Effect of Low-Fat Diet in Obese Mice Lacking Toll-like Receptors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. Histological Examination
2.3. Deep Sequencing of Small RNA
2.4. Determination of Differentially Expressed miRNA
2.5. Target Prediction and Functional Annotation of Differentially Expressed miRNA
2.6. Statistical Analysis
3. Results
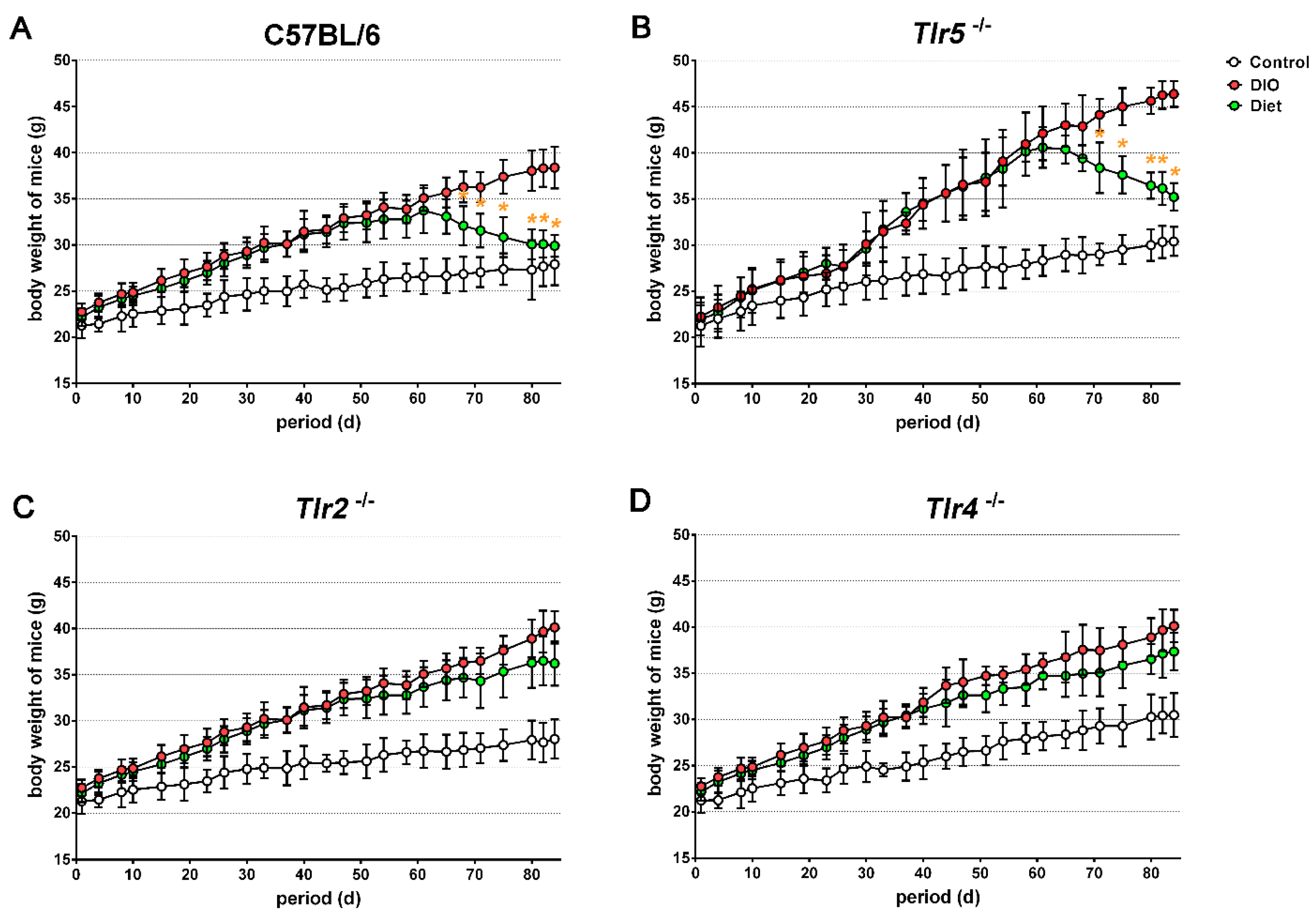
3.1. Body Weight Reduction by Low-Fat Diet (LFD)
3.2. Adiposity in Response to Body Weight Reduction
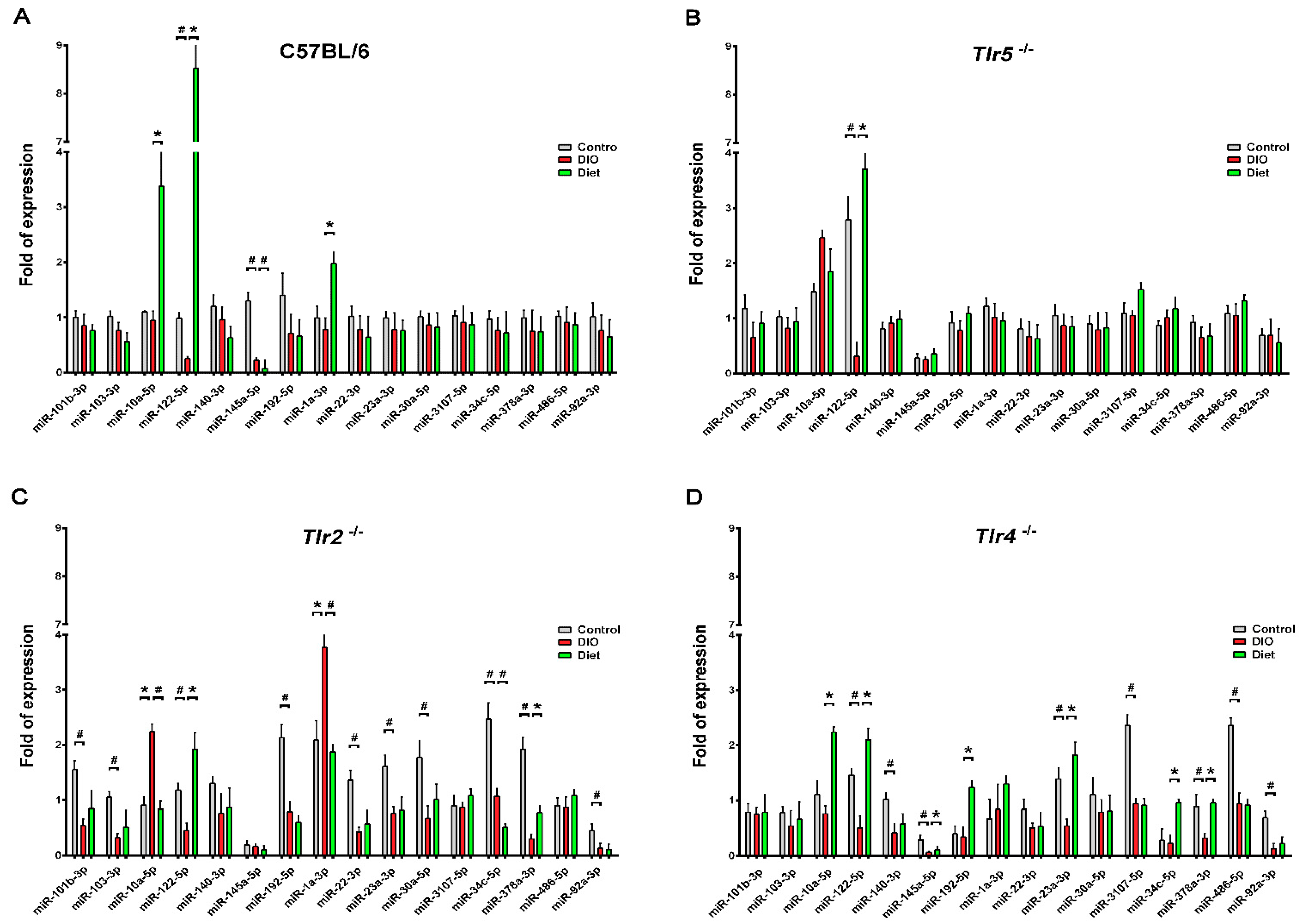
3.3. Expression of miRNAs in the Epididymal Fat
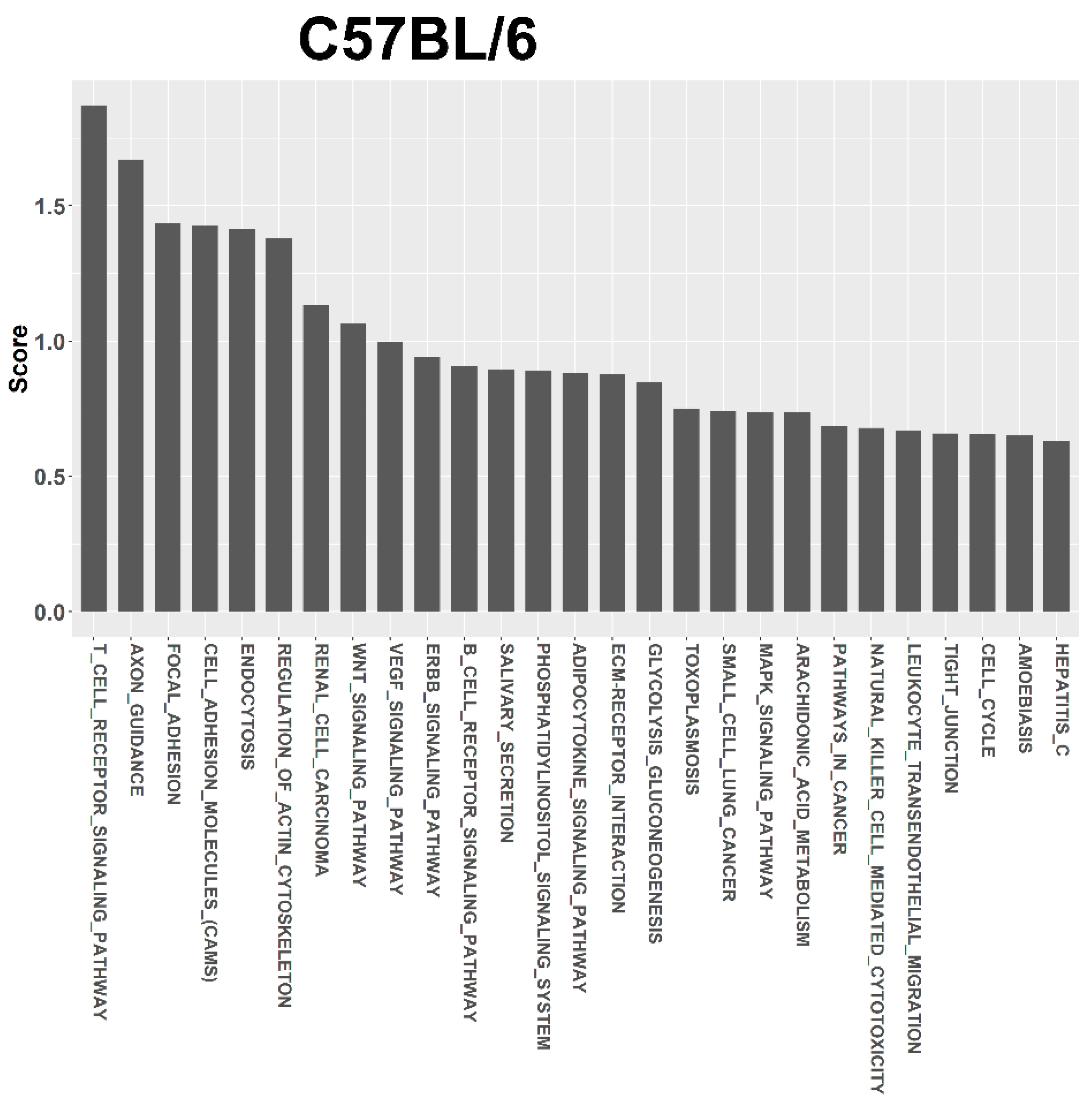
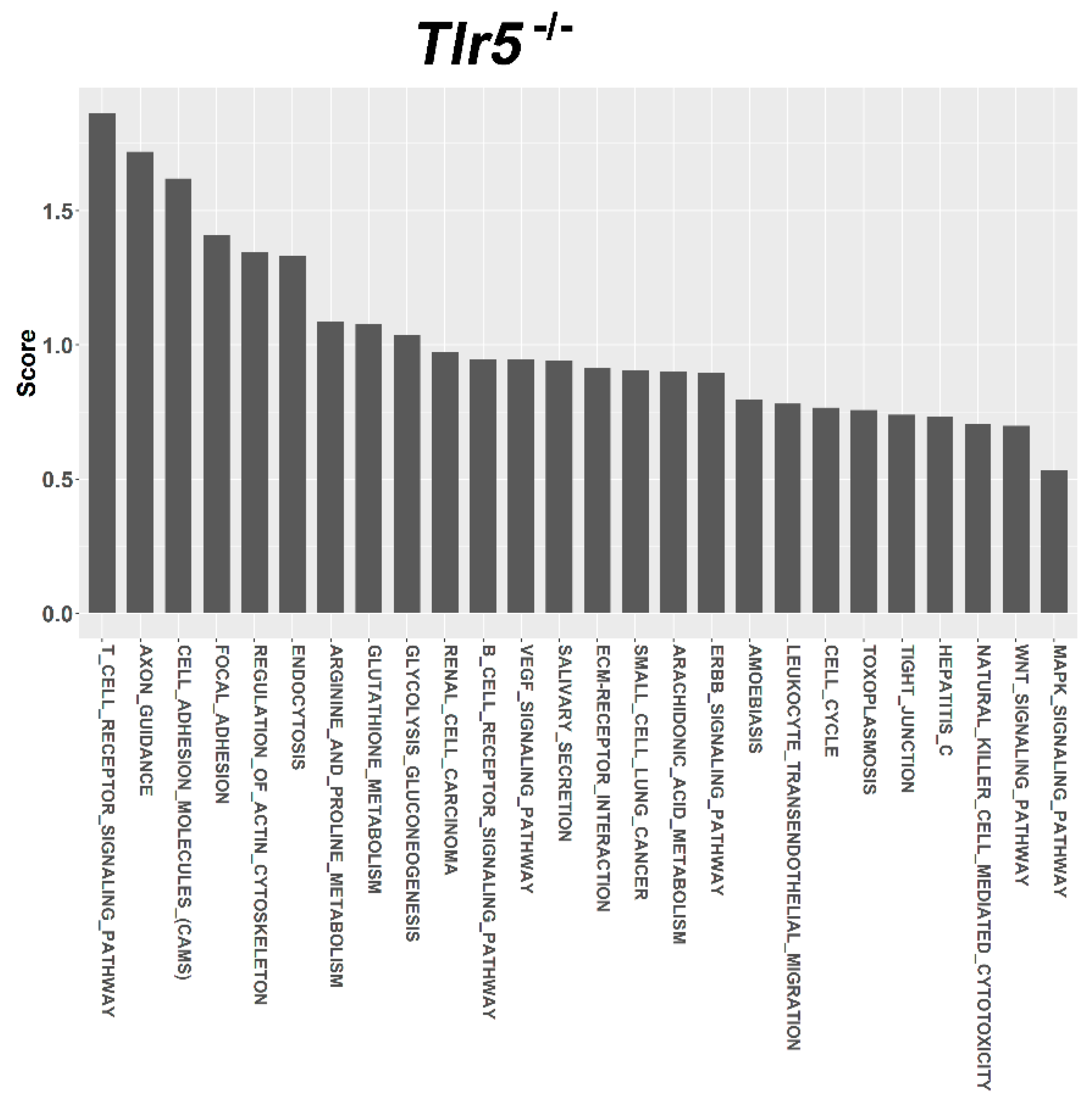
3.4. Target Prediction and Functional Annotation of Dysregulated miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Monteiro, R.; Azevedo, I. Chronic inflammation in obesity and the metabolic syndrome. Mediat. Inflamm. 2010, 2010, 289645. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, R.; Wang, H.; Liang, F. Mechanisms Linking Inflammation to Insulin Resistance. In. J. Endocrinol. 2015, 2015, 508409. [Google Scholar] [CrossRef] [PubMed]
- Engin, A.B. Adipocyte-Macrophage Cross-Talk in Obesity. Adv. Exp. Med. Biol. 2017, 960, 327–343. [Google Scholar] [PubMed]
- Omagari, K.; Kato, S.; Tsuneyama, K.; Inohara, C.; Kuroda, Y.; Tsukuda, H.; Fukazawa, E.; Shiraishi, K.; Mune, M. Effects of a long-term high-fat diet and switching from a high-fat to low-fat, standard diet on hepatic fat accumulation in Sprague-Dawley rats. Dig. Dis. Sci. 2008, 53, 3206–3212. [Google Scholar] [CrossRef] [PubMed]
- JafariNasabian, P.; Inglis, J.E.; Reilly, W.; Kelly, O.J.; Ilich, J.Z. Aging human body: Changes in bone, muscle and body fat with consequent changes in nutrient intake. J. Endocrinol. 2017, 234, R37–R51. [Google Scholar] [CrossRef] [PubMed]
- Pal China, S.; Sanyal, S.; Chattopadhyay, N. Adiponectin signaling and its role in bone metabolism. Cytokine 2018. [Google Scholar] [CrossRef] [PubMed]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- De Taeye, B.M.; Novitskaya, T.; McGuinness, O.P.; Gleaves, L.; Medda, M.; Covington, J.W.; Vaughan, D.E. Macrophage TNF-alpha contributes to insulin resistance and hepatic steatosis in diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E713–E725. [Google Scholar] [CrossRef] [PubMed]
- Rogero, M.M.; Calder, P.C. Obesity, Inflammation, Toll-Like Receptor 4 and Fatty Acids. Nutrients 2018, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Tall, A.R.; Yvan-Charvet, L. Cholesterol, inflammation and innate immunity. Nat. Rev. Immunol. 2015, 15, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anthoney, N.; Foldi, I.; Hidalgo, A. Toll and Toll-like receptor signalling in development. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Arancibia, S.A.; Beltran, C.J.; Aguirre, I.M.; Silva, P.; Peralta, A.L.; Malinarich, F.; Hermoso, M.A. Toll-like receptors are key participants in innate immune responses. Biol. Res. 2007, 40, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. TLR signaling. Cell Death Differ. 2006, 13, 816–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.J.; Choi, Y.; Choi, Y.H.; Park, T. Obesity activates toll-like receptor-mediated proinflammatory signaling cascades in the adipose tissue of mice. J. Nutr. Biochem. 2012, 23, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Favelyukis, S.; Nguyen, A.K.; Reichart, D.; Scott, P.A.; Jenn, A.; Liu-Bryan, R.; Glass, C.K.; Neels, J.G.; Olefsky, J.M. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J. Biol. Chem. 2007, 282, 35279–35292. [Google Scholar] [CrossRef] [PubMed]
