Estimating Biomass of Native Grass Grown under Complex Management Treatments Using WorldView-3 Spectral Derivatives
Abstract
:1. Introduction
2. Methods and Materials
2.1. Study Area Description
2.2. Experimental Design
2.3. Field Campaign
2.4. Remotely Sensed Data
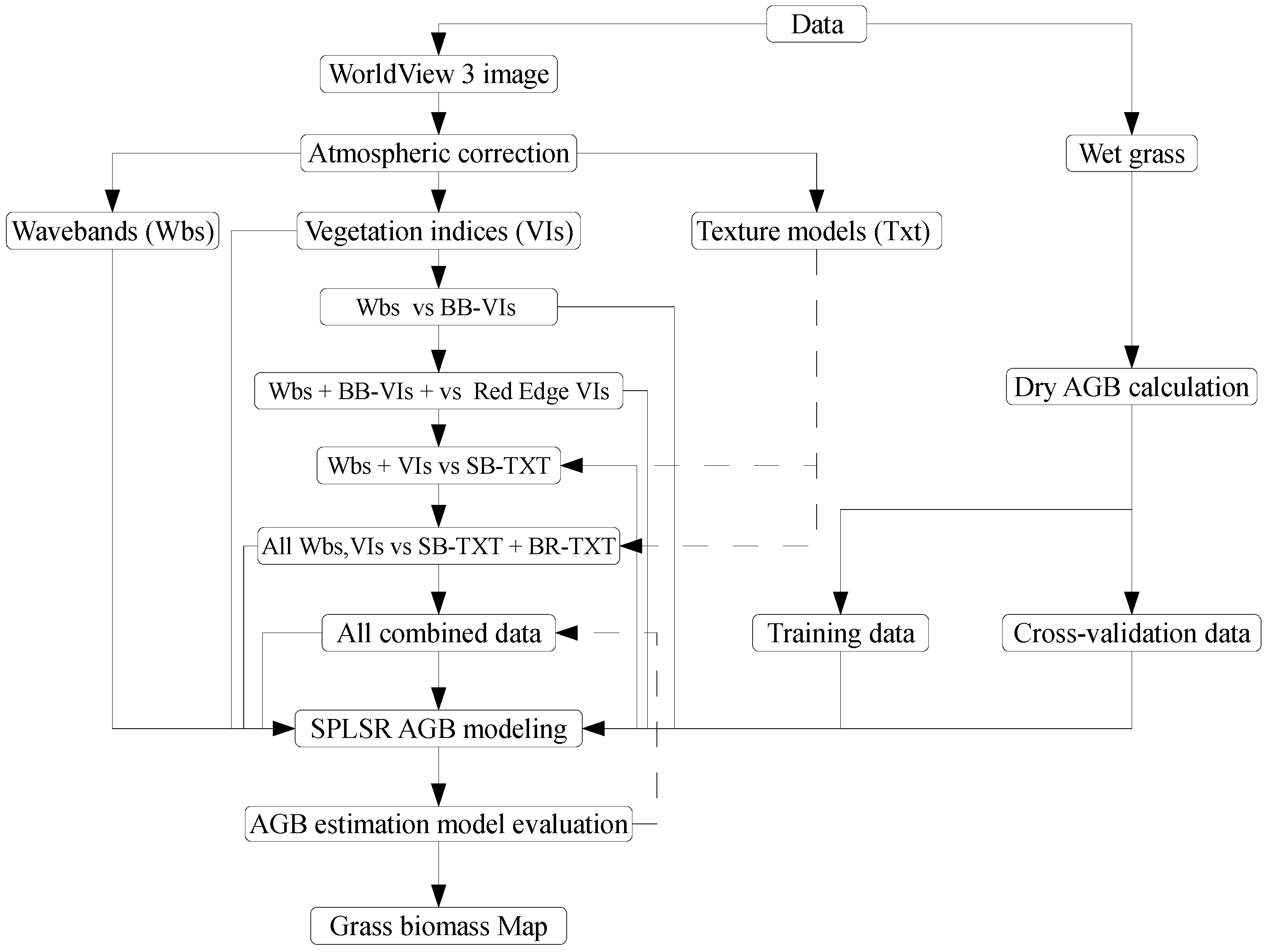
2.5. Modelling Above-Ground Grass Biomass
2.5.1. Statistical Modelling of Above-Ground Grass Biomass
2.5.2. Regression Modelling
