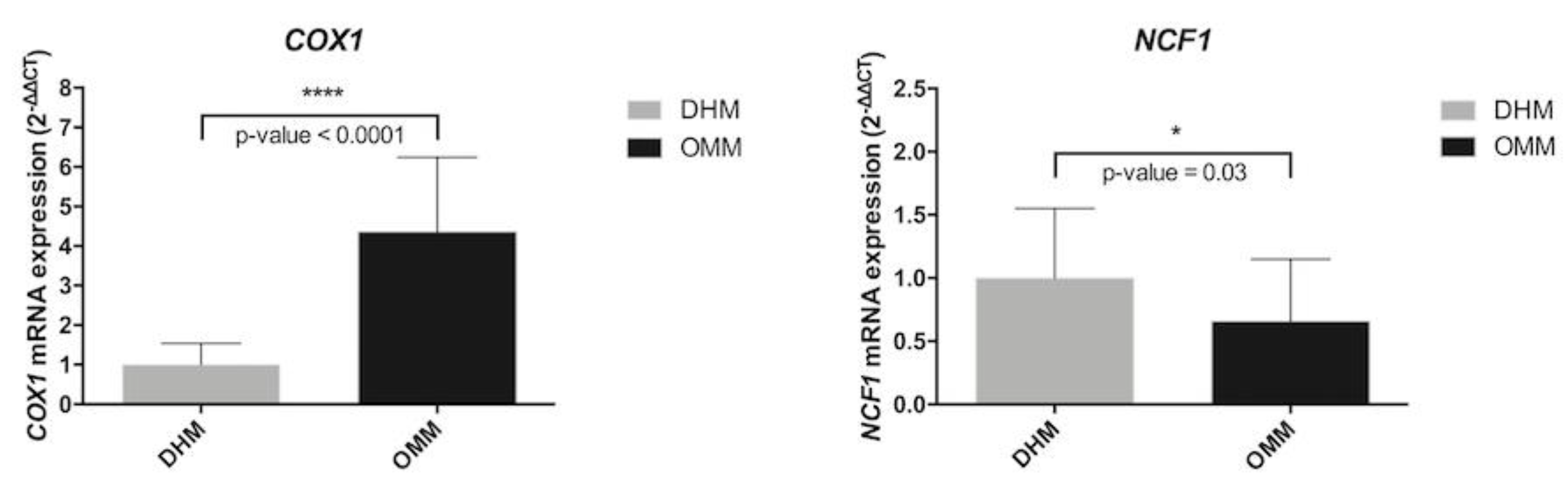
2.2. Real-Time RT-PCR Validation
2.3. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
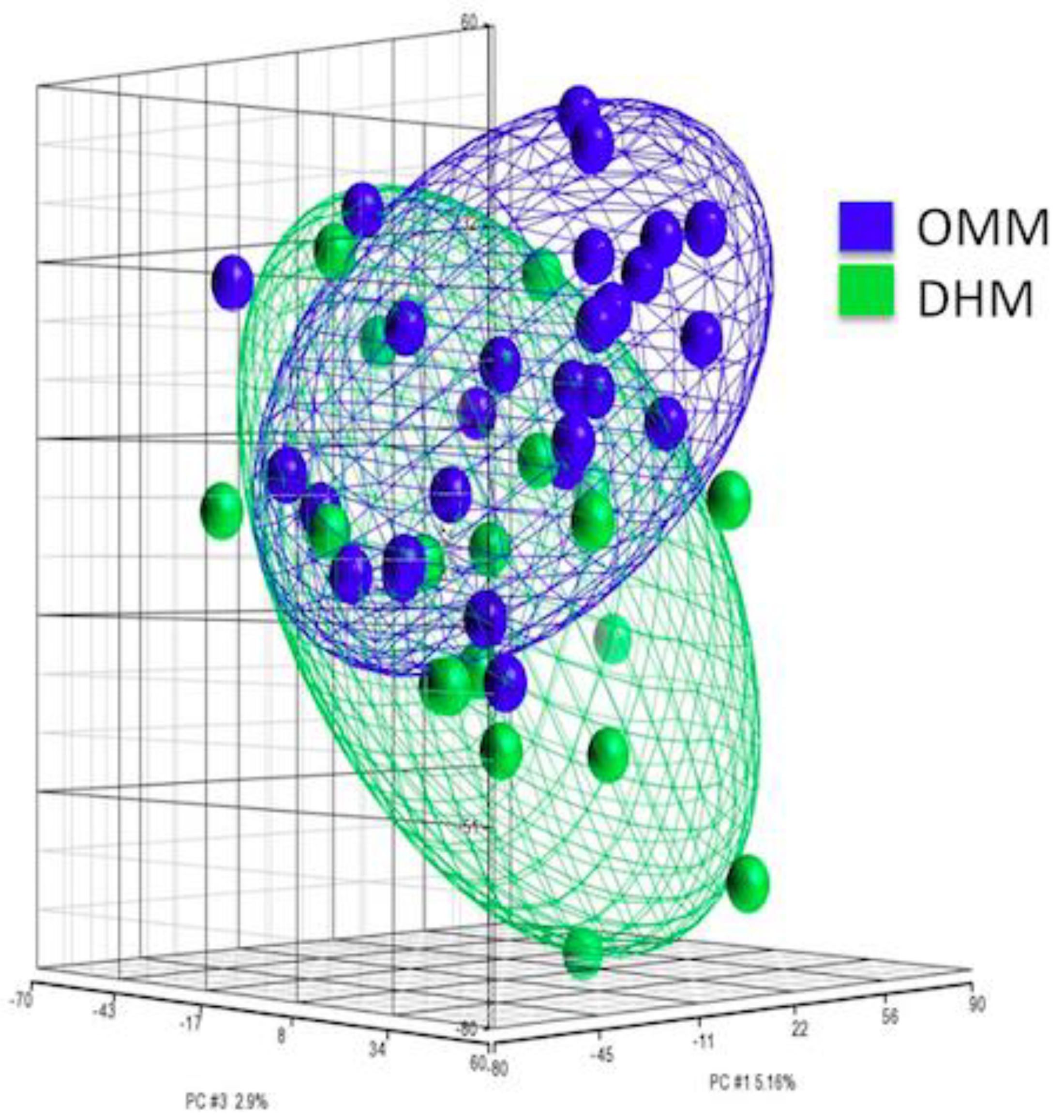
3.2. PCA of Gene Expression from EEIC of Preterm Infants
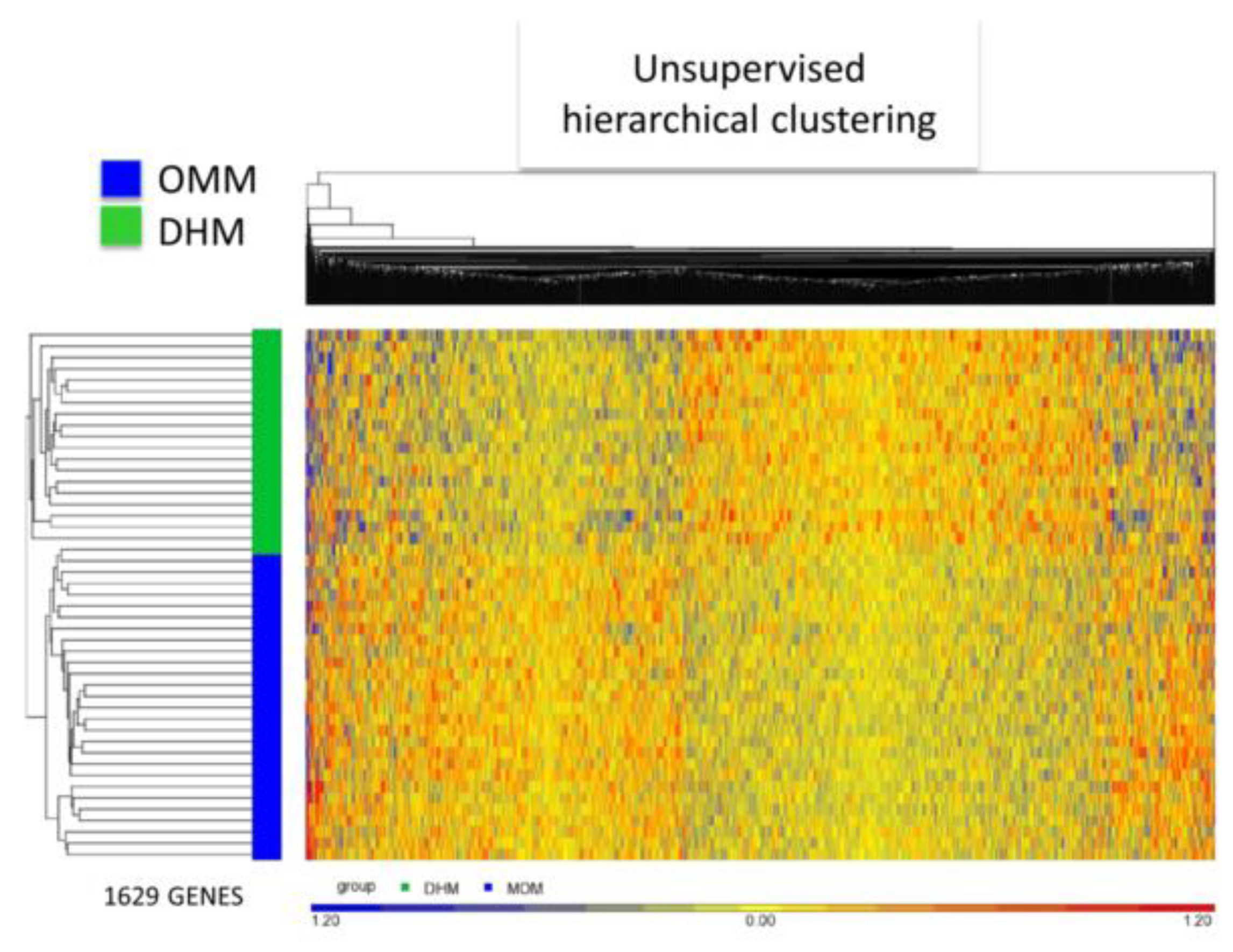
3.3. EEIC Transcriptome Analysis in the OMM Versus DHM Groups
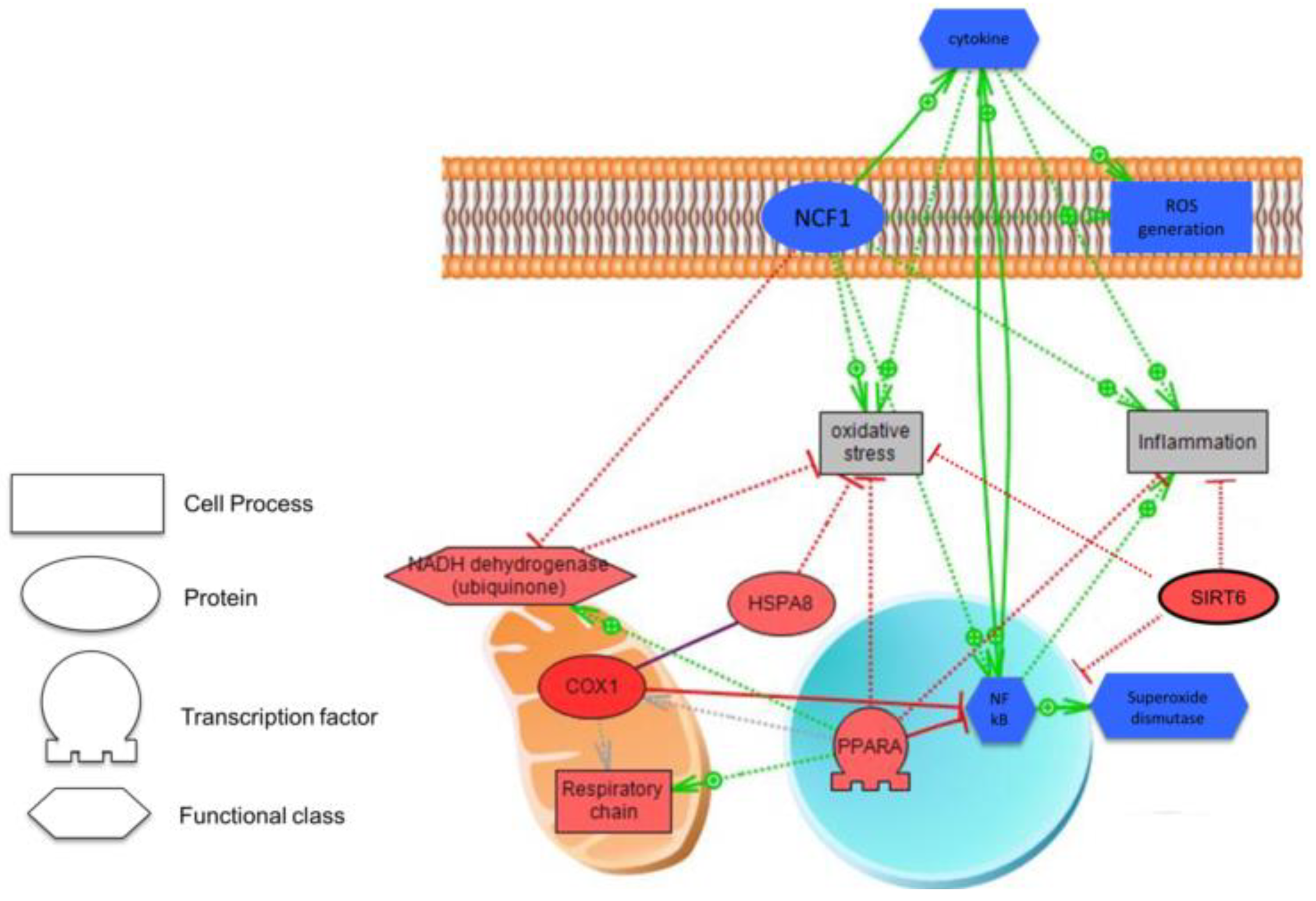
3.4. Biological Analysis
3.5. Biomarkers in Preterm Infants According to Type of Nutrition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations and Acronyms
| ROP | Retinopathy of prematurity |
| BPD | Bronchopulmonary dysplasia |
| NEC | Necrotizing Enterocolitis |
| PVL | Periventricular leucomalacia |
| FR | Free Radicals |
| ROS | Reactive oxygen species |
| OMM | Own Mother’s Milk |
| DHM | Donor Human Milk |
| FM | Formula Milk |
| HM | Human Milk |
| DEG | Differentially expressed genes |
| BW | Birth Weight |
| GA | Gestational Age |
| WHO | World Health Organization |
| EEIC | Exfoliated epithelial intestinal cells |
| COX1 | Cytochrome C oxidase subunit 1 |
| NCF1 | Neutrophil Cytosolic Factor 1 |
References
- Yismaw, A.E.; Gelagay, A.A.; Sisay, M.M. Survival and predictors among preterm neonates admitted at University of Gondar comprehensive specialized hospital neonatal intensive care unit, Northwest Ethiopia. Ital. J. Pediatr. 2019, 45, 4. [Google Scholar] [CrossRef] [PubMed]
- Glass, H.C.; Costarino, A.T.; Stayer, S.A.; Brett, C.M.; Cladis, F.; Davis, P.J. Outcomes for extremely premature infants. Anesth. Analg. 2015, 120, 1337–1351. [Google Scholar] [CrossRef] [PubMed]
- Torres-Cuevas, I.; Parra-Llorca, A.; Sánchez-Illana, A.; Nuñez-Ramiro, A.; Kuligowski, J.; Cháfer-Pericás, C.; Cernada, M.; Escobar, J.; Vento, M. Oxygen and oxidative stress in the perinatal period. Redox Biol. 2017, 12, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Smuder, A.J.; Sollanek, K.J.; Nelson, W.B.; Min, K.; Talbert, E.E.; Kavazis, A.N.; Hudson, M.B.; Sandri, M.; Szeto, H.H.; Powers, S.K. Crosstalk between autophagy and oxidative stress regulates proteolysis in the diaphragm during mechanical ventilation. Free Radic. Biol. Med. 2018, 115, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Ozsurekci, Y.; Aykac, K. Oxidative Stress Related Diseases in Newborns. Oxid. Med. Cell Longev. 