Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Culture Media, and Cultivation
2.2. Carbohydrate Fermentation Assays
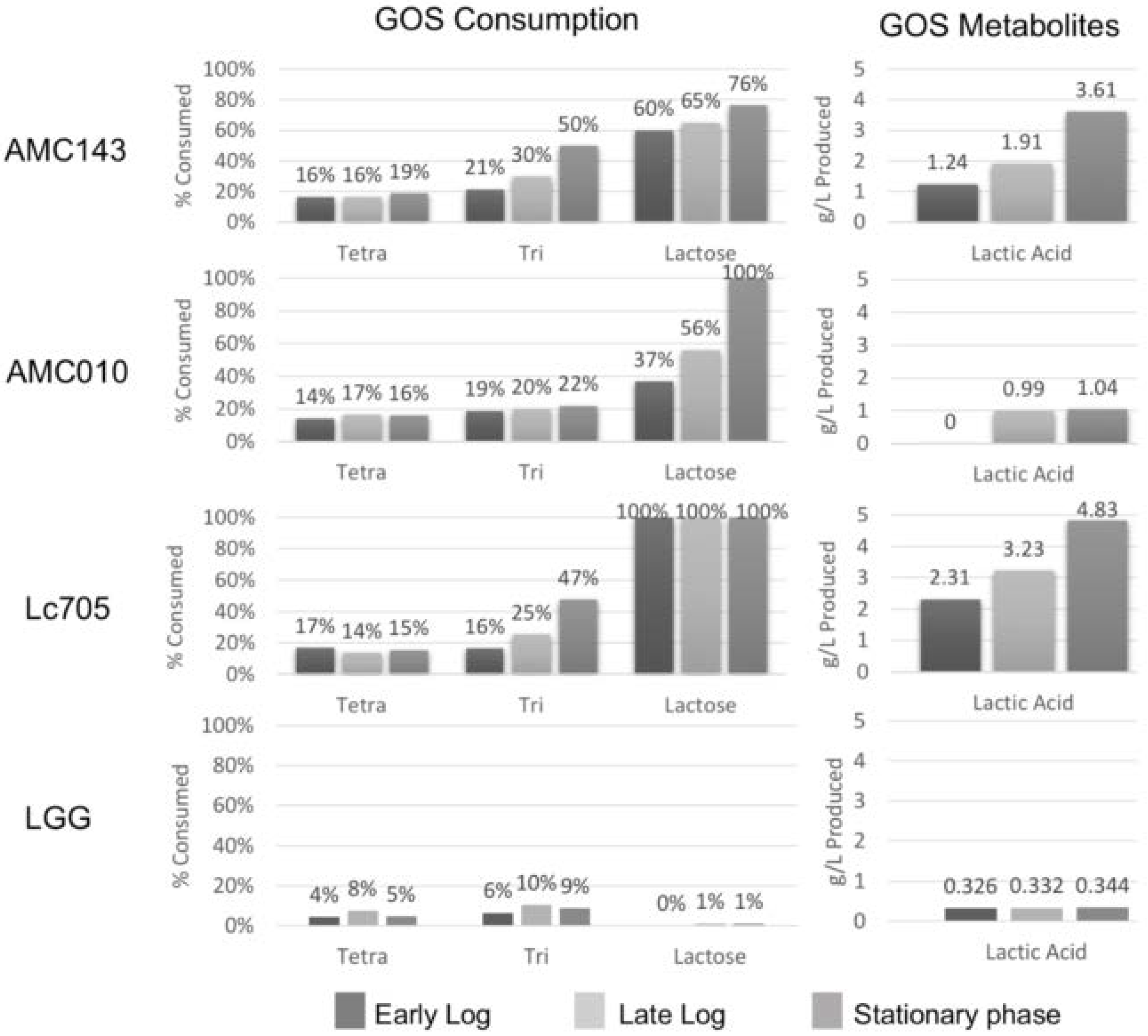
2.3. Determination of GOS Metabolites
2.4. RNA Isolation
2.5. mRNA Sequencing
2.6. mRNA Sequencing Data Analysis
2.7. Generation of GOS Operon Insertion Mutant Strains
3. Results
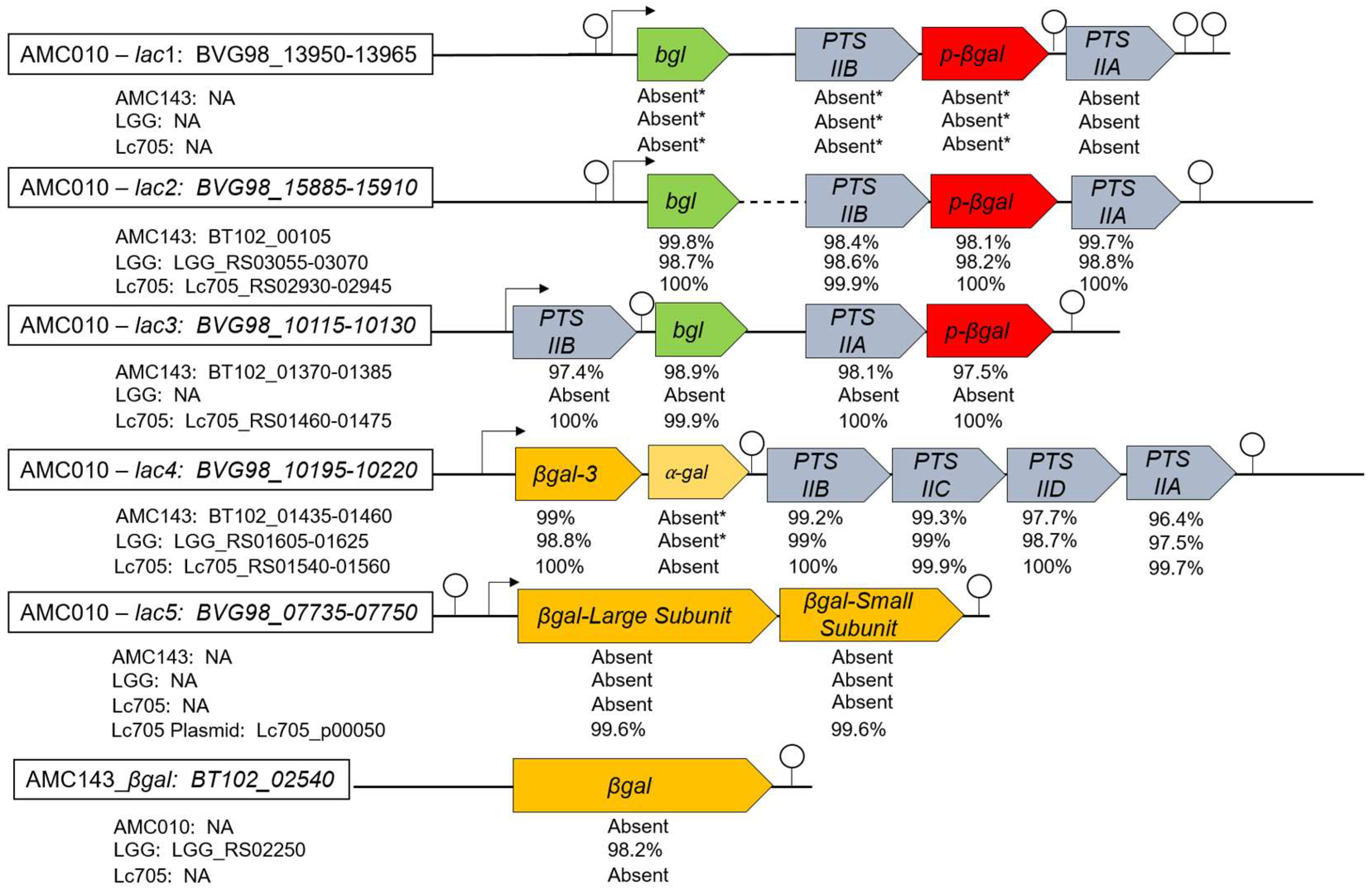
3.1. Comparative Genomic Analysis of GOS Utilization Genes
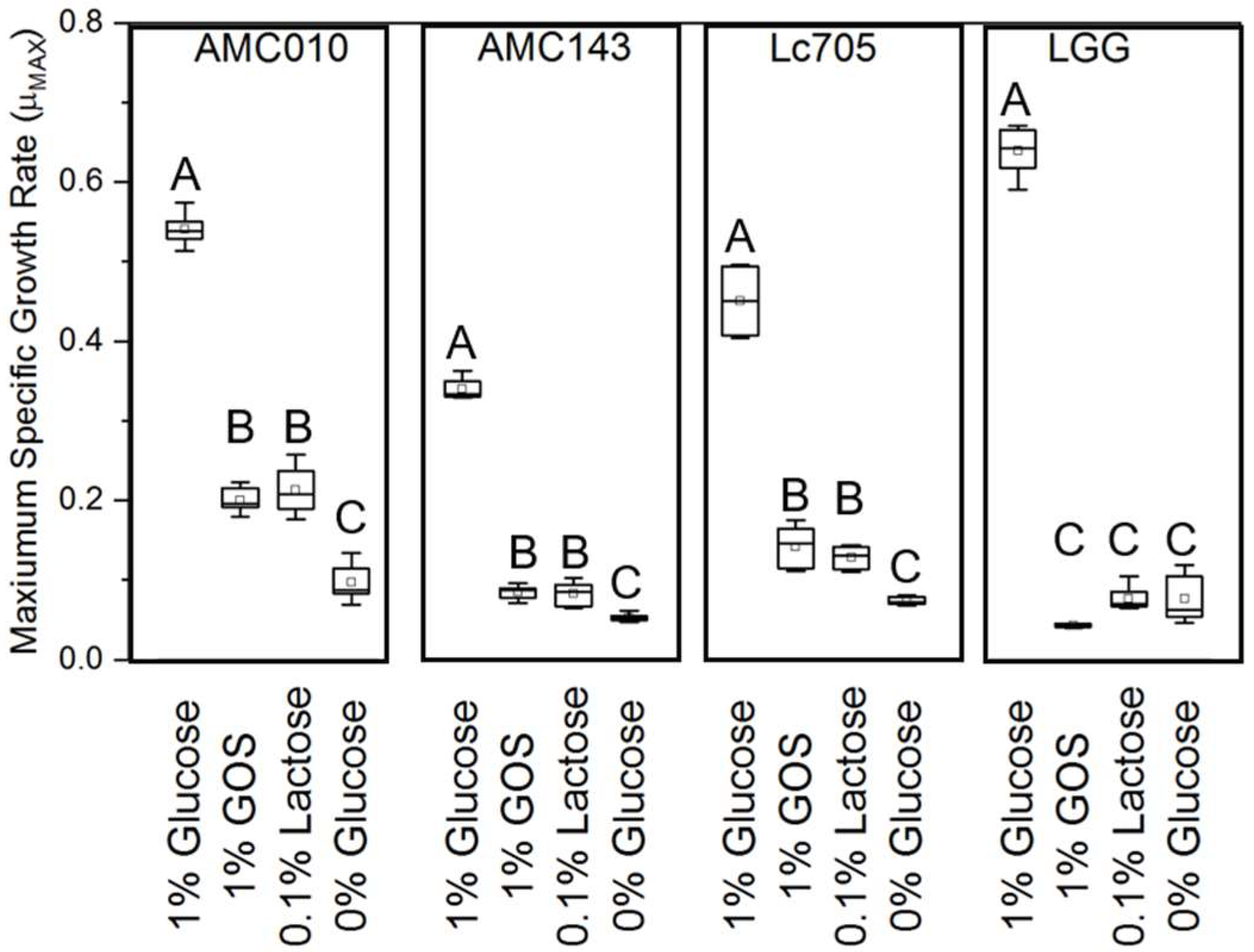
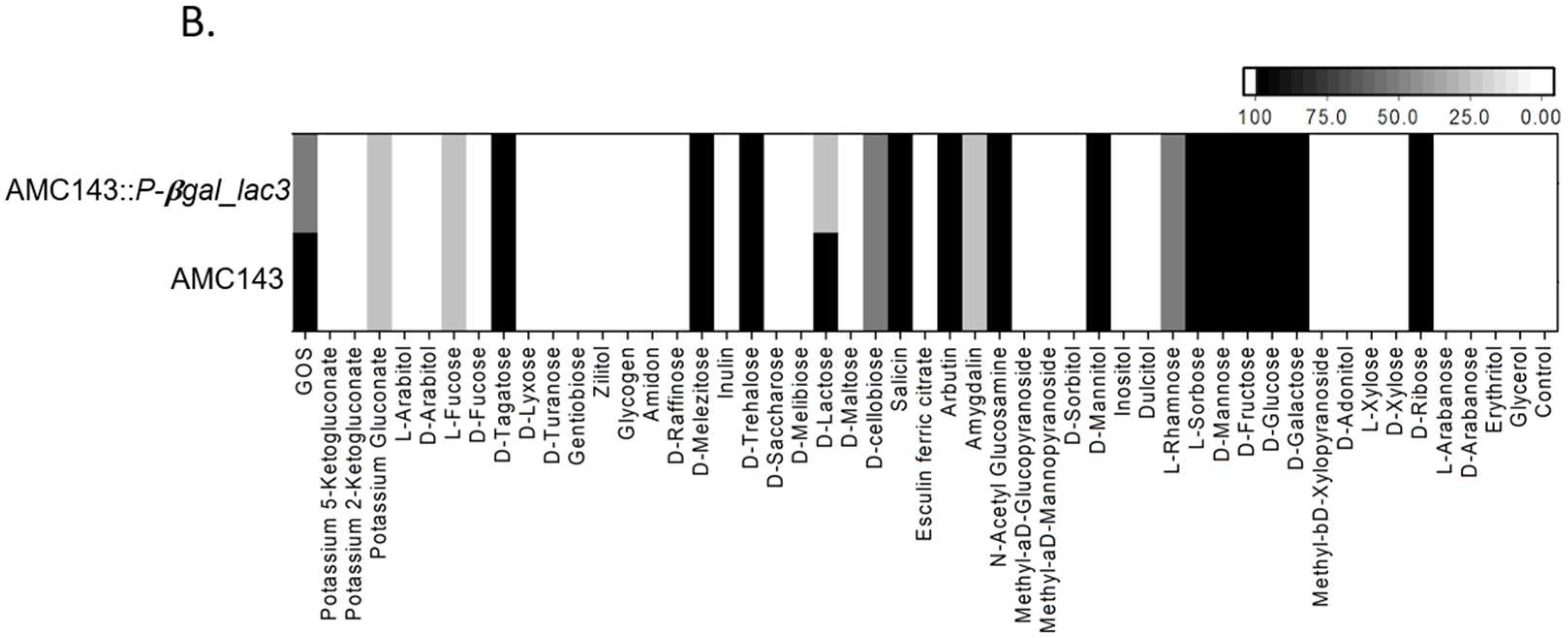
3.2. Carbohydrate Utilization by L. rhamnosus
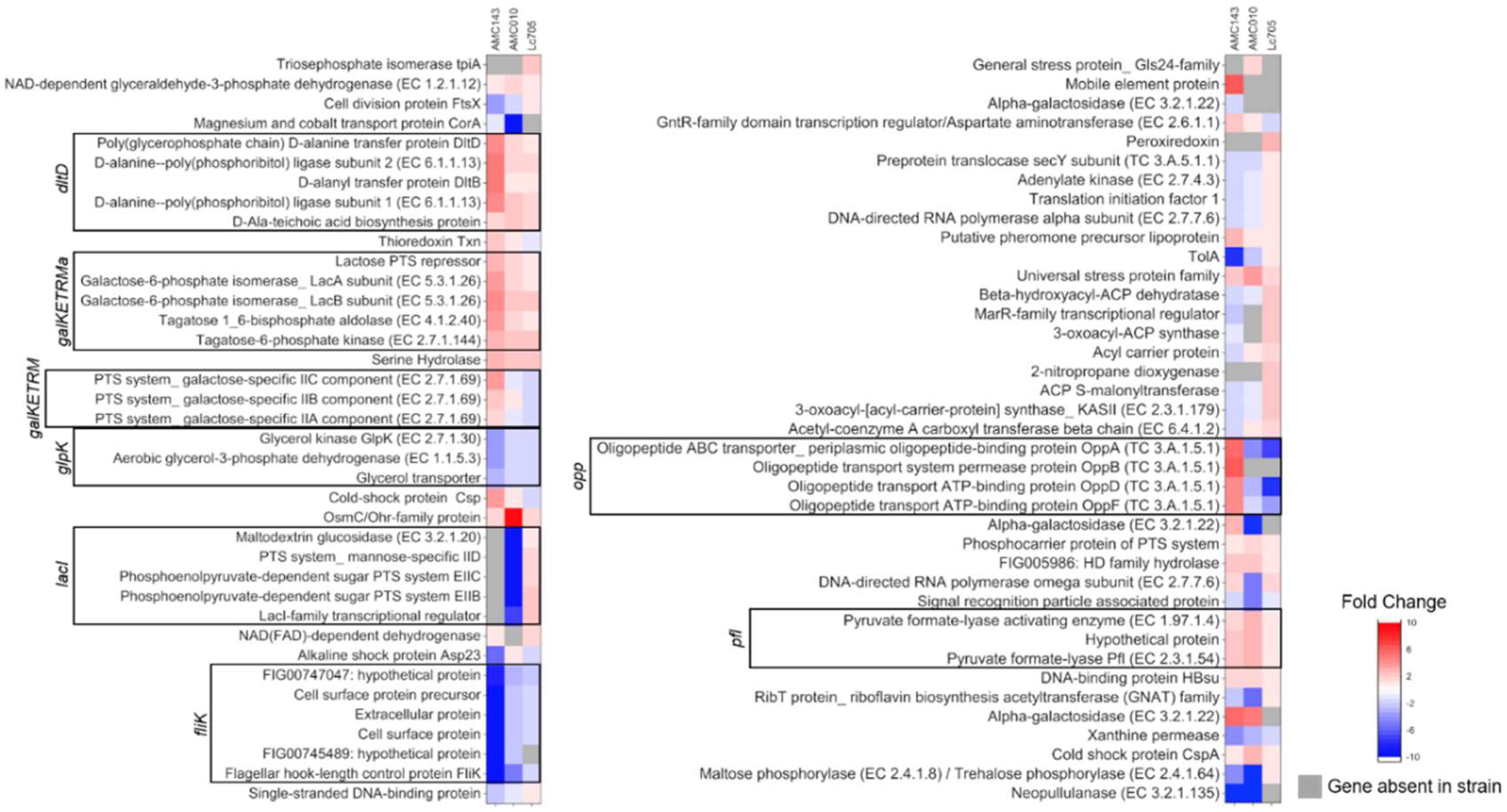
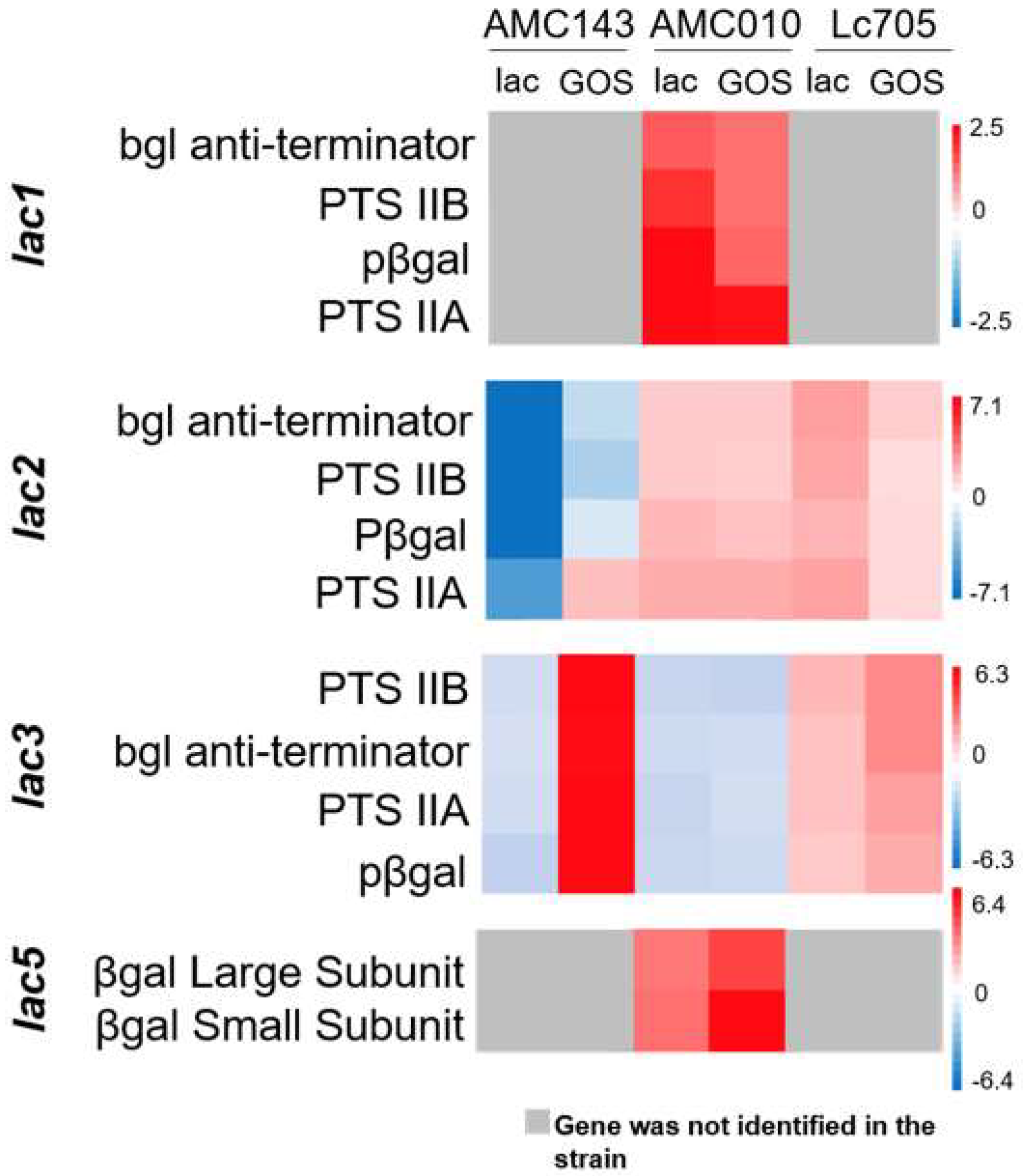
3.3. Gene Expression Analysis of L. rhamnosus in Medium Containing GOS as a Carbon Source
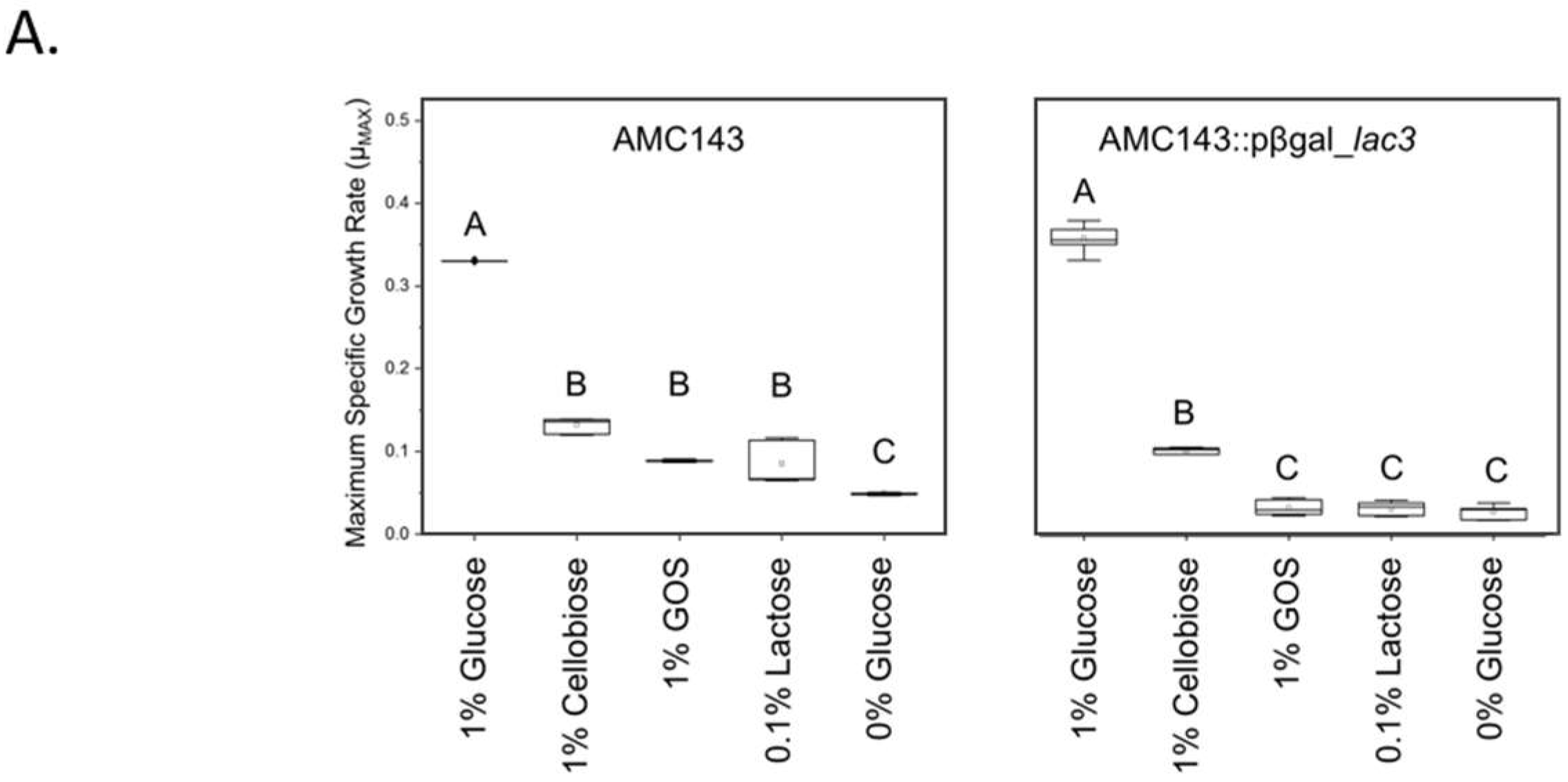
3.4. Functional Characterization of lac3, A GOS-Specific Operon of L. rhamnosus AMC143
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deng, Y.; Misselwitz, B.; Dai, N.; Fox, M. Lactose intolerance in adults: Biological mechanism and dietary management. Nutrients 2015, 7, 8020–8035. [Google Scholar] [CrossRef] [PubMed]
- Oak, S.J.; Jha, R. The effects of probiotics in lactose intolerance: A systematic review. Crit. Rev. Food Sci. Nutr. 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Azcarate-Peril, M.A.; Ritter, A.J.; Savaiano, D.; Monteagudo-Mera, A.; Anderson, C.; Magness, S.T.; Klaenhammer, T.R. Impact of short-chain galactooligosaccharides on the gut microbiome of lactose-intolerant individuals. Proc. Natl. Acad. Sci. USA 2017, 114, E367–E375. [Google Scholar] [CrossRef] [PubMed]
- Savaiano, D.A.; Ritter, A.J.; Klaenhammer, T.R.; James, G.M.; Longcore, A.T.; Chandler, J.R.; Walker, W.A.; Foyt, H.L. Improving lactose digestion and symptoms of lactose intolerance with a novel galacto-oligosaccharide (RP-G28): A randomized, double-blind clinical trial. Nutr. J. 2013, 12, 160. [Google Scholar] [CrossRef] [PubMed]
- Azcarate Peril, M.A.; Savaiano, D.A.; Ritter, A.J.; Klaenhammer, T. Microbiome alterations of lactose intolerant individuals in response to dietary intervention with galacto-oligosaccharides may help negate symptoms of lactose intolerance. Gastroenterology 2013, 144, S-893. [Google Scholar] [CrossRef]
