In Silico Investigation of the Pharmacological Mechanisms of Beneficial Effects of Ginkgo biloba L. on Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
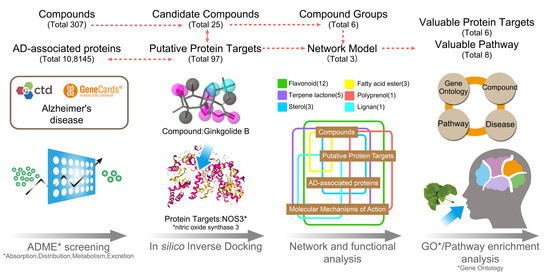
2.1. Data Collection
2.2. Inverse Docking Analysis
2.3. Gene Ontology and KEGG Pathway Enrichment
2.4. Composite Network Integration
3. Results
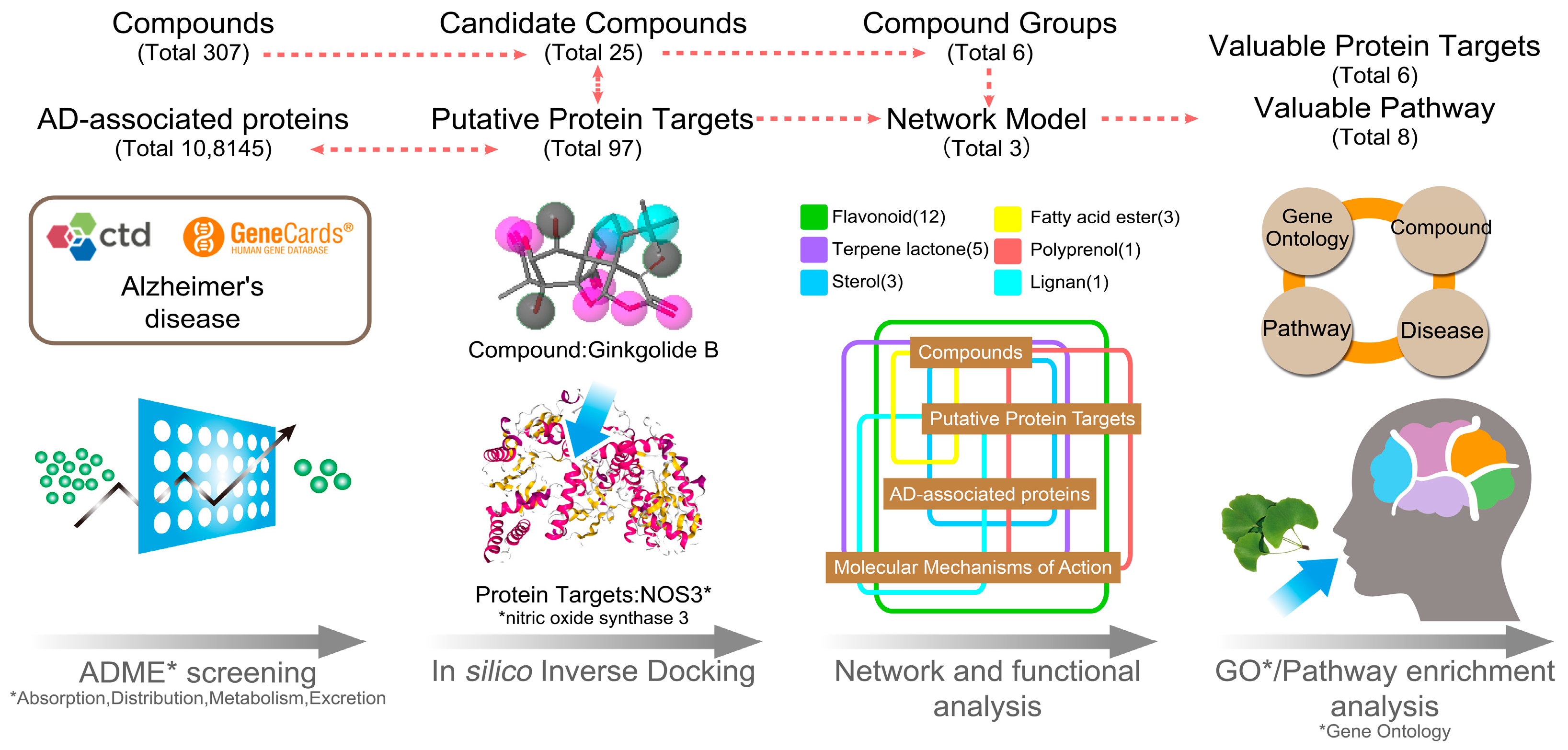
3.1. The Candidate Compounds and Putative Target Proteins
3.2. Exploration of the Molecular Mechanisms of Action
3.3. An Integrated Network Model Analysis
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| AD | Alzheimer’s Disease |
| 2D | 2-dimensional |
| ADME | absorptions, distribution, metabolism, and excretion |
| ALB | serum albumin |
| AR | Androgen receptor |
| ASPL | Average Shortest Path Length |
| Aβ | amyloid β-protein |
| BACE1 | beta-secretase 1 |
| BBB | blood brain barrier |
| BC | Betweenness Centrality |
| CCNA2 | Cyclin-A2 |
| CDK2 | Cyclin-dependent kinase 2 |
| CGTP | Compound-Group-Target-Pathway |
| CT | Compound-Target |
| CTD | Comparative Toxicogenomics Database |
| DAVID | The Database for Annotation, Visualization, and Integrated Discovery |
| DL | drug-likeness |
| ELANE | Neutrophil elastase |
| ERBB2 | Receptor tyrosine-protein kinase erbB-2 |
| ESR1 | Estrogen receptor 1 |
| F2 | prothrombin |
| FDPS | Farnesyl pyrophosphate synthase |
| FDR | false discovery rate |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FOXO | forkhead box O |
| GO | Gene Ontology |
| GSTA1 | Glutathione S-transferase A1 |
| GSTM2 | Glutathione S-transferase Mu 2 |
| GSTP1 | Glutathione S-transferase P |
| HRAS | GTPase HRas |
| HSP90AA1 | Heat shock protein HSP 90-alpha |
| IL2 | interleukin-2 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAOB | amine oxidase B |
| MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 |
| MAPK | mitogen-activated protein kinase |
| MAPK10 | mitogen-activated protein kinase 10 |
| MAPK14 | mitogen-activated protein kinase 14 |
| MFH | medicine food homology |
| MMP3 | matrix metalloproteinase-3 |
| MOA | Molecular Mechanisms of Action |
| NEP | neprilysin |
| NOD | nucleotide bindingoligomerization domain |
| NOS3 | synthase |
| NR3C2 | Mineralocorticoid receptor |
| NSAIDs | nonsteroidal anti-inflammatory drugs |
| OB | bioavailability |
| PI3K | Phosphoinositide 3-kinase |
| PPI | Protein–Protein Interaction |
| PPIA | Peptidyl-prolyl cis-trans isomerase A |
| PTPN1 | Tyrosine-protein phosphatase non-receptor type 1 |
| RBP4 | Retinol-binding protein 4 |
| RNA | ribonucleic acid |
| RnRH | Gonadotropin-releasing hormone |
| SRC | proto-oncogene tyrosine-protein kinase src |
| TCMSP | The Traditional Chinese Medicine System Pharmacology Database and Analysis Platform |
| TNF | Tumor Necrosis Factor |
| TRP | Transient receptor potential |
| TTR | transthyretin |
| USP41-NF36 | United States Pharmacopoeia National Formulary |
| VDR | Vitamin D3 receptor |
| VEGF | vascular endothelial growth factor |
References
- Selkoe, D.J. Preventing Alzheimer’s disease. Science 2012, 337, 1488–1492. [Google Scholar] [CrossRef] [PubMed]