2016, 2016, 2768365. [Google Scholar] [CrossRef] [PubMed]
- Vento, M.; Saugstad, O.D. Oxygen supplementation in the delivery room: Updated information. J. Pediatr. 2011, 158, e5–e7. [Google Scholar] [CrossRef]
- Păduraru, L.; Dimitriu, D.C.; Avasiloaiei, A.L.; Moscalu, M.; Zonda, G.I.; Stamatin, M. Total antioxidant status in fresh and stored human milk from mothers of term and preterm neonates. Pediatr. Neonatol. 2018, 59, 600–605. [Google Scholar] [CrossRef] [Green Version]
- Ledo, A.; Arduini, A.; Asensi, M.A.; Sastre, J.; Escrig, R.; Brugada, M.; Aguar, M.; Saenz, P.; Vento, M. Human milk enhances antioxidant defenses against hydroxyl radical aggression in preterm infants. Am. J. Clin. Nutr. 2009, 89, 210–215. [Google Scholar] [CrossRef]
- Meinzen-Derr, J.; Poindexter, B.; Wrage, L.; Morrow, A.L.; Stoll, B.; Donovan, E.F. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J. Perinatol. 2009, 29, 57–62. [Google Scholar] [CrossRef]
- Peila, C.; Moro, G.E.; Bertino, E.; Cavallarin, L.; Giribaldi, M.; Giulani, F.; Cresi, F.; Coscia, A. The Effect of Holder Pasteurization on Nutrients and Biologically-Active Components in Donor Human Milk: A Review. Nutrients 2016, 8, 477. [Google Scholar] [CrossRef]
- Cernada, M.; Bäuerl, C.; Serna, E.; Collado, M.C.; Martínez, G.P.; Vento, M. Sepsis in preterm infants causes alterations in mucosal gene expression and microbiota profiles compared to non-septic twins. Sci. Rep. 2016, 6, 25497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chapkin, R.S.; Zhao, C.; Ivanov, I.; Davidson, L.A.; Goldsby, J.S.; Lupton, J.R.; Mathai, R.A.; Monaco, M.H.; Rai, D.; Russell, W.M.; et al. Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. Am. J. Physiol. Gastrointest Liver Physiol. 