- Davis, L.M.; Martinez, I.; Walter, J.; Goin, C.; Hutkins, R.W. Barcoded pyrosequencing reveals that consumption of galactooligosaccharides results in a highly specific bifidogenic response in humans. PLoS ONE 2011, 6, e25200. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.P.M.; Gonçalves, M.D.P.F.; Teixeira, J.A.; Rodrigues, L.R. Galacto-oligosaccharides: Production, properties, applications, and significance as prebiotics. Compr. Rev. Food Sci. Food Saf. 2010, 9, 438–454. [Google Scholar] [CrossRef]
- Chey, W.D.; Sandborn, W.J.; Ritter, A.J.; Foyt, H.; Azcarate-Peril, M.A.; Savaiano, D.A. Multiple clinical outcomes improve in lactose intolerance patients after treatment with novel galacto-oligosaccharide, RP-G28. Unpublished work. 2018. [Google Scholar]
- Cardelle-Cobas, A.; Corzo, N.; Olano, A.; Pelaez, C.; Requena, T.; Avila, M. Galactooligosaccharides derived from lactose and lactulose: Influence of structure on Lactobacillus, Streptococcus and Bifidobacterium growth. Int. J. Food Microbiol. 2011, 149, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, R.; Calasso, M.; Calace, L.; Siragusa, S.; Ndagijimana, M.; Vernocchi, P.; Brunetti, L.; Mancino, G.; Tedeschi, G.; Guerzoni, E.; et al. Effect of lactose on gut microbiota and metabolome of infants with cow’s milk allergy. Pediatr. Allergy Immunol. Off. Publ. Eur. Soc. Pediatr. Allergy Immunol. 2012, 23, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, M.E.; Williams, H.J.; Hillerich, B.; Almo, S.C.; Raushel, F.M. L-galactose metabolism in Bacteroides vulgatus from the human gut microbiota. Biochemistry 2014, 53, 4661–4670. [Google Scholar] [CrossRef] [PubMed]
- Do, M.H.; Lee, E.; Oh, M.J.; Kim, Y.; Park, H.Y. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 2018, 10, 761. [Google Scholar] [CrossRef] [PubMed]
- Lebeer, S.; Claes, I.; Tytgat, H.L.; Verhoeven, T.L.; Marien, E.; von Ossowski, I.; Reunanen, J.; Palva, A.; Vos, W.M.; Keersmaecker, S.C.; et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012, 78, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Medina, M.; Izquierdo, E.; Ennahar, S.; Sanz, Y. Differential immunomodulatory properties of Bifidobacterium longum strains: Relevance to probiotic selection and clinical applications. Clin. Exp. Immunol. 2007, 150, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.A.N.; Lawley, T.D. Emerging insights on intestinal dysbiosis during bacterial infections. Curr. Opin. Microbiol. 2014, 17, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Hudault, S.; Lievin, V.; Bernet-Camard, M.F.; Servin, A.L. Antagonistic activity exerted in vitro and in vivo by Lactobacillus casei (strain GG) against Salmonella typhimurium c5 infection. Appl. Environ. Microbiol. 1997, 63, 513–518. [Google Scholar] [PubMed]