- Boni-Schnetzler, M.; Boller, S.; Debray, S.; Bouzakri, K.; Meier, D.T.; Prazak, R.; Kerr-Conte, J.; Pattou, F.; Ehses, J.A.; Schuit, F.C.; et al. Free fatty acids induce a proinflammatory response in islets via the abundantly expressed interleukin-1 receptor I. Endocrinology 2009, 150, 5218–5229. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Peng, Y.; Wu, J.; Wang, Y.; Yao, L. Toll-like receptor 2/4 links to free fatty acid-induced inflammation and beta-cell dysfunction. J. Leukoc. Biol. 2014, 95, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Luo, J.; Ruan, Y.; Xiu, L.; Fang, B.; Zhang, H.; Wang, M.; Chen, H. Free Fatty Acids Activate Renin-Angiotensin System in 3T3-L1 Adipocytes through Nuclear Factor-kappa B Pathway. J. Diabetes Res. 2016, 2016, 1587594. [Google Scholar] [CrossRef] [PubMed]
- Suganami, T.; Tanimoto-Koyama, K.; Nishida, J.; Itoh, M.; Yuan, X.; Mizuarai, S.; Kotani, H.; Yamaoka, S.; Miyake, K.; Aoe, S.; et al. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.E.; Lum, H.; Alvarez, A.; Cipriani, Y.; Garduno-Garcia, J.; Anaya, L.; Dube, J.; Musi, N. A sustained increase in plasma NEFA upregulates the Toll-like receptor network in human muscle. Diabetologia 2014, 57, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Betanzos-Cabrera, G.; Estrada-Luna, D.; Belefant-Miller, H.; Cancino-Diaz, J.C. Mice fed with a high fat diet show a decrease in the expression of “toll like receptor (TLR)2 and TLR6 mRNAs in adipose and hepatic tissues. Nutr. Hosp. 2012, 27, 1196–1203. [Google Scholar] [PubMed]
- Kennedy, A.; Martinez, K.; Chuang, C.C.; LaPoint, K.; McIntosh, M. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: Mechanisms of action and implications. J. Nutr. 2009, 139, 1–4. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, S.J.; Feskens, E.J.; Bos, M.B.; Hoelen, D.W.; Heijligenberg, R.; Bromhaar, M.G.; de Groot, L.C.; de Vries, J.H.; Muller, M.; Afman, L.A. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am. J. Clin. Nutr. 2009, 90, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.E.; Braucher, D.R.; Walker-Daniels, J.; Spurlock, M.E. Absence of Tlr2 protects against high-fat diet-induced inflammation and results in greater insulin-stimulated glucose transport in cultured adipocytes. J. Nutr. Biochem. 2011, 22, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.H.; Tsai, P.J.; Jiang, M.J.; Chuang, Y.L.; Yu, L.; Lai, K.T.; Tsai, Y.S. Toll-like receptor 2 deficiency improves insulin sensitivity and hepatic insulin signalling in the mouse. Diabetologia 2011, 54, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Caricilli, A.M.; Nascimento, P.H.; Pauli, J.R.; Tsukumo, D.M.; Velloso, L.A.; Carvalheira, J.B.; Saad, M.J. Inhibition of toll-like receptor 2 expression improves insulin sensitivity and signaling in muscle and white adipose tissue of mice fed a high-fat diet. J. Endocrinol. 2008, 199, 399–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukumo, D.M.; Carvalho-Filho, M.A.; Carvalheira, J.B.; Prada, P.O.; Hirabara, S.M.; Schenka, A.A.; Araujo, E.P.; Vassallo, J.; Curi, R.; Velloso, L.A.; et al. Statement of Retraction. Loss-of-Function Mutation in Toll-Like Receptor 4 Prevents Diet-Induced Obesity and Insulin Resistance. Diabetes 2016, 65, 1126–1127. [Google Scholar] [CrossRef] [PubMed]