2010, 298, G582–G589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, S.M.; Wang, M.; Monaco, M.H.; Martin, C.R.; Davidson, L.A.; Ivanov, I.; Chapkin, R.S. Noninvasive molecular fingerprinting of host-microbiome interactions in neonates. FEBS Lett. 2014, 588, 4112–4119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, J.M.; Davidson, L.A.; Herman, D.; Martin, C.R.; Goldsby, J.S.; Ivanov, I.V.; Donovan, S.M.; Chapkin, R.S. Non-invasive analysis of intestinal development in preterm and term infants using RNA-Sequencing. Sci. Rep. 2014, 4, 5453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garwolińska, D.; Namieśnik, J.; Kot-Wasik, A.; Hewelt-Belka, W. Chemistry of Human Breast Milk-A Comprehensive Review of the Composition and Role of Milk Metabolites in Child Development. J. Agric. Food Chem. 2018, 66, 11881–11896. [Google Scholar] [CrossRef]
- Cortez, J.; Makker, K.; Kraemer, D.F.; Neu, J.; Sharma, R.; Hudak, M.L. Maternal milk feedings reduce sepsis, necrotizing enterocolitis and improve outcomes of premature infants. J. Perinatol. 2018, 38, 71–74. [Google Scholar] [CrossRef]
- Wilson, E.; Edstedt Bonamy, A.K.; Bonet, M.; Toome, L.; Rodrigues, C.; Howell, E.A.; Cuttini, M.; Zeitlin, J. EPICE Research Group. Room for improvement in breast milk feeding after very preterm birth in Europe: Results from the EPICE cohort. Matern. Child Nutr. 2017, 14. [Google Scholar] [CrossRef]
- ESPGHAN Committee on Nutrition; Arslanoglu, S.; Corpeleijn, W.; Moro, G.; Braegger, C.; Campoy, C.; Colomb, V.; Decsi, T.; Domellöf, M.; Fewtrell, M.; et al. Donor human milk for preterm infants: Current evidence and research directions. J. Pediatr. Gastroenterol. Nutr. 2013, 57, 535–542. [Google Scholar] [CrossRef]
- Quigley, M.; Embleton, N.D.; McGuire, W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst. Rev. 2018, 20, 6:CD002971. [Google Scholar] [CrossRef]