- Lleó, A.; Greenberg, S.M.; Growdon, J.H. Current Pharmacotherapy for Alzheimer’s Disease. Ann. Rev. Med. 2006, 57, 513–533. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Resolving controversies on the path to Alzheimer’s therapeutics. Nat. Med. 2011, 17, 1060. [Google Scholar] [CrossRef] [PubMed]
- Wolffram, S.; Ader, P.; Rimbach, G.; Packer, L.; Maguire, J.J.; Schultz, P.G.; Gohil, K. The in vivo Neuromodulatory Effects of the Herbal Medicine Ginkgo Biloba. Proc. Natl. Acad. Sci. USA 2001, 98, 6577. [Google Scholar]
- Howes, M.J.; Houghton, P.J. Ethnobotanical treatment strategies against Alzheimer’s disease. Curr. Alzheimer Res. 2012, 9, 67–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Zhi, D.G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins Struct. Funct. Bioinform. 2001, 43, 217–226. [Google Scholar] [CrossRef]
- Wang, X.; Shen, Y.; Wang, S.; Li, S.; Zhang, W.; Liu, X.; Lai, L.; Pei, J.; Li, H. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017, 45, W356–W360. [Google Scholar] [CrossRef] [PubMed]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C.; Li, P.; Guo, Z.; Tao, W.; Yang, Y. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Stein, T.I.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database’s 10th year anniversary: Update 2015. Nucleic Acids Res. 2015, 43, D914–D920. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, J.; Wu, F.; Yew, D.T. Ginkgo biloba Extract in Alzheimer’s Disease: From Action Mechanisms to Medical Practice. Int. J. Mol. Sci. 2010, 11, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Tapan Kumar, M.; Yasinalli, T.; Zubaidha, P.K. Phytochemical and medicinal importance of Ginkgo biloba L. Nat. Prod. Res. 2014, 28, 746–752. [Google Scholar]
- Ahlemeyer, B.; Krieglstein, J. Neuroprotective effects of Ginkgo biloba extract. Cell. Mol. Life Sci. 2003, 60, 1779–1792. [Google Scholar] [CrossRef] [PubMed]
- Shi, C. Protective effects of Ginkgo biloba extract (EGb761) and its constituents quercetin and ginkgolide B against β-amyloid peptide-induced toxicity in SH-SY5Y cells. Chem. Biol. Interact. 2009, 181, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.V.; Luo, Y. Studies on molecular mechanisms of Ginkgo biloba extract. Appl. Microbiol. Biotechnol. 2004, 64, 465–472. [Google Scholar] [PubMed]
- Maclennan, K.M.; Darlington, C.L.; Smith, P.F. The CNS effects of Ginkgo biloba extracts and ginkgolide B. Prog. Neurobiol. 2002, 67, 235–257. [Google Scholar] [CrossRef]
- Smith, J.V.; Luo, Y. Elevation of oxidative free radicals in Alzheimer’s disease models can be attenuated by Ginkgo biloba extract EGb 761. J. Alzheimers Dis. 2003, 5, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Bastianetto, S.; Ramassamy, C.; Doré, S.; Christen, Y.; Poirier, J.; Quirion, R. The Ginkgo biloba extract (EGb 761) protects hippocampal neurons against cell death induced by beta-amyloid. Eur. J. Neurosci. 2000, 12, 1882–1890. [Google Scholar] [CrossRef] [PubMed]