- Tang, J. Microbial metabolomics. Curr. Genom. 2011, 12, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Ursell, L.K.; Haiser, H.J.; Van Treuren, W.; Garg, N.; Reddivari, L.; Vanamala, J.; Dorrestein, P.C.; Turnbaugh, P.J.; Knight, R. The intestinal metabolome: An intersection between microbiota and host. Gastroenterology 2014, 146, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R. Nutritional modulation of gene expression: Might this be of benefit to individuals with crohn’s disease? Front. Immunol. 2015, 6, 467. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.J.; Guinane, C.M.; O’Toole, P.W.; Cotter, P.D. Beneficial modulation of the gut microbiota. FEBS Lett. 2014, 588, 4120–4130. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.U. Clinical uses of probiotics. Medicine 2016, 95, e2658. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; Jass, J.; Sebulsky, M.T.; McCormick, J.K. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 2003, 16, 658–672. [Google Scholar] [CrossRef] [PubMed]
- Corzo, N.; Alonso, J.L.; Azpiroz, F.; Calvo, M.A.; Cirici, M.; Leis, R.; Lombo, F.; Mateos-Aparicio, I.; Plou, F.J.; Ruas-Madiedo, P.; et al. Prebiotics: Concept, properties and beneficial effects. Nutr. Hosp. 2015, 31 (Suppl. 1), 99–118. [Google Scholar] [PubMed]
- Reid, G.; Sanders, M.E.; Gaskins, H.R.; Gibson, G.R.; Mercenier, A.; Rastall, R.; Roberfroid, M.; Rowland, I.; Cherbut, C.; Klaenhammer, T.R. New scientific paradigms for probiotics and prebiotics. J. Clin. Gastroenterol. 2003, 37, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Taylor, C.; Lawley, B.; Loach, D.; Gould, M.; Dunn, A.C.; McLellan, A.D.; Black, M.A.; McNoe, L.; Dekker, J.; et al. Altered transcription of murine genes induced in the small bowel by administration of probiotic strain Lactobacillus rhamnosus HN001. Appl. Environ. Microbiol. 2014, 80, 2851–2859. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.M.; Gibson, G.R.; Rowland, I. In vitro evaluation of single- and multi-strain probiotics: Inter-species inhibition between probiotic strains, and inhibition of pathogens. Anaerobe 2012, 18, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Hutt, P.; Shchepetova, J.; Loivukene, K.; Kullisaar, T.; Mikelsaar, M. Antagonistic activity of probiotic lactobacilli and bifidobacteria against entero- and uropathogens. J. Appl. Microbiol. 2006, 100, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- De Vrese, M.; Schrezenmeir, J. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 2008, 111, 1–66. [Google Scholar] [PubMed]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C. Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Boil. Rev. 2008, 72, 728–764. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Andriantsoanirina, V.; Allano, S.; Butel, M.J.; Aires, J. Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe 2013, 21, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Papadimitriou, K.; Alegria, A.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.W.; Simpson, J.B.; Roach, J.; Kwintkiewicz, J.; Azcarate-Peril, M.A. Intra-species genomic and physiological variability impact stress resistance in strains of probiotic potential. Front. Microbiol. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Markowiak, P.; Śliżewska, K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients 2017, 9, 2021. [Google Scholar]
- Zhang, C.; Zhao, L. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med. 2016, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Greenblum, S.; Carr, R.; Borenstein, E. Extensive strain-level copy-number variation across human gut microbiome species. Cell 2015, 160, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.M.; Barrangou, R.; Abou Hachem, M.; Lahtinen, S.; Goh, Y.J.; Svensson, B.; Klaenhammer, T.R. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 2011, 108, 17785–17790. [Google Scholar] [CrossRef] [PubMed]
- Arnold, J.W.; Monteagudo-Mera, A.; Altermann, E.; Cadenas, M.B.; Thompson, A.L.; Azcarate-Peril, M.A. Genome sequences of potential probiotic isolates from human infants. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Thompson, A.L.; Monteagudo-Mera, A.; Cadenas, M.B.; Lampl, M.L.; Azcarate-Peril, M. Milk-and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front. Cell. Infect. Microbiol. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Jacobus, N.V.; Deneke, C.; Gorbach, S.L. Antimicrobial substance from a human Lactobacillus strain. Antimicrob. Agents Chemother. 1987, 31, 1231–1233. [Google Scholar] [CrossRef] [PubMed]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; von Ossowski, I.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef] [PubMed]
- Bruno-Barcena, J.M.; Azcarate-Peril, M.A. Galacto-oligosaccharides and colorectal cancer: Feeding our intestinal probiome. J. Funct. Foods 2015, 12, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Dagher, S.F.; Azcarate-Peril, M.A.; Bruno-Barcena, J.M. Heterologous expression of a bioactive β-hexosyltransferase, an enzyme producer of prebiotics, from Sporobolomyces singularis. Appl. Environ. Microbiol. 2013, 79, 1241–1249. [Google Scholar] [CrossRef] [PubMed]
- Welker, D.L.; Hughes, J.E.; Steele, J.L.; Broadbent, J.R. High efficiency electrotransformation of Lactobacillus casei. FEMS Microbiol. Lett. 2015, 362, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Han, K.S.; Oh, S.; You, S.; Kim, S.H. Optimization of technical conditions for the transformation of lactobacillus acidophilus strains by electroporation. J. Appl. Microbiol. 2005, 99, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Thongaram, T.; Hoeflinger, J.L.; Chow, J.; Miller, M.J. Prebiotic galactooligosaccharide metabolism by probiotic lactobacilli and bifidobacteria. J. Agric. Food Chem. 2017, 65, 4184–4192. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Swarnkar, M.K.; Kumar, S.; Singh, A.K.; Gupta, M. Complete genome sequence of potential probiotic Lactobacillus sp. Hfc8, isolated from human gut using PacBio SMRT sequencing. Genome Announc. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.; Salamov, A. Automatic annotation of microbial genomes and metagenomic sequences in metagenomics and its applications in agriculture. Biomed. Environ. Stud. 2011, 18, 61–78. [Google Scholar]
- Gautheret, D.; Lambert, A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001, 313, 1003–1011. [Google Scholar] [CrossRef] [PubMed]
- Bettenbrock, K.; Siebers, U.; Ehrenreich, P.; Alpert, C.A. Lactobacillus casei 64h contains a phosphoenolpyruvate-dependent phosphotransferase system for uptake of galactose, as confirmed by analysis of ptsh and different gal mutants. J. Bacteriol. 1999, 181, 225–230. [Google Scholar] [PubMed]