- Tramullas, M.; Finger, B.C.; Dinan, T.G.; Cryan, J.F. Obesity Takes Its Toll on Visceral Pain: High-Fat Diet Induces Toll-Like Receptor 4-Dependent Visceral Hypersensitivity. PLoS ONE 2016, 11, e0155367. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.K.; Hill, A.A.; Hasty, A.H. Stearic acid accumulation in macrophages induces toll-like receptor 4/2-independent inflammation leading to endoplasmic reticulum stress-mediated apoptosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1687–1695. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Wang, Y.; Wang, W.; Lv, X.; Wang, M.; Yin, H. The mAb against adipocyte fatty acid-binding protein 2E4 attenuates the inflammation in the mouse model of high-fat diet-induced obesity via toll-like receptor 4 pathway. Mol. Cell. Endocrinol. 2015, 403, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pal, D.; Dasgupta, S.; Kundu, R.; Maitra, S.; Das, G.; Mukhopadhyay, S.; Ray, S.; Majumdar, S.S.; Bhattacharya, S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012, 18, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.J.; Kim, S.P.; Zhang, D.; Sun, H.; Cao, Q.; Lu, X.; Ying, Z.; Li, L.; Henry, R.R.; Ciaraldi, T.P.; et al. Deletion of interleukin 1 receptor-associated kinase 1 (Irak1) improves glucose tolerance primarily by increasing insulin sensitivity in skeletal muscle. J. Biol. Chem. 2017, 292, 12339–12350. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Nahid, M.A.; Satoh, M.; Chan, E.K. MicroRNA in TLR signaling and endotoxin tolerance. Cell. Mol. Immunol. 2011, 8, 388–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Shi, X. MicroRNAs in the regulation of TLR and RIG-I pathways. Cell. Mol. Immunol. 2013, 10, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Aalaei-andabili, S.H.; Rezaei, N. Toll like receptor (TLR)-induced differential expression of microRNAs (MiRs) promotes proper immune response against infections: A systematic review. J. Infect. 2013, 67, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Murri, M.; El Azzouzi, H. MicroRNAs as regulators of mitochondrial dysfunction and obesity. Am. J. Physiol. Heart Circ. Physiol. 2018, 315, H291–H302. [Google Scholar] [CrossRef] [PubMed]
- Maurizi, G.; Babini, L.; Della Guardia, L. Potential role of microRNAs in the regulation of adipocytes liposecretion and adipose tissue physiology. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Zaravinos, A.; Ziros, P.G.; Iskrenova, R.P.; Psyrogiannis, A.I.; Kyriazopoulou, V.E.; Habeos, I.G. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 2012, 7, e34872. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009, 58, 1050–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takanabe, R.; Ono, K.; Abe, Y.; Takaya, T.; Horie, T.; Wada, H.; Kita, T.; Satoh, N.; Shimatsu, A.; Hasegawa, K. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem. Biophys. Res. Commun. 2008, 376, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Baselga-Escudero, L.; Pascual-Serrano, A.; Ribas-Latre, A.; Casanova, E.; Salvado, M.J.; Arola, L.; Arola-Arnal, A.; Blade, C. Long-term supplementation with a low dose of proanthocyanidins normalized liver miR-33a and miR-122 levels in high-fat diet-induced obese rats. Nutr. Res. 2015, 35, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol. Cell. Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [PubMed]
- Lavery, C.A.; Kurowska-Stolarska, M.; Holmes, W.M.; Donnelly, I.; Caslake, M.; Collier, A.; Baker, A.H.; Miller, A.M. miR-34a(-/-) mice are susceptible to diet-induced obesity. Obesity 2016, 24, 1741–1751. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Ren, H.; Xu, P.; Cheng, J.; Huang, S.; Zhou, R.; Mu, Y.; Yang, S.; Qi, D.; Wang, Y.; et al. Adipocyte miR-200b/a/429 ablation in mice leads to high-fat-diet-induced obesity. Oncotarget 2016, 7, 67796–67807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtue, A.; Johnson, C.; Lopez-Pastrana, J.; Shao, Y.; Fu, H.; Li, X.; Li, Y.F.; Yin, Y.; Mai, J.; Rizzo, V.; et al. MicroRNA-155 Deficiency Leads to Decreased Atherosclerosis, Increased White Adipose Tissue Obesity, and Non-alcoholic Fatty Liver Disease: A novel mouse model of obesity paradox. J. Biol. Chem. 2017, 292, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.; Wang, Y.X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 4725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrer, M.; Liu, N.; Grueter, C.E.; Williams, A.H.; Frisard, M.I.; Hulver, M.W.; Bassel-Duby, R.; Olson, E.N. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc. Natl. Acad. Sci. USA 2012, 109, 15330–15335. [Google Scholar] [CrossRef] [PubMed]
- Sartipy, P.; Loskutoff, D.J. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc. Natl. Acad. Sci. USA 2003, 100, 7265–7270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, E.P.; Racette, S.B.; Villareal, D.T.; Fontana, L.; Steger-May, K.; Schechtman, K.B.; Klein, S.; Holloszy, J.O. Improvements in glucose tolerance and insulin action induced by increasing energy expenditure or decreasing energy intake: A randomized controlled trial. Am. J. Clin. Nutr. 2006, 84, 1033–1042. [Google Scholar] [PubMed]
- Muurling, M.; Jong, M.C.; Mensink, R.P.; Hornstra, G.; Dahlmans, V.E.; Pijl, H.; Voshol, P.J.; Havekes, L.M. A low-fat diet has a higher potential than energy restriction to improve high-fat diet-induced insulin resistance in mice. Metab. Clin. Exp. 2002, 51, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Podrini, C.; Cambridge, E.L.; Lelliott, C.J.; Carragher, D.M.; Estabel, J.; Gerdin, A.K.; Karp, N.A.; Scudamore, C.L.; Ramirez-Solis, R.; White, J.K. High-fat feeding rapidly induces obesity and lipid derangements in C57BL/6N mice. Mamm. Genome 2013, 24, 240–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, S.; Martin, T.L.; Surwit, R.S.; Robidoux, J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: Physiological and molecular characteristics. Physiol. Behav. 2004, 81, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Report of the American Institute of Nurtition ad hoc Committee on Standards for Nutritional Studies. J. Nutr. 1977, 107, 1340–1348. [CrossRef] [PubMed]