- Parra-Llorca, A.; Gormaz, M.; Alcántara, C.; Cernada, M.; Nuñez-Ramiro, A.; Vento, M.; Collado, M.C. Preterm gut microbiome depending on feeding type: Significance of donor human milk. Front. Microbiol. 2018, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Baro, C.; Giribaldi, M.; Arslanoglu, S.; Giuffrida, M.G.; Dellavalle, G.; Conti, A.; Tonetto, P.; Biasini, A.; Coscia, A.; Fabris, C.; et al. Effect of two pasteurization methods on the protein content of human milk. Front. Biosci. 2001, 3, 818–829. [Google Scholar] [CrossRef]
- Sousa, S.G.; Santos, M.D.; Fidalgo, L.G.; Delgadillo, I.; Saraiva, J.A. Effect of thermal pasteurization and high-pressure processing on immunoglobulin content and lysozyme and lactoperoxidase activity in human colostrum. Food Chem. 2014, 151, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Martos, I.; Montilla, A.; de Segura, A.G.; Escuder, D.; Bustos, G.; Pallás, C.; Rodríguez, J.M.; Corzo, N.; Fernández, L. Bacteriological, biochemical, and immunological modifications in human colostrum after Holder pasteurization. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 560–568. [Google Scholar] [CrossRef] [PubMed]
- Untalan, P.B.; Keeney, S.E.; Palkowetz, K.H.; Rivera, A.; Goldman, A.S. Heat susceptibility of interleukin-10 and other cytokines in donor human milk. Breastfeed Med. 2009, 4, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Bertino, E.; Coppa, G.V.; Giuliani, F.; Coscia, A.; Gabrielli, O.; Sabatino, G.; Sgarrella, M.; Testa, T.; Zampini, L.; Fabris, C. Effects of Holder pasteurization on human milk oligosaccharides. Int. J. Immunopathol. Pharmacol. 2008, 21, 381–385. [Google Scholar] [CrossRef]
- Coscia, A.; Peila, C.; Bertino, E.; Coppa, G.V.; Moro, G.E.; Gabrielli, O.; Zampini, L.; Galeazzi, T.; Maccari, F.; Volpi, N. Effect of Holder pasteurization on human milk glycosaminoglycans. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 127–130. [Google Scholar] [CrossRef]
- Garcia-Rodenas, C.L.; De Castro, C.A.; Jenni, R.; Thakkar, S.K.; Beauport, L.; Tolsa, J.F.; Fisher-Fumeaux, C.J.; Affolter, M. Temporal changes of major protein concentrations in preterm and term human milk. A prospective cohort study. Clin. Nutr. 2019, 38, 1844–1852. [Google Scholar] [CrossRef]