- Honda, H.; Nagaoka, S.; Kawai, Y.; Kemperman, R.; Kok, J.; Yamazaki, Y.; Tateno, Y.; Kitazawa, H.; Saito, T. Purification and characterization of two phospho-β-galactosidases, lacg1 and lacg2, from Lactobacillus gasseri ATCC33323(t). J. Gen. Appl. Microbiol. 2012, 58, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Takala, T.M.; Saris, P.E.; Tynkkynen, S.S. Food-grade host/vector expression system for Lactobacillus casei based on complementation of plasmid-associated phospho-β-galactosidase gene lacg. Appl. Microbiol. Biotechnol. 2003, 60, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Suzuki, M.; Konno, K.; Kitazawa, H.; Kawai, Y.; Itoh, T.; Kamio, Y. Molecular cloning and sequencing of two phospho-β-galactosidase I and II genes of Lactobacillus gasseri JCM1031 isolated from human intestine. Biosci. Biotechnol. Biochem. 1998, 62, 2318–2327. [Google Scholar] [CrossRef] [PubMed]
- Bidart, G.N.; Rodriguez-Diaz, J. The lactose operon from Lactobacillus casei is involved in the transport and metabolism of the human milk oligosaccharide core-2 n-acetyllactosamine. Sci. Rep. 2018, 8, 7152. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Saito, T.; Itoh, T. Coexistence of two kinds of 6-phospho-β-galactosidase in the cytosol of Lactobacillus gasseri JCM1031--purification and characterization of 6-phospho-β-galactosidase II. Biosci. Biotechnol. Biochem. 1996, 60, 708–710. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Saito, T.; Itoh, T. Purification and characterization of 6-phospho-β-galactosidase from Lactobacillus gasseri JCM1031. Biosci. Biotechnol. Biochem. 1996, 60, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Bruno-Barcena, J.M.; Azcarate-Peril, M.A.; Klaenhammer, T.R.; Hassan, H.M. Marker-free chromosomal integration of the manganese superoxide dismutase gene (soda) from streptococcus thermophilus into Lactobacillus gasseri. FEMS Microbiol. Lett. 2005, 246, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.H.; Choi, S.H.; Park, S.W.; Choi, N.J.; Kim, Y.; Kim, S.H. Synbiotic impact of tagatose on viability of Lactobacillus rhamnosus strain GG mediated by the phosphotransferase system (PTS). Food Microbiol. 2013, 36, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ganzle, M.G.; Follador, R. Metabolism of oligosaccharides and starch in lactobacilli: A review. Front. Microbiol. 2012, 3, 340. [Google Scholar] [CrossRef] [PubMed]