- Parekh, P.I.; Petro, A.E.; Tiller, J.M.; Feinglos, M.N.; Surwit, R.S. Reversal of diet-induced obesity and diabetes in C57BL/6J mice. Metabolism 1998, 47, 1089–1096. [Google Scholar] [CrossRef]
- Petro, A.E.; Cotter, J.; Cooper, D.A.; Peters, J.C.; Surwit, S.J.; Surwit, R.S. Fat, carbohydrate, and calories in the development of diabetes and obesity in the C57BL/6J mouse. Metabolism 2004, 53, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.H.; Rau, C.S.; Wu, S.C.; Yang, J.C.; Wu, Y.C.; Lu, T.H.; Tzeng, S.L.; Wu, C.J.; Lin, C.W. Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC Genomics 2015, 16, 699. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.T.; Tsai, K.W.; Hung, T.M.; Lin, W.C.; Pan, C.Y.; Yu, H.R.; Li, S.C. miRSeq: A user-friendly standalone toolkit for sequencing quality evaluation and miRNA profiling. BioMed Res. Int. 2014, 2014, 462135. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.P.; Lee, C.Y.; Tsai, M.H.; Chiu, Y.C.; Hsiao, C.K.; Lai, L.C.; Chuang, E.Y. miRSystem: An integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS ONE 2012, 7, e42390. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Bertin, E.; Nguyen, P.; Guenounou, M.; Durlach, V.; Potron, G.; Leutenegger, M. Plasma levels of tumor necrosis factor-alpha (TNF-alpha) are essentially dependent on visceral fat amount in type 2 diabetic patients. Diabetes Metab. 2000, 26, 178–182. [Google Scholar] [PubMed]
- You, T.; Nicklas, B.J.; Ding, J.; Penninx, B.W.; Goodpaster, B.H.; Bauer, D.C.; Tylavsky, F.A.; Harris, T.B.; Kritchevsky, S.B. The metabolic syndrome is associated with circulating adipokines in older adults across a wide range of adiposity. J. Gerontol. A Biol. Sci. Med. Sci. 2008, 63, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Gnacinska, M.; Malgorzewicz, S.; Stojek, M.; Lysiak-Szydlowska, W.; Sworczak, K. Role of adipokines in complications related to obesity: A review. Adv. Med. Sci. 2009, 54, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, O.; Tobia, C.; Ebersole, J.; Novak, M.J. Caloric restriction and chronic inflammatory diseases. Oral Dis. 2012, 18, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Redman, L.M.; Ravussin, E. Caloric restriction in humans: Impact on physiological, psychological, and behavioral outcomes. Antioxid. Redox Signal. 2011, 14, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Keller, J.N. Regulation of energy metabolism by inflammation: A feedback response in obesity and calorie restriction. Aging 2010, 2, 361–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, R.; de Castro, P.M.; Calhau, C.; Azevedo, I. Adipocyte size and liability to cell death. Obes. Surg. 2006, 16, 804–806. [Google Scholar] [CrossRef] [PubMed]