2.5.3. Assessing the Accuracy of Above-Ground Grass Biomass Models
2.5.4. Phases of Estimating Above-Ground Grass Biomass
3. Results
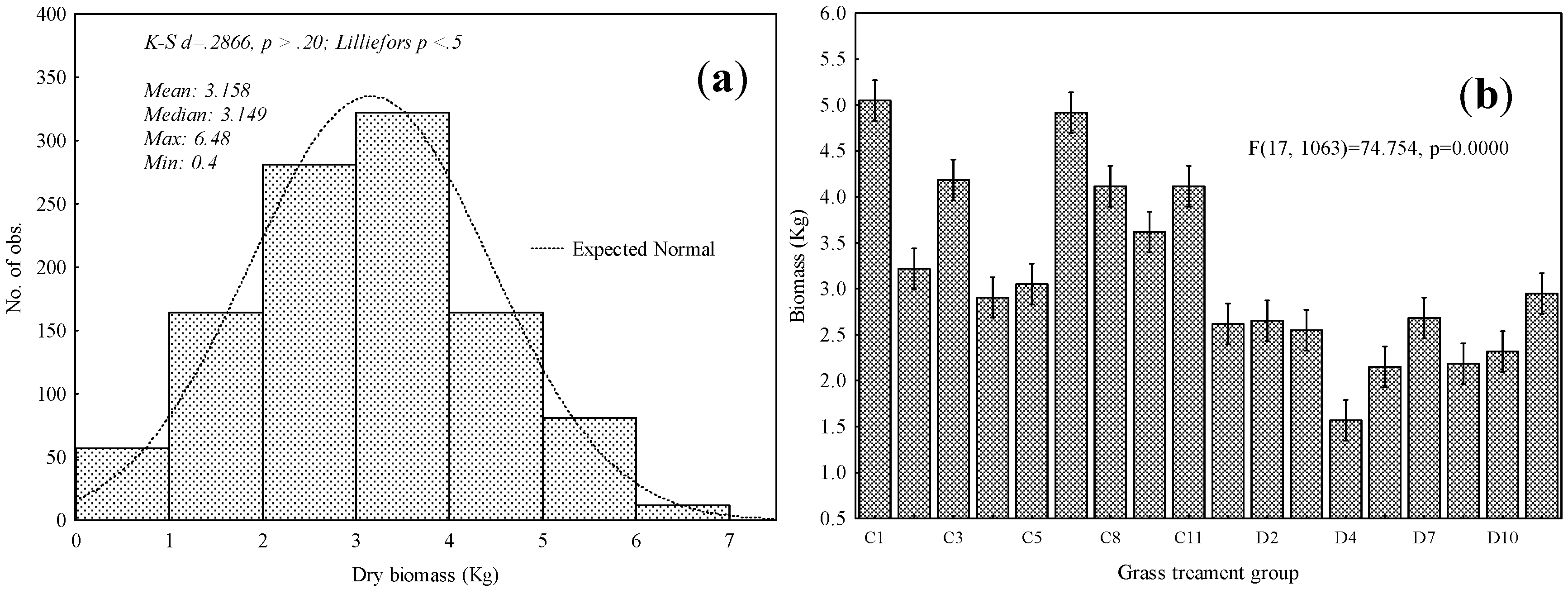
3.1. Descriptive Statistical Analysis and ANOVA Tests
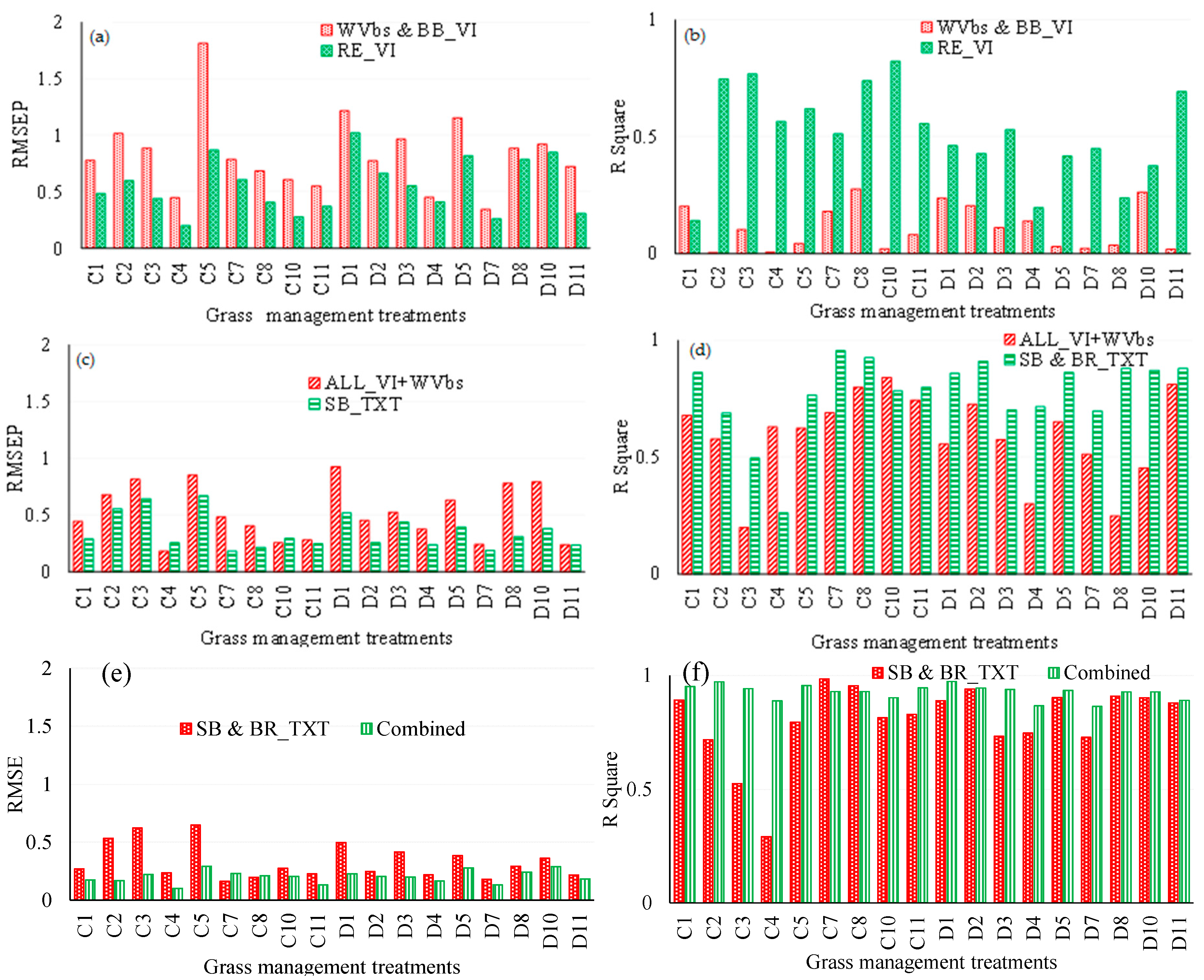