- Lonnerdal, B. Nutritional and physiologic significance of human milk proteins. Am. J. Clin. Nutr. 2003, 77, 1537S–1543S. [Google Scholar] [CrossRef]
- Lonnerdal, B. Bioactive proteins in breast milk. J. Paediatr. Child Health 2013, 49, 1–7. [Google Scholar] [CrossRef]
- Beck, K.L.; Weber, D.; Phinney, B.S.; Smilowitz, J.T.; Hinde, K.; Lonnerdal, B.; Korf, I.; Lemay, D.G. Comparative proteomics of human and macaque milk reveals species-specific nutrition during postnatal development. J. Proteome Res. 2015, 14, 2143–2157. [Google Scholar] [CrossRef] [PubMed]
- Prentice, A. Constituents of human milk. Food Nutr. Bull. 1996, 17, 305–312. [Google Scholar] [CrossRef]
- Mimouni, F.B.; Lubetzky, R.; Yochpaz, S.; Mandel, D. Preterm Human Milk Macronutrient and Energy Composition: A Systematic Review and Meta-Analysis. Clin. Perinatol. 2017, 44, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Goenka, A.; Kollmann, T.R. Development of immunity in early life. J. Infect. 2015, 71, S112–S120. [Google Scholar] [CrossRef]
- Kollmann, T.R.; Levy, O.; Montgomery, R.R.; Goriely, S. Innate immune function by Toll-like receptors: Distinct responses in newborns and the elderly. Immunity 2012, 37, 771–783. [Google Scholar] [CrossRef]
- Nathan, C.; Shiloh, M.U. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8841–8848. [Google Scholar] [CrossRef] [Green Version]
- Sordillo, L.M.; Aitken, S.L. Impact of oxidative stress on the health and immune function of dairy cattle. Vet. Immunol. Immunopathol. 2009, 128, 104–109. [Google Scholar] [CrossRef]
- Saugstad, O.D.; Oei, J.L.; Lakshminrusimha, S.; Vento, M. Oxygen therapy of the newborn from molecular understanding to clinical practice. Pediatr. Res. 2019, 85, 20–29. [Google Scholar] [CrossRef]