- Berrocal, M.; Marcos, D.; Sepúlveda, M.R.; Pérez, M.; Avila, J.; Mata, A.M. Altered Ca2+ dependence of synaptosomal plasma membrane Ca2+-ATPase in human brain affected by Alzheimer’s disease. FASEB J. 2009, 23, 1826–1834. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Ren, Y.; Wu, W.; Wang, Y.; Cao, M.; Zhu, Z.; Wang, M.; Li, W. Protective effects of bilobalide on Aβ(25-35) induced learning and memory impairments in male rats. Pharmacol. Biochem. Behav. 2013, 106, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Doreulee, N.; Sergeeva, O.A.; Yanovsky, Y.; Chepkova, A.N.; Selbach, O.; Gödecke, A.; Schrader, J.; Haas, H.L. Cortico-striatal synaptic plasticity in endothelial nitric oxide synthase deficient mice. Brain Res. 2003, 964, 159–163. [Google Scholar] [CrossRef]
- Austin, S.A.; Santhanam, A.V.; Hinton, D.J.; Choi, D.S.; Katusic, Z.S. Endothelial nitric oxide deficiency promotes Alzheimer’s disease pathology. J. Neurochem. 2013, 127, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Austin, S.A.; Santhanam, A.V.; Katusic, Z.S. Endothelial Nitric Oxide Modulates Expression and Processing of Amyloid Precursor ProteinNovelty and Significance. Circ. Res. 2010, 107, 1498–1502. [Google Scholar] [CrossRef] [PubMed]
- Haul, S.; Gödecke, A.; Schrader, J.; Haas, H.L.; Luhmann, H.J. Impairment of neocortical long-term potentiation in mice deficient of endothelial nitric oxide synthase. J. Neurophysiol. 1999, 81, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Austin, S.A.; D’Uscio, L.V.; Katusic, Z.S. Supplementation of nitric oxide attenuates AβPP and BACE1 protein in cerebral microcirculation of eNOS-deficient mice. J. Alzheimers Dis. 2013, 33, 29–33. [Google Scholar] [PubMed]
- Howell, S.; Nalbantoglu, J.; Crine, P. Neutral endopeptidase can hydrolyze β-amyloid(1–40) but shows no effect on β-amyloid precursor protein metabolism. Peptides 1995, 16, 647–652. [Google Scholar] [CrossRef]
- Takaki, Y.; Iwata, N.; Tsubuki, S.; Taniguchi, S.; Toyoshima, S.; Lu, B.; Gerard, N.P.; Gerard, C.; Lee, H.J.; Shirotani, K. Biochemical identification of the neutral endopeptidase family member responsible for the catabolism of amyloid beta peptide in the brain. J. Biochem. 2000, 128, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Colciaghi, F.; Borroni, B.; Zimmermann, M.; Bellone, C.; Longhi, A.; Padovani, A.; Cattabeni, F.; Christen, Y.; Di, L.M. Amyloid precursor protein metabolism is regulated toward alpha-secretase pathway by Ginkgo biloba extracts. Neurobiol. Dis. 2004, 16, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.D.; Cross, D.J.; Minoshima, S.; Belongia, D.; Watson, G.S.; Craft, S. Insulin resistance and Alzheimer-like reductions in regional cerebral glucose metabolism for cognitively normal adults with prediabetes or early type 2 diabetes. Arch. Neurol. 2011, 68, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Subhadra, B.; Schaller, K.; Seeds, N.W. Neuroserpin up-regulation in the Alzheimer’s disease brain is associated with elevated thyroid hormone receptor-β1 and HuD expression. Neurochem. Int. 2013, 63, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Vassar, R. Targeting the β secretase BACE1 for Alzheimer’s disease therapy. Lancet Neurol. 2014, 13, 319–329. [Google Scholar] [CrossRef]