- Silvestroni, A.; Connes, C.; Sesma, F.; De Giori, G.S.; Piard, J.C. Characterization of the mela locus for α-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 2002, 68, 5464–5471. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.A.; Phillips, R.S.; Kilgore, P.B.; Smith, G.L.; Hoover, T.R. A mannose family phosphotransferase system permease and associated enzymes are required for utilization of fructoselysine and glucoselysine in Salmonella enterica serovar typhimurium. J. Bacteriol. 2015, 197, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Vallino, J.J.; Hopkinson, C.S.; Hobbie, J.E. Modeling bacterial utilization of dissolved organic matter: Optimization replaces monod growth kinetics. Limnol. Oceanogr. 1996, 41, 1591–1609. [Google Scholar] [CrossRef]
- Ceapa, C.; Davids, M.; Ritari, J.; Lambert, J.; Wels, M.; Douillard, F.P.; Smokvina, T.; de Vos, W.M.; Knol, J.; Kleerebezem, M. The variable regions of Lactobacillus rhamnosus genomes reveal the dynamic evolution of metabolic and host-adaptation repertoires. Genome Boil. Evol. 2016, 8, 1889–1905. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guedon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [PubMed]
- Chassy, B.M.; Alpert, C.A. Molecular characterization of the plasmid-encoded lactose-pts of Lactobacillus casei. FEMS Microbiol. Rev. 1989, 5, 157–165. [Google Scholar] [CrossRef]
- Henrich, A.; Kuhlmann, N.; Eck, A.W.; Kramer, R.; Seibold, G.M. Maltose uptake by the novel abc transport system musEFGK2I causes increased expression of ptsG in Corynebacterium glutamicum. J. Bacteriol. 2013, 195, 2573–2584. [Google Scholar] [CrossRef] [PubMed]
- Krumbeck, J.A.; Rasmussen, H.E.; Hutkins, R.W.; Clarke, J.; Shawron, K.; Keshavarzian, A.; Walter, J. Probiotic Bifidobacterium strains and galactooligosaccharides improve intestinal barrier function in obese adults but show no synergism when used together as synbiotics. Microbiome 2018, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Förster-Fromme, K.; Damms-Machado, A.; Scheurenbrand, T.; Biskup, S.; Huson, D.H.; Bischoff, S.C. Analysis of the intestinal microbiota using solid 16S rRNA gene sequencing and solid shotgun sequencing. BMC Genom. 2013, 14, S16. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