- Alrob, O.A.; Khatib, S.; Naser, S.A. MicroRNAs 33, 122, and 208: A potential novel targets in the treatment of obesity, diabetes, and heart-related diseases. J. Physiol. Biochem. 2017, 73, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hong, J.; Cao, Y.; Shi, J.; Gu, W.; Ning, G.; Zhang, Y.; Wang, W. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur. J. Endocrinol. 2015, 172, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Danielson, K.M.; Benton, M.C.; Ziegler, O.; Shah, R.; Stubbs, R.S.; Das, S.; Macartney-Coxson, D. miRNA Signatures of Insulin Resistance in Obesity. Obesity 2017, 25, 1734–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, N.; Wang, J.; Xie, W.; Lyu, Q.; Wu, J.; He, J.; Qiu, W.; Xu, N.; Zhang, Y. MiR-378a-3p enhances adipogenesis by targeting mitogen-activated protein kinase 1. Biochem. Biophys. Res. Commun. 2015, 457, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidou, A.; Mougios, V.; Sidossis, L.S. Alphacute Exercise Alters the Levels of Human Saliva miRNAs Involved in Lipid Metabolism. Int. J. Sports Med. 2016, 37, 584–588. [Google Scholar] [PubMed]
- Kulyte, A.; Lorente-Cebrian, S.; Gao, H.; Mejhert, N.; Agustsson, T.; Arner, P.; Ryden, M.; Dahlman, I. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E267–E274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frias, F.T.; Rocha, K.C.E.; de Mendonca, M.; Murata, G.M.; Araujo, H.N.; de Sousa, L.G.O.; de Sousa, E.; Hirabara, S.M.; Leite, N.C.; Carneiro, E.M.; et al. Fenofibrate reverses changes induced by high-fat diet on metabolism in mice muscle and visceral adipocytes. J. Cell. Physiol. 2018, 233, 3515–3528. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Bartolome, J.; Llaurado, G.; Portero-Otin, M.; Altuna-Coy, A.; Rojo-Martinez, G.; Vendrell, J.; Jorba, R.; Rodriguez-Gallego, E.; Chacon, M.R. Altered Expression of miR-181a-5p and miR-23a-3p Is Associated With Obesity and TNFalpha-Induced Insulin Resistance. J. Clin. Endocrinol. Metab. 2018, 103, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Li, X.; Yin, Y.; Wu, Z.; Liu, C.; Tekwe, C.D.; Wu, G. Regulatory roles for L-arginine in reducing white adipose tissue. Front. Biosci. (Landmark Ed.) 2012, 17, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Jobgen, W.S.; Fried, S.K.; Fu, W.J.; Meininger, C.J.; Wu, G. Regulatory role for the arginine-nitric oxide pathway in metabolism of energy substrates. J. Nutr. Biochem. 2006, 17, 571–588. [Google Scholar] [CrossRef] [PubMed]
- Araujo, T.R.; Freitas, I.N.; Vettorazzi, J.F.; Batista, T.M.; Santos-Silva, J.C.; Bonfleur, M.L.; Balbo, S.L.; Boschero, A.C.; Carneiro, E.M.; Ribeiro, R.A. Benefits of L-alanine or L-arginine supplementation against adiposity and glucose intolerance in monosodium glutamate-induced obesity. Eur. J. Nutr. 2017, 56, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Monti, L.D.; Setola, E.; Lucotti, P.C.; Marrocco-Trischitta, M.M.; Comola, M.; Galluccio, E.; Poggi, A.; Mammi, S.; Catapano, A.L.; Comi, G.; et al. Effect of a long-term oral l-arginine supplementation on glucose metabolism: A randomized, double-blind, placebo-controlled trial. Diabetes Obes. Metab. 2012, 14, 893–900. [Google Scholar] [CrossRef] [PubMed]