- Vassar, R.; Kovacs, D.M.; Yan, R.; Wong, P.C. The β-Secretase Enzyme BACE in Health and Alzheimer’s Disease: Regulation, Cell Biology, Function, and Therapeutic Potential. J. Neurosci. 2009, 29, 12787–12794. [Google Scholar] [CrossRef] [PubMed]
- White, H.L.; Scates, P.W.; Cooper, B.R. Extracts of Ginkgo biloba leaves inhibit monoamine oxidase. Life Sci. 1996, 58, 1315–1321. [Google Scholar] [CrossRef]
- Pardon, M.C.; Joubert, C.; Perez-Diaz, F.; Christen, Y.; Launay, J.M.; Cohen-Salmon, C. In vivo regulation of cerebral monoamine oxidase activity in senescent controls and chronically stressed mice by long-term treatment with Ginkgo biloba extract (EGb 761). Mech. Aging Dev. 2000, 113, 157–168. [Google Scholar] [CrossRef]
- Chaurasiya, N.D.; Ganesan, S.; Nanayakkara, N.P.; Dias, L.R.; Walker, L.A.; Tekwani, B.L. Inhibition of human monoamine oxidase A and B by 5-phenoxy 8-aminoquinoline analogs. Bioorg. Med. Chem. Lett. 2012, 22, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Aliev, G.; Seyidova, D.; Lamb, B.T.; Obrenovich, M.E.; Siedlak, S.L.; Vinters, H.V.; Friedland, R.P.; Lamanna, J.C.; Smith, M.A.; Perry, G. Mitochondria and vascular lesions as a central target for the development of Alzheimer’s disease and Alzheimer disease-like pathology in transgenic mice. Neurol. Res. 2003, 25, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Biere, A.L.; Ostaszewski, B.; Stimson, E.R.; Hyman, B.T.; Maggio, J.E.; Selkoe, D.J. Amyloid beta-peptide is transported on lipoproteins and albumin in human plasma. J. Biol. Chem. 1996, 271, 32916–32922. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Arora, P.D.; Chen, Y.; Mcculloch, C.A.; Liu, P. Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 2012, 32, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Atwood, C.S.; Meethal, S.V.; Liu, T.; Wilson, A.C.; Gallego, M.; Smith, M.A.; Bowen, R.L. Dysregulation of the hypothalamic-pituitary-gonadal axis with menopause and andropause promotes neurodegenerative senescence. J. Neuropathol. Exp. Neurol. 2005, 64, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Meethal, S.V.; Smith, M.A.; Bowen, R.L.; Atwood, C.S. The gonadotropin connection in Alzheimer’s disease. Endocrine 2005, 26, 317–325. [Google Scholar] [CrossRef]
- Pedrós, I.; Petrov, D.; Artiach, G.; Abad, S.; Ramon-Duaso, C.; Sureda, F.; Pallàs, M.; Beas-Zarate, C.; Folch, J.; Camins, A. Adipokine pathways are altered in hippocampus of an experimental mouse model of Alzheimer’s disease. J. Nutr. Health Aging 2015, 19, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Dye, R.V.; Miller, K.J.; Singer, E.J.; Levine, A.J. Hormone Replacement Therapy and Risk for Neurodegenerative Diseases. Int. J. Alzheimers Dis. 2012, 2012, 258454. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; He, P.; Shen, Y.; Li, R. New evidence of mitochondria dysfunction in the female Alzheimer’s brain: Deficiency of estrogen receptor-β. J. Alzheimers Dis. 2012, 30, 545–558. [Google Scholar] [PubMed]