- Rönnau, C.; Liebermann, H.E.; Helbig, F.; Staudt, A.; Felix, S.B.; Ewert, R.; Landsberger, M. The bio-complex "reaction pattern in vertebrate cells" reduces cytokine-induced cellular adhesion molecule mRNA expression in human endothelial cells by attenuation of NF-kappaB translocation. BMB Rep. 2009, 42, 106–112. [Google Scholar] [CrossRef]
- Schwab, M.; Reynders, V.; Loitsch, S.; Steinhilber, D.; Stein, J.; Schröder, O. Involvement of different nuclear hormone receptors in butyrate-mediated inhibition of inducible NF kappa B signalling. Mol. Immunol. 2007, 44, 3625–3632. [Google Scholar] [CrossRef]
- Guan, Y.; Breyer, M.D. Peroxisome proliferator-activated receptors (PPARs): Novel therapeutic targets in renal disease. Kidney Int. 2001, 60, 14–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besson, V.C.; Chen, X.R.; Plotkine, M.; Marchand-Verrecchia, C. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, exerts neuroprotective effects in traumatic brain injury. Neurosci. Lett. 2005, 388, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Poynter, M.E.; Daynes, R.A. Peroxisome proliferator-activated receptor alpha activation modulates cellular redox status, represses nuclear factor-kappaB signaling, and reduces inflammatory cytokine production in aging. J. Biol. Chem. 1998, 273, 32833–32841. [Google Scholar] [CrossRef] [PubMed]
- Chiueh, C.C. Neuroprotective properties of nitric oxide. Ann. N. Y. Acad. Sci. 1999, 890, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Fatehi-Hassanabad, Z.; Chan, C.B.; Furman, B.L. Reactive oxygen species and endothelial function in diabetes. Eur. J. Pharmacol. 2010, 636, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.W. Free radicals and lipid peroxidation mediated injury in burn trauma: The role of antioxidant therapy. Toxicology 2003, 189, 75–88. [Google Scholar] [CrossRef]
- Aceti, A.; Beghetti, I.; Martini, S.; Faldella, G.; Corvaglia, L. Oxidative Stress and Necrotizing Enterocolitis: Pathogenetic Mechanisms, Opportunities for Intervention, and Role of Human Milk. Oxid. Med. Cell Longev. 2018, 2, 7397659. [Google Scholar] [CrossRef]