- Alamshah, A.; McGavigan, A.K.; Spreckley, E.; Kinsey-Jones, J.S.; Amin, A.; Tough, I.R.; O’Hara, H.C.; Moolla, A.; Banks, K.; France, R.; et al. L-arginine promotes gut hormone release and reduces food intake in rodents. Diabetes Obes. Metab. 2016, 18, 508–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jobgen, W.; Meininger, C.J.; Jobgen, S.C.; Li, P.; Lee, M.J.; Smith, S.B.; Spencer, T.E.; Fried, S.K.; Wu, G. Dietary L-arginine supplementation reduces white fat gain and enhances skeletal muscle and brown fat masses in diet-induced obese rats. J. Nutr. 2009, 139, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Invest. 2004, 114, 1752–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquilano, K.; Baldelli, S.; Ciriolo, M.R. Glutathione: New roles in redox signaling for an old antioxidant. Front. Pharmacol. 2014, 5, 196. [Google Scholar] [CrossRef] [PubMed]
- Di Renzo, L.; Galvano, F.; Orlandi, C.; Bianchi, A.; Di Giacomo, C.; La Fauci, L.; Acquaviva, R.; De Lorenzo, A. Oxidative stress in normal-weight obese syndrome. Obesity 2010, 18, 2125–2130. [Google Scholar] [CrossRef] [PubMed]
- Renaud, H.J.; Cui, J.Y.; Lu, H.; Klaassen, C.D. Effect of diet on expression of genes involved in lipid metabolism, oxidative stress, and inflammation in mouse liver-insights into mechanisms of hepatic steatosis. PLoS ONE 2014, 9, e88584. [Google Scholar] [CrossRef] [PubMed]
- Okuno, Y.; Fukuhara, A.; Hashimoto, E.; Kobayashi, H.; Kobayashi, S.; Otsuki, M.; Shimomura, I. Oxidative Stress Inhibits Healthy Adipose Expansion Through Suppression of SREBF1-Mediated Lipogenic Pathway. Diabetes 2018, 67, 1113–1127. [Google Scholar] [CrossRef] [PubMed]
- Lettieri Barbato, D.; Tatulli, G.; Maria Cannata, S.; Bernardini, S.; Aquilano, K.; Ciriolo, M.R. Glutathione Decrement Drives Thermogenic Program in Adipose Cells. Sci. Rep. 2015, 5, 13091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Findeisen, H.M.; Gizard, F.; Zhao, Y.; Qing, H.; Jones, K.L.; Cohn, D.; Heywood, E.B.; Bruemmer, D. Glutathione depletion prevents diet-induced obesity and enhances insulin sensitivity. Obesity 2011, 19, 2429–2432. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Karmakar, S.; Babu, S.P. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, L.; Chen, S. Endogenous toll-like receptor ligands and their biological significance. J. Cell. Mol. Med. 2010, 14, 2592–2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Akira, S. Microbial recognition by Toll-like receptors. J. Dermatol. Sci. 2004, 34, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Underhill, D.M.; Ozinsky, A.; Hajjar, A.M.; Stevens, A.; Wilson, C.B.; Bassetti, M.; Aderem, A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 1999, 401, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Valentini, M.; Piermattei, A.; Di Sante, G.; Migliara, G.; Delogu, G.; Ria, F. Immunomodulation by gut microbiota: Role of Toll-like receptor expressed by T cells. J. Immunol. Res. 2014, 2014, 586939. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Lund, P.K. Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 328–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of high-fat diet on gut microbiota: A driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care 2015. [Google Scholar] [CrossRef] [PubMed]




| Term | Term ID | Total Genes of the Term | Union Targets in the Term | Mirnas in the Term | Score |
|---|---|---|---|---|---|
| C57BL/6 | |||||
| T CELL RECEPTOR SIGNALING PATHWAY | 4660 | 109 | 7 | 3 | 1.869 |
| AXON GUIDANCE | 4360 | 131 | 10 | 4 | 1.671 |
| FOCAL ADHESION | 4510 | 197 | 11 | 3 | 1.436 |