- Jackson, H.M.; Soto, I.; Graham, L.C.; Carter, G.W.; Howell, G.R. Clustering of transcriptional profiles identifies changes to insulin signaling as an early event in a mouse model of Alzheimer’s disease. BMC Genom. 2013, 14, 831. [Google Scholar] [CrossRef] [PubMed]
- Burbach, J.P.; Voorhuis, T.A.; van Tol, H.H.; Ivell, R. In situ hybridization of oxytocin messenger RNA: Macroscopic distribution and quantitation in rat hypothalamic cell groups. Biochem. Biophys. Res. Commun. 1987, 145, 10–14. [Google Scholar] [CrossRef]
- Hollowell, J.G.; Staehling, N.W.; Flanders, W.D.; Hannon, W.H.; Gunter, E.W.; Spencer, C.A.; Braverman, L.E. Serum TSH, T(4), and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J. Clin. Endocr. Metab. 2002, 87, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Riederer, P.; Bartl, J.; Laux, G.; Grünblatt, E. Diabetes type II: A risk factor for depression-Parkinson-Alzheimer? Neurotox. Res. 2011, 19, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Mouri, A.; Kokubo, H.; Nakajima, R.; Suemoto, T.; Higuchi, M.; Staufenbiel, M.; Noda, Y.; Yamaguchi, H.; Nabeshima, T. Neprilysin-sensitive synapse-associated amyloid-beta peptide oligomers impair neuronal plasticity and cognitive function. J. Biol. Chem. 2006, 281, 17941–17951. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yao, J.; Mao, Z.; Chen, S.; Wang, Y.; Brinton, R.D. 17β-Estradiol regulates insulin-degrading enzyme expression via an ERβ/PI3-K pathway in hippocampus: Relevance to Alzheimer’s prevention. Neurobiol. Aging 2011, 32, 1949–1963. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Andersson, S.; Warner, M.; Gustafsson, J.A. Morphological abnormalities in the brains of estrogen receptor beta knockout mice. Proc. Natl. Acad. Sci. USA 2001, 98, 2792–2796. [Google Scholar] [CrossRef] [PubMed]
- Waters, E.M.; Yildirim, M.; Janssen, W.G.M.; Lou, W.Y.W.; Mcewen, B.S.; Morrison, J.H.; Milner, T.A. Estrogen and aging affect the synaptic distribution of estrogen receptor α-immunoreactivity in the CA1 region of female rat hippocampus. Brain Res. 2011, 1379, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Z.; Schneider, L.S.; Brinton, R.D. Estrogen receptor β-selective phytoestrogenic formulation prevents physical and neurological changes in a preclinical model of human menopause. Menopause 2011, 18, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Kalgutkar, A.S.; Dalvie, D.K., Jr.; Castagnoli, N.; Taylor, T.J. Interactions of nitrogen-containing xenobiotics with monoamine oxidase (MAO) isozymes A and B: SAR studies on MAO substrates and inhibitors. Chem. Res. Toxicol. 2001, 14, 1139–1162. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Miklossy, J.; Klegeris, A.; Guo, J.P.; Mcgeer, P.L. Thrombin and prothrombin are expressed by neurons and glial cells and accumulate in neurofibrillary tangles in Alzheimer disease brain. J. Neuropathol. Exp. Neurol. 2006, 65, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Era, S.; Kuwata, K.; Imai, H.; Nakamura, K.; Hayashi, T.; Sogami, M. Age-related change in redox state of human serum albumin. Biochim. Biophys. Acta 1995, 1247, 12–16. [Google Scholar] [CrossRef]