- Holmdahl, R.; Sareila, O.; Olsson, L.M.; Bäckdahl, L.; Wing, K. Ncf1 polymorphism reveals oxidative regulation of autoimmune chronic inflammation. Immunol. Rev. 2016, 269, 228–247. [Google Scholar] [CrossRef]
- Rao, M.; Wong, C.; Kanetsky, P.; Girndt, M.; Stenvinkel, P.; Reilly, M.; Raj, D.S. Cytokine gene polymorphism and progression of renal and cardiovascular diseases. Kidney Int. 2007, 72, 549–556. [Google Scholar] [CrossRef] [Green Version]
- Mino, T.; Takeuchi, O. Post-transcriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation. Biotechnol. Genet. Eng. Rev. 2013, 29, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Prahalad, S.; Glass, D.N. A comprehensive review of the genetics of juvenile idiopathic arthritis. Pediatr. Rheumatol. Online J. 2008, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Hojo, Y.; Shimada, K. Role of cytokines in acute coronary syndrome. Nihon Rinsho. 1998, 56, 2500–2503. [Google Scholar] [PubMed]
- Lloyd, R.E.; Foster, P.G.; Guille, M.; Littlewood, D.T. Next generation sequencing and comparative analyses of Xenopus mitogenomes. BMC Genom. 2012, 13, 496. [Google Scholar] [CrossRef] [PubMed]
| OMM (N = 27) | DHM (N = 20) | p-Value | |
|---|---|---|---|
| GA weeks, (median; 5–95% CI) | 29 (28–30) | 30 (29–31) | 0.07 |
| Antenatal Steroids full course, n (%) | 25 (92.6) | 20 (100) | 0.21 |
| Sex male, n (%) | 21 (77.7) | 11 (55) | 0.1 |
| Type of delivery, n (%) | |||
| - Vaginal | 13 (48.1) | 10 (50) | 0.9 |
| - C- Section | 14 (51.8) | 10 (50) | |
| Birth weight (g), mean (SD) | 1206 (264) | 1315 (193) | 0.22 |
| Apgar 1 min (median; 5–95% CI) | 7 (6–9) | 7 (6–9) | 0.48 |
| Apgar 5 min (median; 5–95% CI) | 9 (8–19) | 9 (8–9) | 0.28 |
| Age (d) at sample collection, (median; 5–95% CI) | 9 (7–11) | 8 (6–10) | 0.52 |
| Chorioamnionitis, n (%) | 4 (14.8) | 2 (10) | 0.62 |
| Persistent ductus arteriosus, n (%) | 8 (29.6) | 6 (30) | 0.98 |
| Antibiotic therapy, n (%) | 11 (40.7) | 6 (30) | 0.45 |
| Bronchopulmonary Dysplasia, n (%) | 1 (3.7) | 0 (0) | 0.38 |