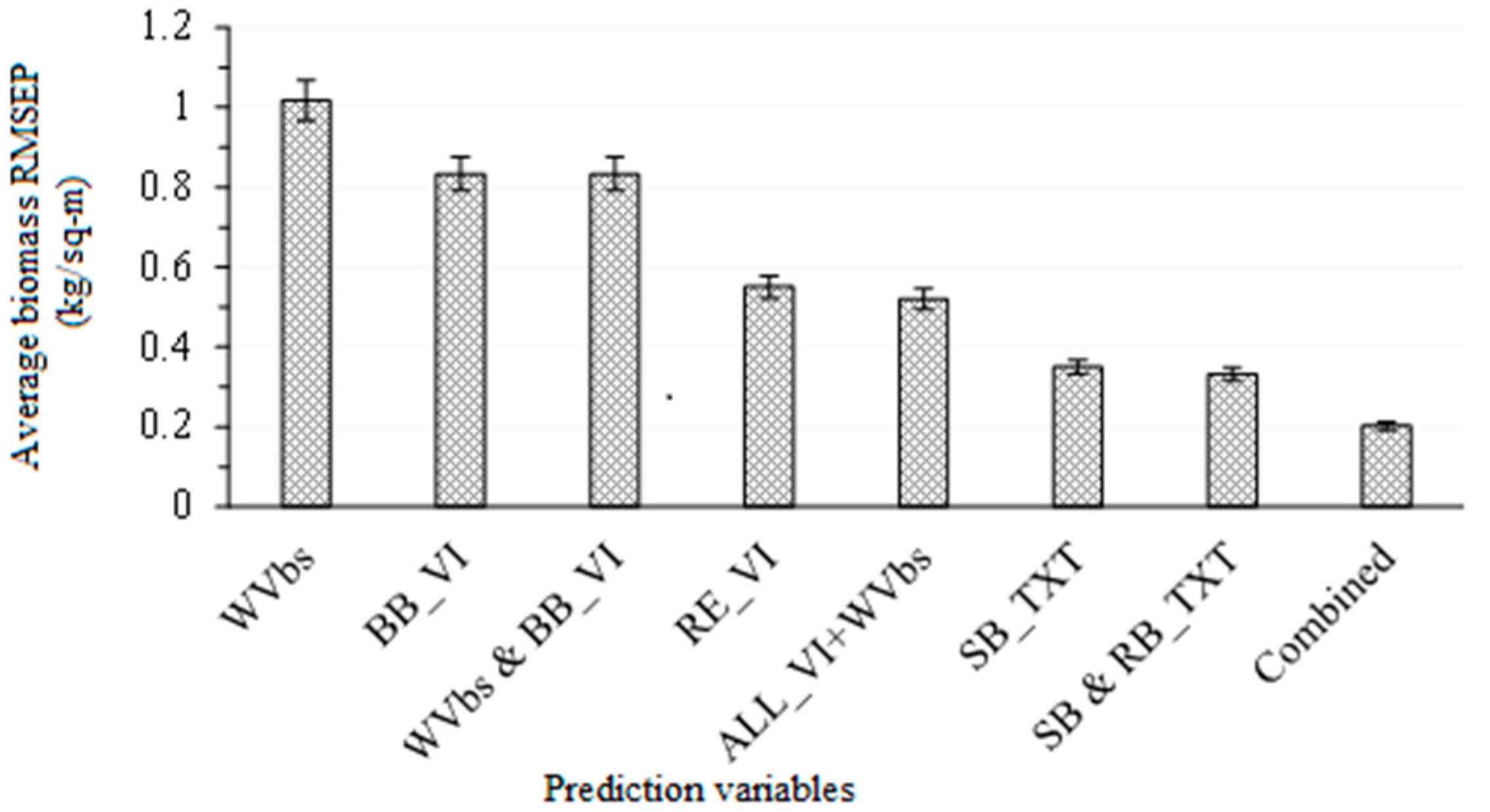
3.2. Comparing the Performance of WorldView-3 Wavebands Combined with Broadband Vegetation Indices (Vis) and Red-Edge VIs in Estimating Above-Ground Grass Biomass