| CELL ADHESION MOLECULES (CAMS) | 4514 | 149 | 4 | 3 | 1.428 |
| ENDOCYTOSIS | 4144 | 219 | 12 | 4 | 1.416 |
| REGULATION OF ACTIN CYTOSKELETON | 4810 | 215 | 11 | 4 | 1.379 |
| RENAL CELL CARCINOMA | 5211 | 71 | 7 | 2 | 1.133 |
| WNT SIGNALING PATHWAY | 4310 | 153 | 9 | 4 | 1.065 |
| VEGF SIGNALING PATHWAY | 4370 | 76 | 4 | 3 | 0.998 |
| ERBB SIGNALING PATHWAY | 4012 | 87 | 5 | 3 | 0.941 |
| TLR5-KO | |||||
| T CELL RECEPTOR SIGNALING PATHWAY | 4660 | 109 | 2 | 1 | 1.864 |
| AXON GUIDANCE | 4360 | 131 | 2 | 1 | 1.719 |
| CELL ADHESION MOLECULES (CAMS) | 4514 | 149 | 2 | 1 | 1.62 |
| FOCAL ADHESION | 4510 | 197 | 2 | 1 | 1.41 |
| REGULATION OF ACTIN CYTOSKELETON | 4810 | 215 | 2 | 1 | 1.346 |
| ENDOCYTOSIS | 4144 | 219 | 2 | 1 | 1.333 |
| ARGININE AND PROLINE METABOLISM | 330 | 53 | 1 | 1 | 1.087 |
| GLUTATHIONE METABOLISM | 480 | 54 | 1 | 1 | 1.079 |
| GLYCOLYSIS GLUCONEOGENESIS | 10 | 60 | 1 | 1 | 1.038 |
| RENAL CELL CARCINOMA | 5211 | 71 | 1 | 1 | 0.973 |
| TLR2-KO | |||||
| ARGININE AND PROLINE METABOLISM | 330 | 53 | 2 | 2 | 1.171 |
| GLUTATHIONE METABOLISM | 480 | 54 | 2 | 2 | 1.164 |
| T CELL RECEPTOR SIGNALING PATHWAY | 4660 | 109 | 8 | 4 | 1.106 |
| AXON GUIDANCE | 4360 | 131 | 9 | 4 | 1.098 |
| FOCAL ADHESION | 4510 | 197 | 13 | 3 | 1.049 |
| REGULATION OF ACTIN CYTOSKELETON | 4810 | 215 | 14 | 4 | 1.018 |
| ENDOCYTOSIS | 4144 | 219 | 11 | 4 | 0.975 |
| RENAL CELL CARCINOMA | 5211 | 71 | 8 | 3 | 0.888 |
| CELL ADHESION MOLECULES (CAMS) | 4514 | 149 | 7 | 4 | 0.875 |
| WNT SIGNALING PATHWAY | 4310 | 153 | 10 | 4 | 0.749 |
| TLR4-KO | |||||
| ADHERENS JUNCTION | 4520 | 74 | 12 | 4 | 2.242 |
| ARGININE AND PROLINE METABOLISM | 330 | 53 | 2 | 2 | 1.931 |
| GLUTATHIONE METABOLISM | 480 | 54 | 2 | 2 | 1.864 |
| PATHWAYS IN CANCER | 5200 | 323 | 21 | 4 | 1.808 |
| WNT SIGNALING PATHWAY | 4310 | 153 | 14 | 5 | 1.802 |
| AXON GUIDANCE | 4360 | 131 | 11 | 5 | 1.669 |
| REGULATION OF ACTIN CYTOSKELETON | 4810 | 215 | 15 | 5 | 1.504 |
| PROSTATE CANCER | 5215 | 89 | 10 | 3 | 1.363 |
| FOCAL ADHESION | 4510 | 197 | 12 | 4 | 1.265 |
| MAPK SIGNALING PATHWAY | 4010 | 271 | 16 | 5 | 1.22 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rau, C.-S.; Wu, S.-C.; Lu, T.-H.; Wu, Y.-C.; Wu, C.-J.; Chien, P.-C.; Kuo, P.-J.; Lin, C.-W.; Tsai, C.-W.; Hsieh, C.-H. Effect of Low-Fat Diet in Obese Mice Lacking Toll-like Receptors. Nutrients 2018, 10, 1464. https://doi.org/10.3390/nu10101464
Rau C-S, Wu S-C, Lu T-H, Wu Y-C, Wu C-J, Chien P-C, Kuo P-J, Lin C-W, Tsai C-W, Hsieh C-H. Effect of Low-Fat Diet in Obese Mice Lacking Toll-like Receptors. Nutrients. 2018; 10(10):1464. https://doi.org/10.3390/nu10101464
Chicago/Turabian StyleRau, Cheng-Shyuan, Shao-Chun Wu, Tsu-Hsiang Lu, Yi-Chan Wu, Chia-Jung Wu, Peng-Chen Chien, Pao-Jen Kuo, Chia-Wei Lin, Chia-Wen Tsai, and Ching-Hua Hsieh. 2018. "Effect of Low-Fat Diet in Obese Mice Lacking Toll-like Receptors" Nutrients 10, no. 10: 1464. https://doi.org/10.3390/nu10101464
APA StyleRau, C.-S., Wu, S.-C., Lu, T.-H., Wu, Y.-C., Wu, C.-J., Chien, P.-C., Kuo, P.-J., Lin, C.-W., Tsai, C.-W., & Hsieh, C.-H. (2018). Effect of Low-Fat Diet in Obese Mice Lacking Toll-like Receptors. Nutrients, 10(10), 1464. https://doi.org/10.3390/nu10101464