| Intraventricular hemorrhage, n (%) | 6 (22.2) | 3 (15) | 0.53 |
| Necrotizing enterocolitis, n (%) | 1 (3.7) | 0 (0) | 0.38 |
| Retinopathy of prematurity, n (%) | 1 (3.7) | 0 (0) | 0.38 |
| Gene Symbol | Gene Name | Fold-Change | p-Value |
|---|---|---|---|
| LALBA | Lactalbumin alpha | 2.9 | 0.002 |
| CSN3 | Casein kappa | 2.6 | 0.002 |
| CSN2 | Casein beta | 2.1 | 0.009 |
| COX1 | Cytochrome C oxidase subunit1 | 2.1 | 0.030 |
| CSN1S1 | Casein alpha-s1 | 1.7 | 0.008 |
| NCF1 | Neutrophil cytosolic factor 1 | −1.6 | 0.040 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra-Llorca, A.; Gormaz, M.; Lorente-Pozo, S.; Cernada, M.; García-Robles, A.; Torres-Cuevas, I.; Kuligowski, J.; Collado, M.C.; Serna, E.; Vento, M. Impact of Donor Human Milk in the Preterm Very Low Birth Weight Gut Transcriptome Profile by Use of Exfoliated Intestinal Cells. Nutrients 2019, 11, 2677. https://doi.org/10.3390/nu11112677
Parra-Llorca A, Gormaz M, Lorente-Pozo S, Cernada M, García-Robles A, Torres-Cuevas I, Kuligowski J, Collado MC, Serna E, Vento M. Impact of Donor Human Milk in the Preterm Very Low Birth Weight Gut Transcriptome Profile by Use of Exfoliated Intestinal Cells. Nutrients. 2019; 11(11):2677. https://doi.org/10.3390/nu11112677
Chicago/Turabian StyleParra-Llorca, Anna, María Gormaz, Sheila Lorente-Pozo, Maria Cernada, Ana García-Robles, Isabel Torres-Cuevas, Julia Kuligowski, Maria Carmen Collado, Eva Serna, and Máximo Vento. 2019. "Impact of Donor Human Milk in the Preterm Very Low Birth Weight Gut Transcriptome Profile by Use of Exfoliated Intestinal Cells" Nutrients 11, no. 11: 2677. https://doi.org/10.3390/nu11112677
APA StyleParra-Llorca, A., Gormaz, M., Lorente-Pozo, S., Cernada, M., García-Robles, A., Torres-Cuevas, I., Kuligowski, J., Collado, M. C., Serna, E., & Vento, M. (2019). Impact of Donor Human Milk in the Preterm Very Low Birth Weight Gut Transcriptome Profile by Use of Exfoliated Intestinal Cells. Nutrients, 11(11), 2677. https://doi.org/10.3390/nu11112677