- Guerin-Dubourg, A.; Catan, A.; Bourdon, E.; Rondeau, P. Structural modifications of human albumin in diabetes. Diabetes Metab. 2012, 38, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Johansson, P.; Almqvist, E.G.; Johansson, J.O.; Mattsson, N.; Hansson, O.; Wallin, A.; Blennow, K.; Zetterberg, H.; Svensson, J. Reduced cerebrospinal fluid level of thyroxine in patients with Alzheimer’s disease. Psychoneuroendocrinology 2013, 38, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Ray, I.; Chauhan, A.; Wegiel, J.; Chauhan, V. Gelsolin inhibits the fibrillization of amyloid beta-protein, and also defibrillizes its preformed fibrils. Brain Res. 2000, 853, 344–351. [Google Scholar] [CrossRef]
- Hirko, A.C.; Meyer, E.M.; King, M.A.; Hughes, J.A. Peripheral Transgene Expression of Plasma Gelsolin Reduces Amyloid in Transgenic Mouse Models of Alzheimer’s Disease. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 1623–1629. [Google Scholar] [CrossRef] [PubMed]

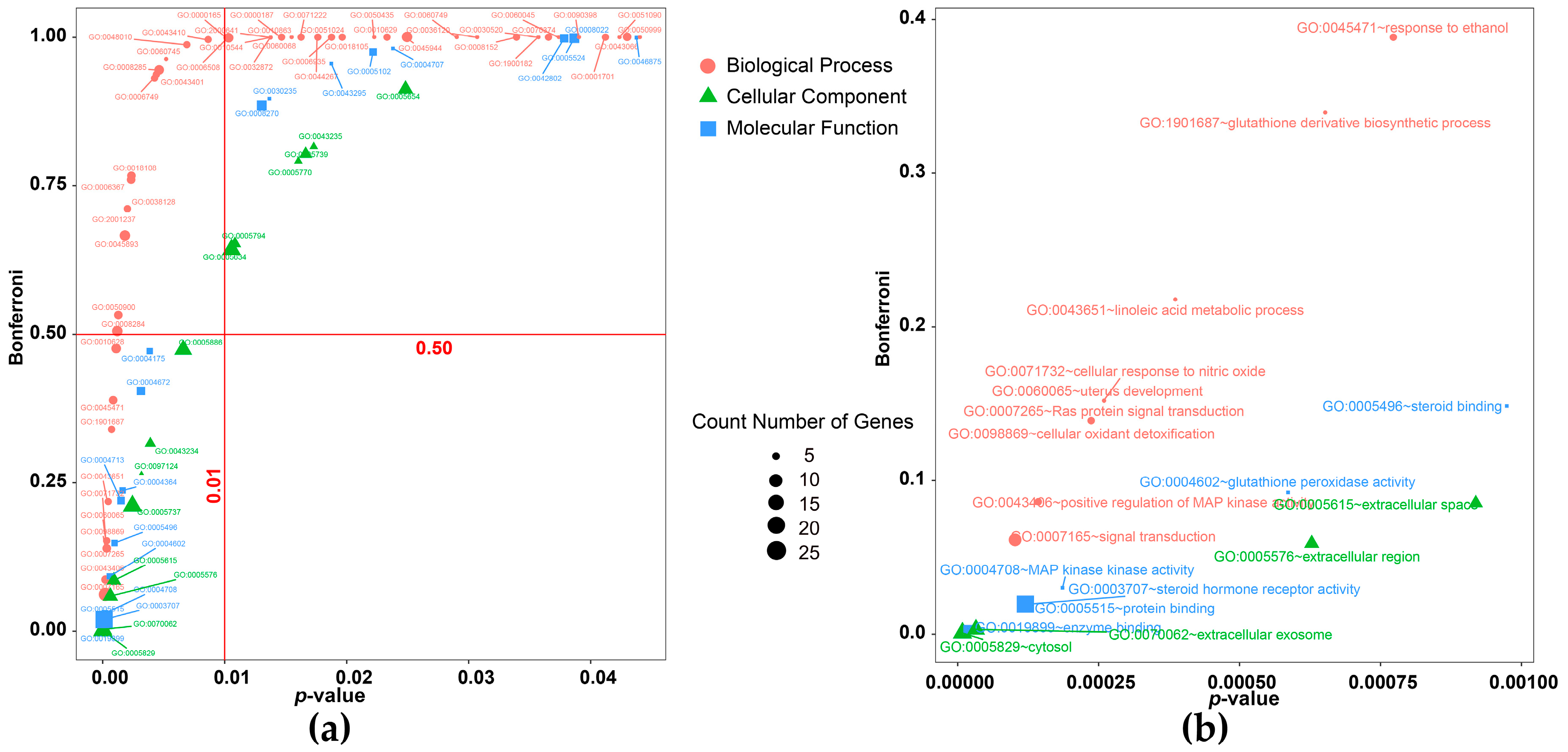
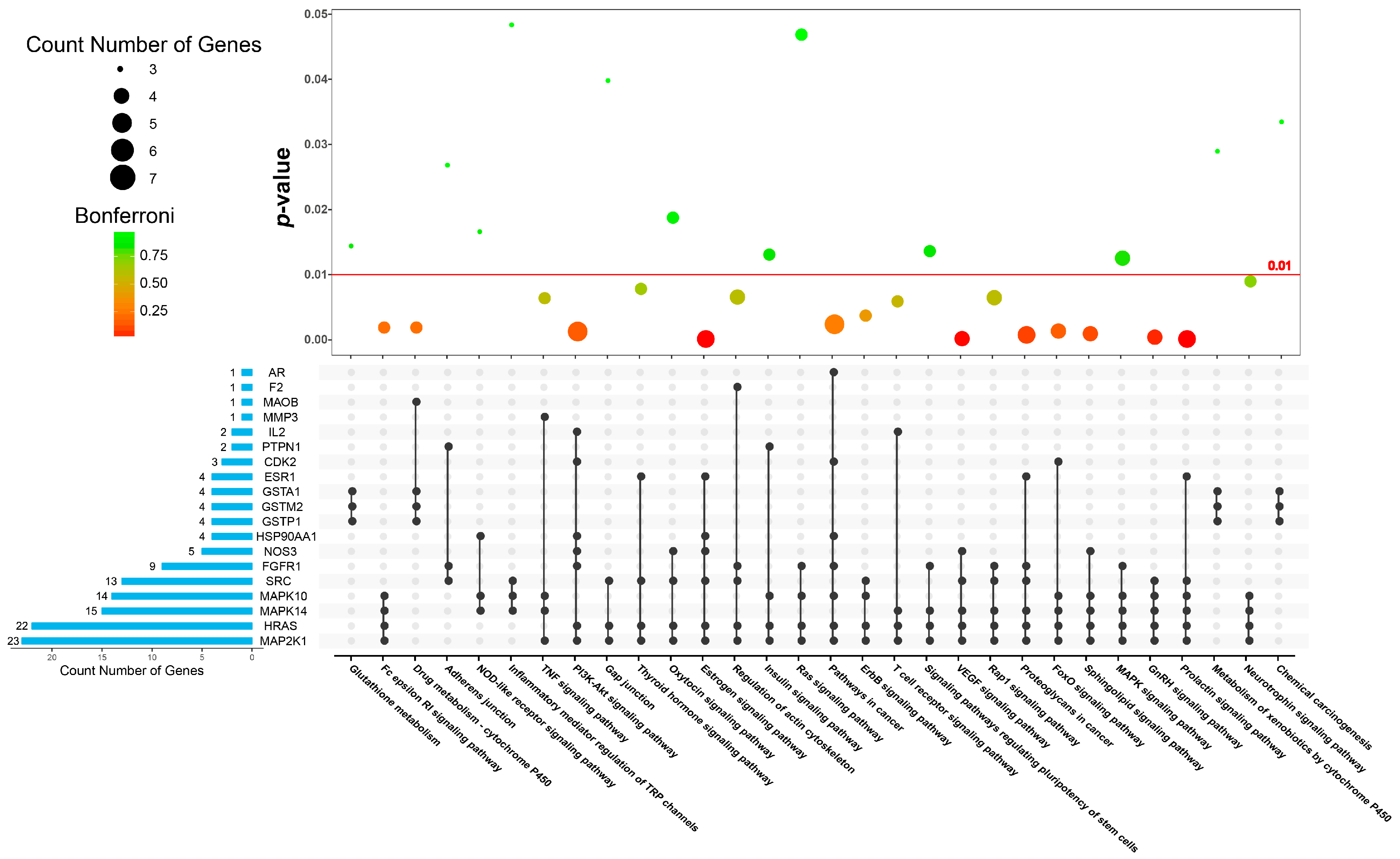
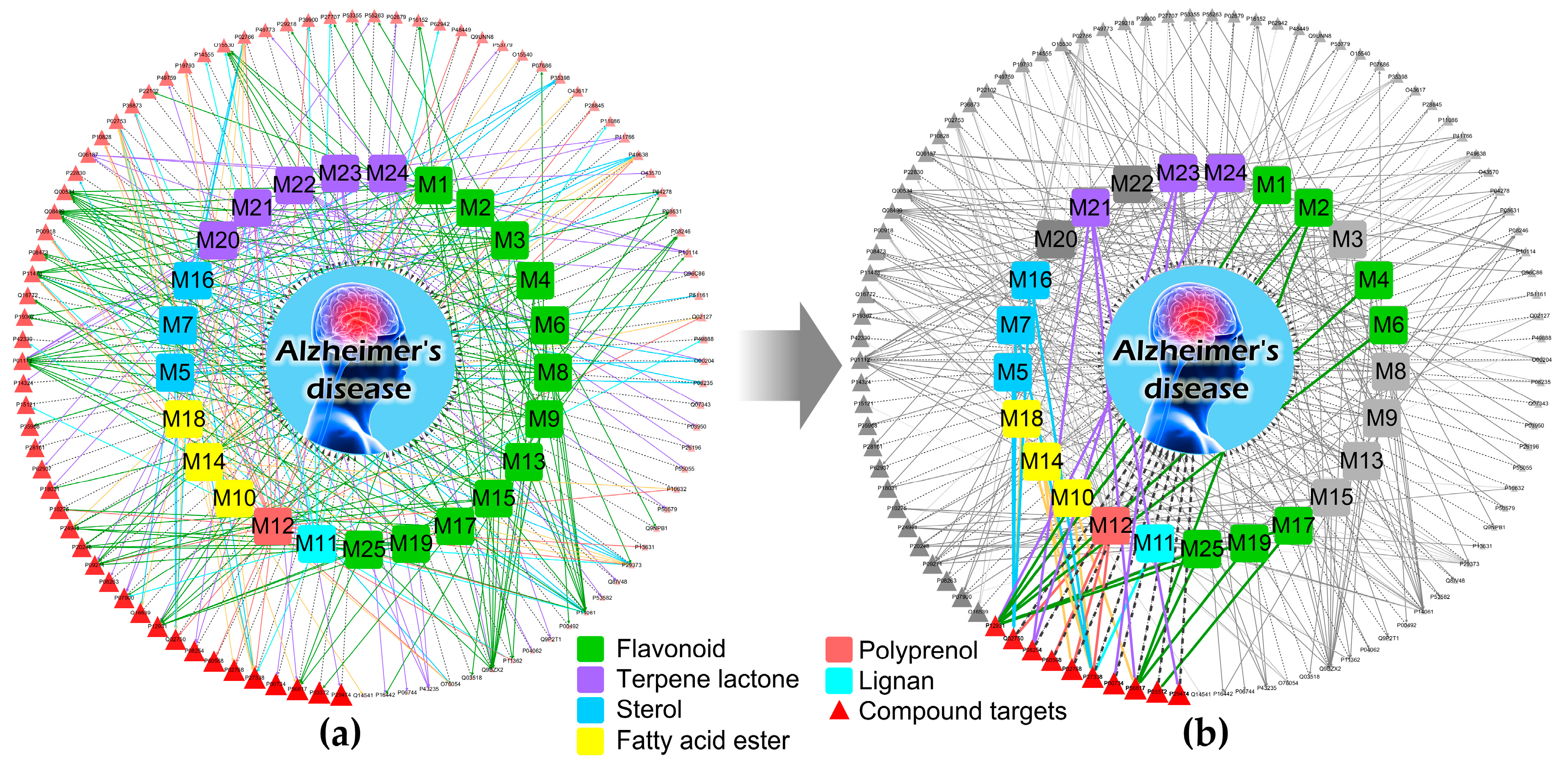
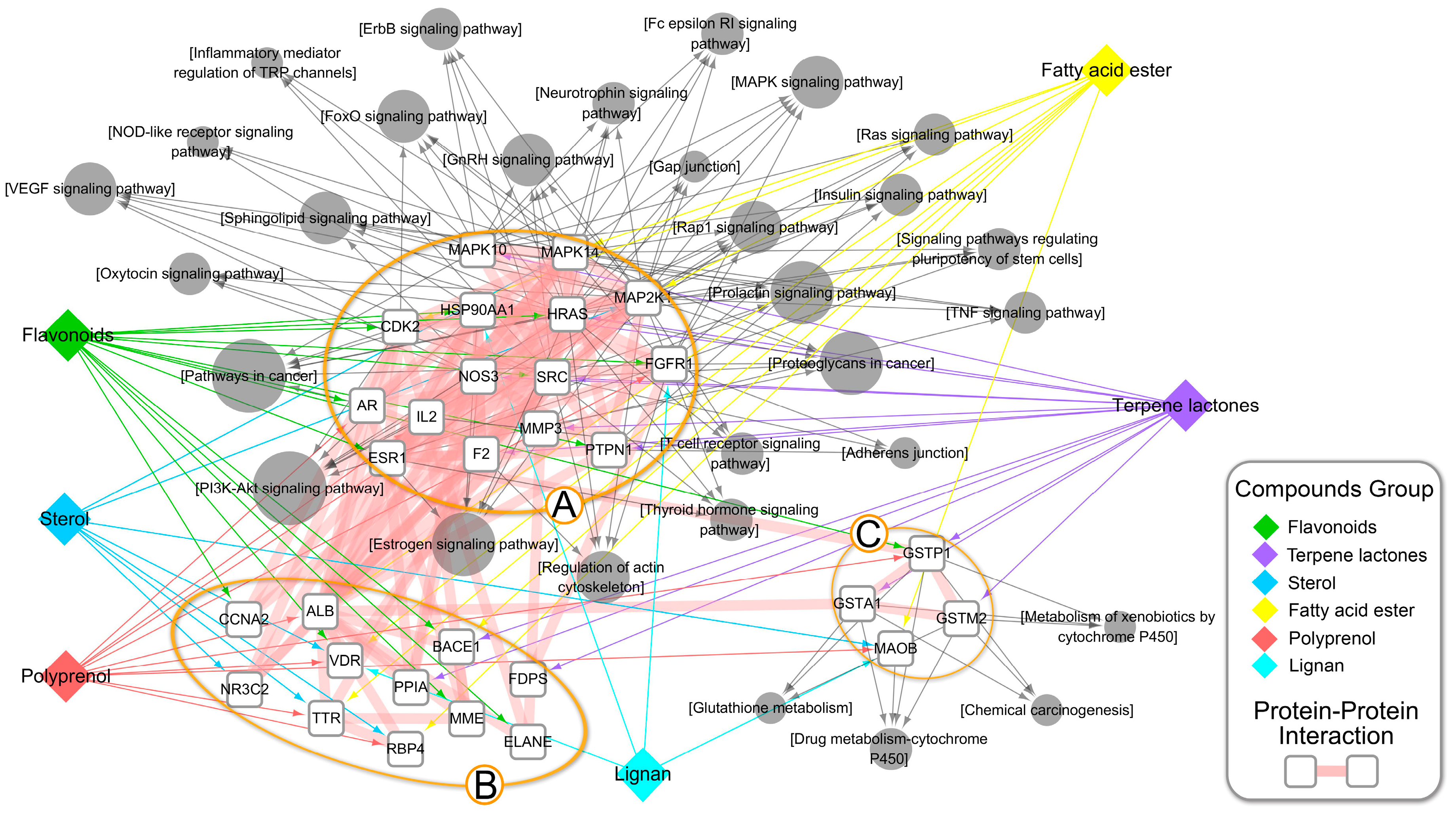
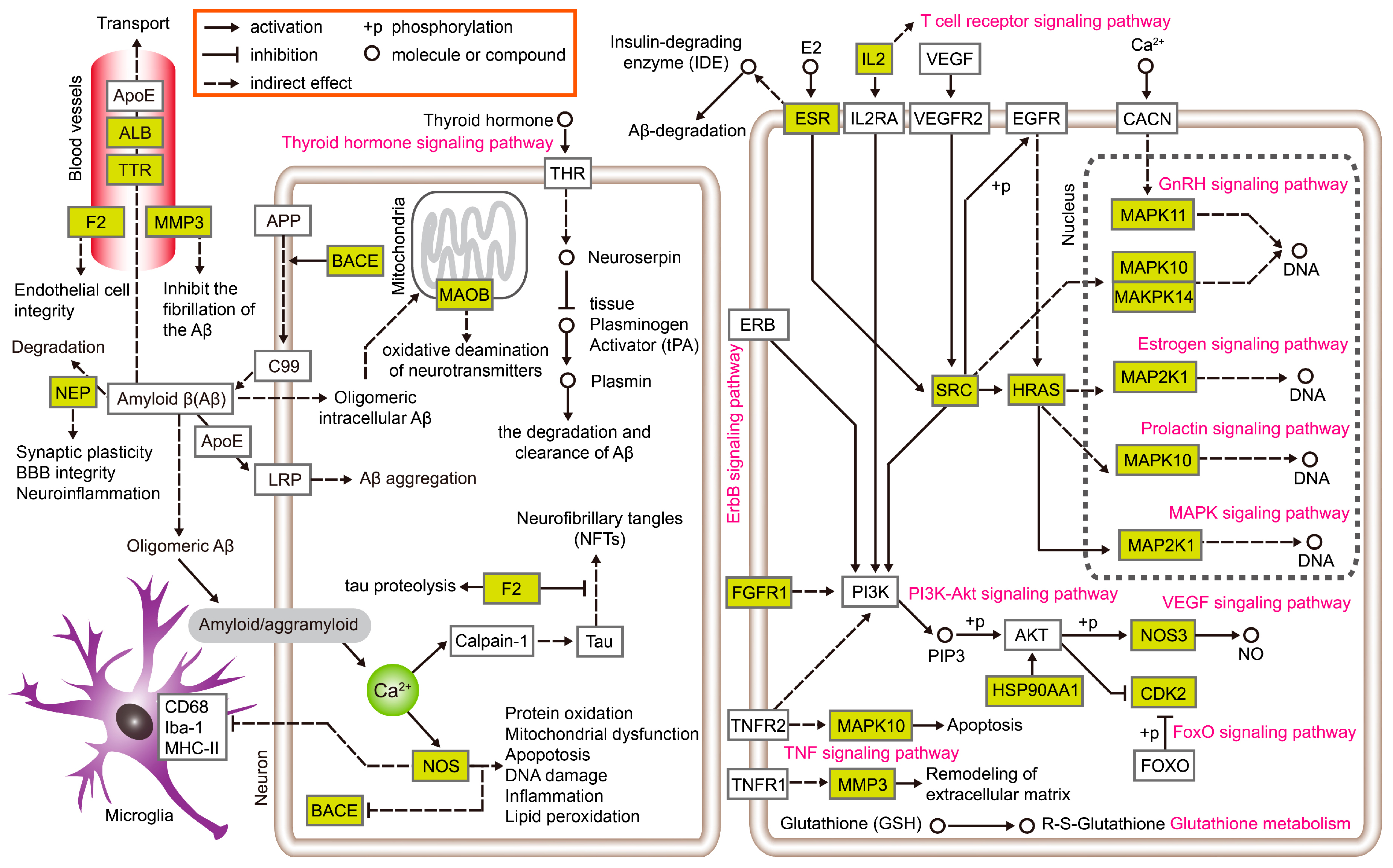
| Category | Biological Process Terms | p-Value | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|
| signal transduction | GO:0007165~signal transduction | 0.0001 | 0.0611 | 0.0611 | 0.1472 |
| GO:0043406~positive regulation of MAP kinase activity | 0.0001 | 0.0860 | 0.0439 | 0.2099 | |
| GO:0007265~Ras protein signal transduction | 0.0002 | 0.1388 | 0.0486 | 0.3487 | |
| GO:0010628~positive regulation of gene expression | 0.0010 | 0.4753 | 0.0774 | 1.4961 | |
| GO:0045944~positive regulation of transcription from RNA polymerase II promoter | 0.0248 | 1.0000 | 0.3845 | 31.2264 | |
| GO:0006367~transcription initiation from RNA polymerase II promoter | 0.0022 | 0.7597 | 0.1039 | 3.2779 | |
| GO:0018105~peptidyl-serine phosphorylation | 0.0196 | 1.0000 | 0.3428 | 25.4995 | |
| GO:0018108~peptidyl-tyrosine phosphorylation | 0.0023 | 0.7660 | 0.0986 | 3.3386 | |
| synthesis and metabolism | GO:0043651~linoleic acid metabolic process | 0.0004 | 0.2177 | 0.0479 | 0.5724 |
| GO:1901687~glutathione derivative biosynthetic process | 0.0007 | 0.3396 | 0.0668 | 0.9652 | |
| GO:0006749~glutathione metabolic process | 0.0042 | 0.9309 | 0.1632 | 6.0551 | |
| hormone related | GO:0043401~steroid hormone mediated signaling pathway | 0.0043 | 0.9371 | 0.1588 | 6.2622 |
| GO:0030520~intracellular estrogen receptor signaling pathway | 0.0357 | 1.0000 | 0.4649 | 41.7726 | |
| protein metabolic | GO:0006508~proteolysis | 0.0102 | 0.9986 | 0.2573 | 14.1863 |
| GO:0050435~beta-amyloid metabolic process | 0.0222 | 1.0000 | 0.3699 | 28.4455 | |
| GO:0044267~cellular protein metabolic process | 0.0176 | 1.0000 | 0.3417 | 23.1933 | |
| inflammatory cascade reaction | GO:0050900~leukocyte migration | 0.0012 | 0.5319 | 0.0731 | 1.7588 |
| GO:0048010~vascular endothelial growth factor receptor signaling pathway | 0.0068 | 0.9873 | 0.2053 | 9.7017 | |