| Strain | Origin | Reference |
|---|---|---|
| L. rhamnosus GG | Healthy human isolate | [41] |
| L. rhamnosus Lc705 | Fermented dairy product | [41] |
| L. rhamnosus AMC010 | Human infant stool | [38] |
| L. rhamnosus AMC143 | Human infant stool | [38] |
| L. rhamnosus AMC143::P-βgal_lac3 | This study |
| Strain | Carbohydrate Transported | |||||||
|---|---|---|---|---|---|---|---|---|
| α-Glucoside | Ascorbate | β-Glucoside | Cellobiose | Fructose | Galactitol | Galactosamine | Galactose | |
| LC705 | 1 | 8 | 5 | 9 | 18 | 5 | 0 | 0 |
| LGG | 0 | 5 | 3 | 10 | 12 | 9 | 0 | 0 |
| AMC143 | 0 | 0 | 5 | 24 | 8 | 3 | 4 | 6 |
| AMC010 | 0 | 0 | 4 | 14 | 10 | 5 | 4 | 6 |
| Glucitol | Hyaluronate-Oligosaccharide | Uncharacterized | Glucose | Lactose | Maltose | Mannitol | Mannose | |
| LC705 | 1 | 0 | 10 | 4 | 6 | 1 | 2 | 8 |
| LGG | 1 | 0 | 7 | 3 | 6 | 0 | 2 | 13 |
| AMC143 | 4 | 2 | 7 | 0 | 5 | 2 | 4 | 15 |
| AMC010 | 4 | 2 | 3 | 0 | 8 | 4 | 4 | 13 |
| Mannose/Fructose/Sorbose | N-acetyl galactosamine | N-acetyl glucosamine | Sorbitol | Sorbose | Trehalose | Sucrose | Total | |
| LC705 | 2 | 3 | 3 | 2 | 3 | 1 | 0 | 46 |
| LGG | 1 | 0 | 0 | 2 | 2 | 1 | 1 | 39 |
| AMC143 | 0 | 2 | 0 | 0 | 3 | 1 | 4 | 50 |
| AMC010 | 0 | 2 | 0 | 0 | 3 | 2 | 4 | 43 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnold, J.W.; Simpson, J.B.; Roach, J.; Bruno-Barcena, J.M.; Azcarate-Peril, M.A. Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus. Nutrients 2018, 10, 1517. https://doi.org/10.3390/nu10101517
Arnold JW, Simpson JB, Roach J, Bruno-Barcena JM, Azcarate-Peril MA. Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus. Nutrients. 2018; 10(10):1517. https://doi.org/10.3390/nu10101517
Chicago/Turabian StyleArnold, Jason W., Joshua B. Simpson, Jeffery Roach, Jose M. Bruno-Barcena, and M. Andrea Azcarate-Peril. 2018. "Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus" Nutrients 10, no. 10: 1517. https://doi.org/10.3390/nu10101517
APA StyleArnold, J. W., Simpson, J. B., Roach, J., Bruno-Barcena, J. M., & Azcarate-Peril, M. A. (2018). Prebiotics for Lactose Intolerance: Variability in Galacto-Oligosaccharide Utilization by Intestinal Lactobacillus rhamnosus. Nutrients, 10(10), 1517. https://doi.org/10.3390/nu10101517