| GO:0010544~negative regulation of platelet activation | 0.0137 | 0.9999 | 0.3182 | 18.6122 | |
| GO:0071222~cellular response to lipopolysaccharide | 0.0162 | 1.0000 | 0.3296 | 21.5741 | |
| GO:0051024~positive regulation of immunoglobulin secretion | 0.0188 | 1.0000 | 0.3415 | 24.6631 | |
| cell apoptosis | GO:0098869~cellular oxidant detoxification | 0.0002 | 0.1388 | 0.0486 | 0.3487 |
| GO:0071732~cellular response to nitric oxide | 0.0003 | 0.1520 | 0.0404 | 0.3845 | |
| GO:0038128~ERBB2 signaling pathway | 0.0019 | 0.7112 | 0.0983 | 2.8617 | |
| GO:2001237~negative regulation of extrinsic apoptotic signaling pathway | 0.0019 | 0.7112 | 0.0983 | 2.8617 | |
| GO:0050999~regulation of nitric-oxide synthase activity | 0.0440 | 1.0000 | 0.4785 | 48.8128 | |
| GO:0043410~positive regulation of MAPK cascade | 0.0086 | 0.9959 | 0.2399 | 12.0357 |
| Category | KEGG Pathway Terms | p-Value | Bonferroni | Benjamini | FDR |
|---|---|---|---|---|---|
| inflammation-generating process | hsa04370: VEGF signaling pathway | 0.0001 | 0.0083 | 0.0017 | 0.0712 |
| hsa04664: Fc epsilon RI signaling pathway | 0.0018 | 0.2196 | 0.0205 | 2.0929 | |
| hsa04660: T cell receptor signaling pathway | 0.0058 | 0.5537 | 0.0524 | 6.6494 | |
| hsa04668: TNF signaling pathway | 0.0063 | 0.5827 | 0.0532 | 7.1833 | |
| hsa04015: Rap1 signaling pathway | 0.0063 | 0.5848 | 0.0504 | 7.2230 | |
| hsa04621: NOD-like receptor signaling pathway | 0.0166 | 0.9007 | 0.0850 | 17.8802 | |
| hsa04750: Inflammatory mediator regulation of TRP channels | 0.0483 | 0.9989 | 0.1571 | 44.1896 | |
| apoptosis | hsa04151: PI3K-Akt signaling pathway | 0.0012 | 0.1492 | 0.0160 | 1.3682 |
| hsa04068: FoxO signaling pathway | 0.0012 | 0.1581 | 0.0155 | 1.4573 | |
| hsa04010: MAPK signaling pathway | 0.0124 | 0.8221 | 0.0755 | 13.6915 | |
| hsa00480: Glutathione metabolism | 0.0144 | 0.8645 | 0.0768 | 15.6705 | |
| hsa05205: Proteoglycans in cancer | 0.0006 | 0.0846 | 0.0110 | 0.7506 | |
| hsa05200: Pathways in cancer | 0.0023 | 0.2710 | 0.0240 | 2.6595 | |
| hsa04014: Ras signaling pathway | 0.0468 | 0.9987 | 0.1559 | 43.0804 | |
| hsa04012: ErbB signaling pathway | 0.0036 | 0.3945 | 0.0352 | 4.1891 | |
| hsa05204: Chemical carcinogenesis | 0.0334 | 0.9908 | 0.1255 | 32.9823 | |
| hormone synthesis and transport | hsa04917: Prolactin signaling pathway | 0.0000 | 0.0006 | 0.0006 | 0.0053 |
| hsa04915: Estrogen signaling pathway | 0.0000 | 0.0032 | 0.0008 | 0.0272 | |
| hsa04912: GnRH signaling pathway | 0.0003 | 0.0390 | 0.0057 | 0.3386 | |
| hsa04910: Insulin signaling pathway | 0.0130 | 0.8356 | 0.0755 | 14.2708 | |
| hsa04921: Oxytocin signaling pathway | 0.0187 | 0.9256 | 0.0918 | 19.8791 | |
| hsa04919: Thyroid hormone signaling pathway | 0.0077 | 0.6569 | 0.0547 | 8.7191 | |
| hsa04722: Neurotrophin signaling pathway | 0.0089 | 0.7084 | 0.0598 | 9.9765 | |
| hsa04071: Sphingolipid signaling pathway | 0.0008 | 0.1077 | 0.0126 | 0.9668 | |
| hsa04921: Oxytocin signaling pathway | 0.0187 | 0.9256 | 0.0918 | 19.8791 | |
| drug metabolism | hsa00982: Drug metabolism-cytochrome P450 | 0.0018 | 0.2196 | 0.0205 | 2.0929 |
| hsa00980: Metabolism of xenobiotics by cytochrome P450 | 0.0290 | 0.9827 | 0.1156 | 29.2357 | |
| ontogeny process | hsa04540: Gap junction | 0.0398 | 0.9963 | 0.1371 | 37.9955 |
| hsa04810: Regulation of actin cytoskeleton | 0.0065 | 0.5910 | 0.0484 | 7.3404 | |
| hsa04520: Adherens junction | 0.0268 | 0.9765 | 0.1106 | 27.3786 |
| Protein Targets | AD-Related Mechanisms/Etiology | Docking Compounds | Research Related to G. biloba for Anti-AD |
|---|---|---|---|
| Neprilysin (NEP) | (1) Aβ degradation enzymes [28,29] | (+)-Catechin Diosmetin Genkwanin | No research |
| (2) maintain blood–brain barrier (BBB) integrity | |||
| (3) participate in neuroinflammation [50] | |||
| Estrogen receptor (ESR) | (1) upregulated insulin-degrading enzyme (IDE) [51] | Genkwanin | No research |
| (2) maintaining steroid homeostasis [52] | |||
| (3) altering synaptic plasticity [53,54] | |||
| (4) participate in neurons oxidative stress-mediated injury [55] | |||
| Prothrombin (F2) | (1) coagulation cascade and endothelial cell integrity [38] | Ginkgolide J | No research |
| (2) ideal molecular-biological indicator for AD [56] | |||
| (3) proteolyzes the microtubule-associated protein tau [56] | |||
| (4) inhibits phosphorylation of tau [56] | |||
| Serum albumin (ALB) | (1) bounded and transported Aβ, maintaining a constant concentration level in the brain [39] | Ethyl oleate Flavoxanthin | No research |
| (2) Aβ excretion from the brain to the blood [57,58] | |||
| Thyroid hormone (TTR) | (1) up-regulation of expression of neuroserpin in neurons [32] | Beta-sitosterol Stigmasterol Mandenol Ethyl oleate Flavoxanthin | No research |
| (2) hyperthyroidism increases the risk of AD [59] | |||
| Matrix metalloproteinase 3 (MMP3) | (1) main Plasma gelsolin (GSN)-degrading enzyme [40] | Ginkgolide B Ginkgolide J | No research |
| (2) inhibits the fibrillation of the Aβ [60] | |||
| (3) a diagnostic biomarker for AD [61] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Sun, X.; Yu, F.; Xu, L.; Miu, J.; Xiao, P. In Silico Investigation of the Pharmacological Mechanisms of Beneficial Effects of Ginkgo biloba L. on Alzheimer’s Disease. Nutrients 2018, 10, 589. https://doi.org/10.3390/nu10050589
Li H, Sun X, Yu F, Xu L, Miu J, Xiao P. In Silico Investigation of the Pharmacological Mechanisms of Beneficial Effects of Ginkgo biloba L. on Alzheimer’s Disease. Nutrients. 2018; 10(5):589. https://doi.org/10.3390/nu10050589
Chicago/Turabian StyleLi, Hongxiang, Xiaoyuan Sun, Fan Yu, Lijia Xu, Jianhua Miu, and Peigen Xiao. 2018. "In Silico Investigation of the Pharmacological Mechanisms of Beneficial Effects of Ginkgo biloba L. on Alzheimer’s Disease" Nutrients 10, no. 5: 589. https://doi.org/10.3390/nu10050589
APA StyleLi, H., Sun, X., Yu, F., Xu, L., Miu, J., & Xiao, P. (2018). In Silico Investigation of the Pharmacological Mechanisms of Beneficial Effects of Ginkgo biloba L. on Alzheimer’s Disease. Nutrients, 10(5), 589. https://doi.org/10.3390/nu10050589