3.3. Comparing the Performance of Single-Band Texture Models with All WV-3 VIs and Band Reflectance Values in Estimating Above-Ground Grass Biomass
3.4. Comparing the Performance of Combined Single-Band and Band-Ratio Texture Models with the Combination of All WV-3 VIs, Band Reflectance Values and Single-Band Texture Models in Estimating Above-Ground Grass Biomass
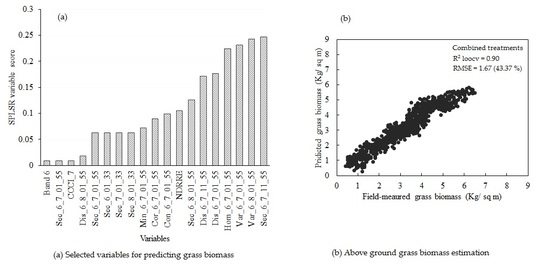
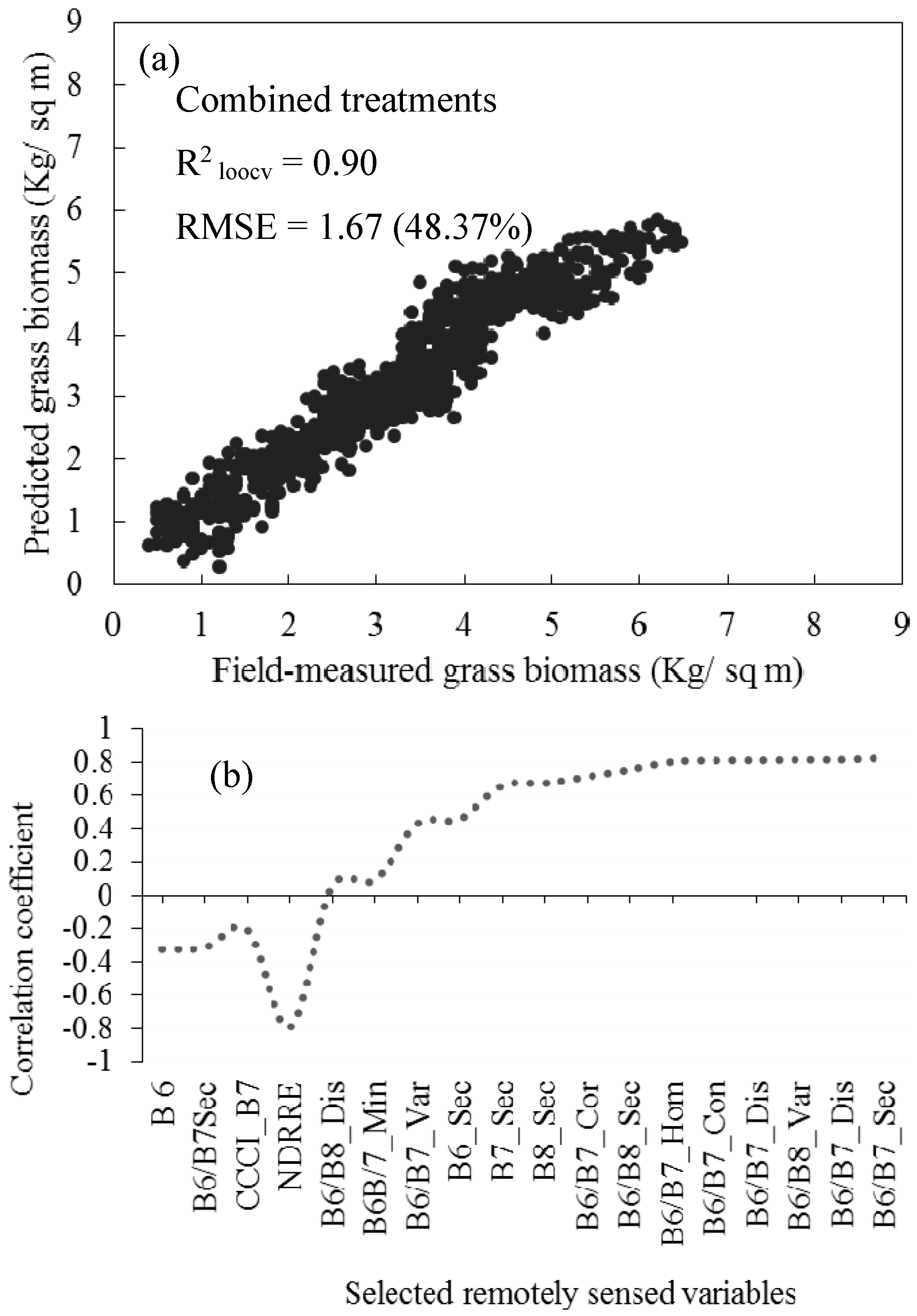
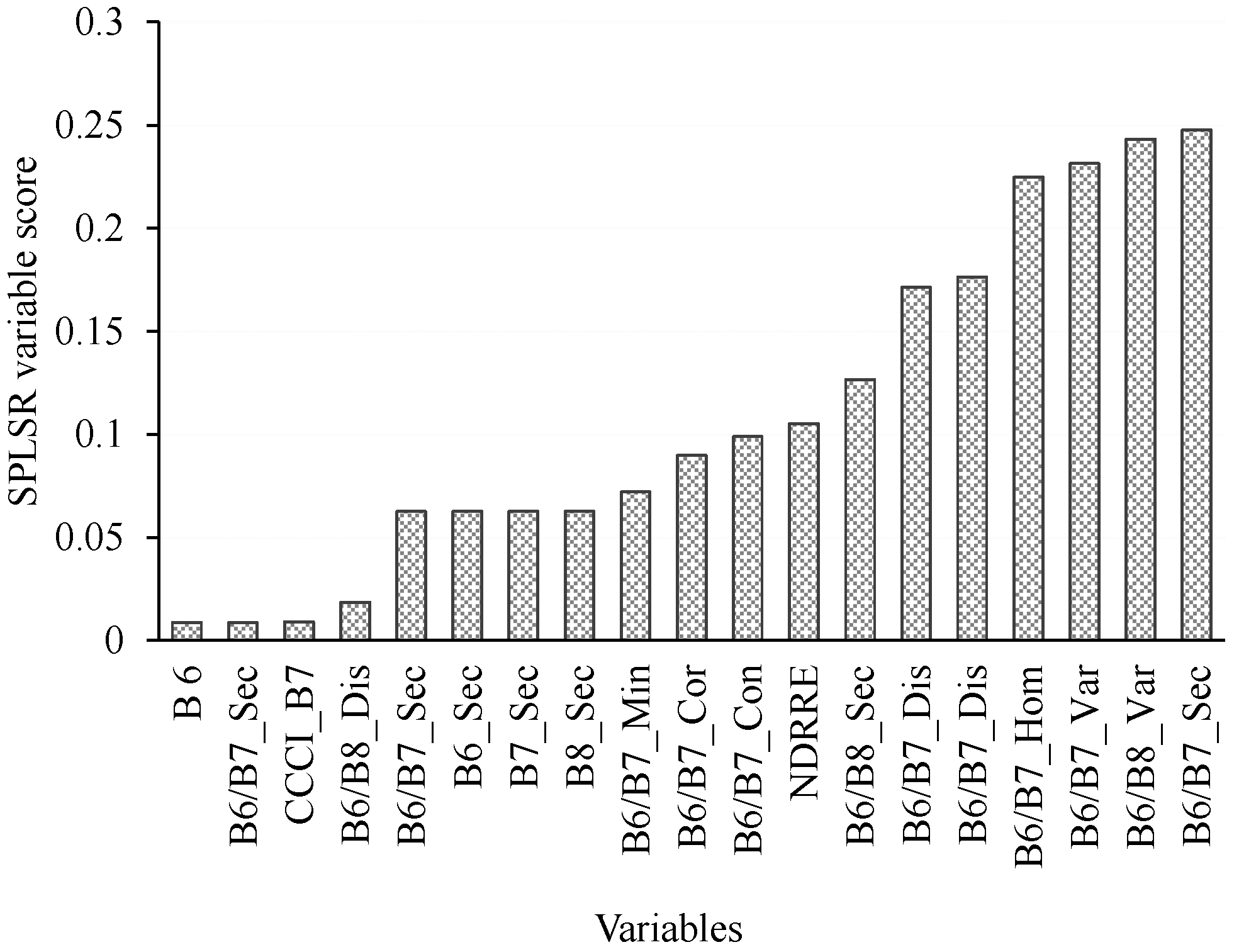
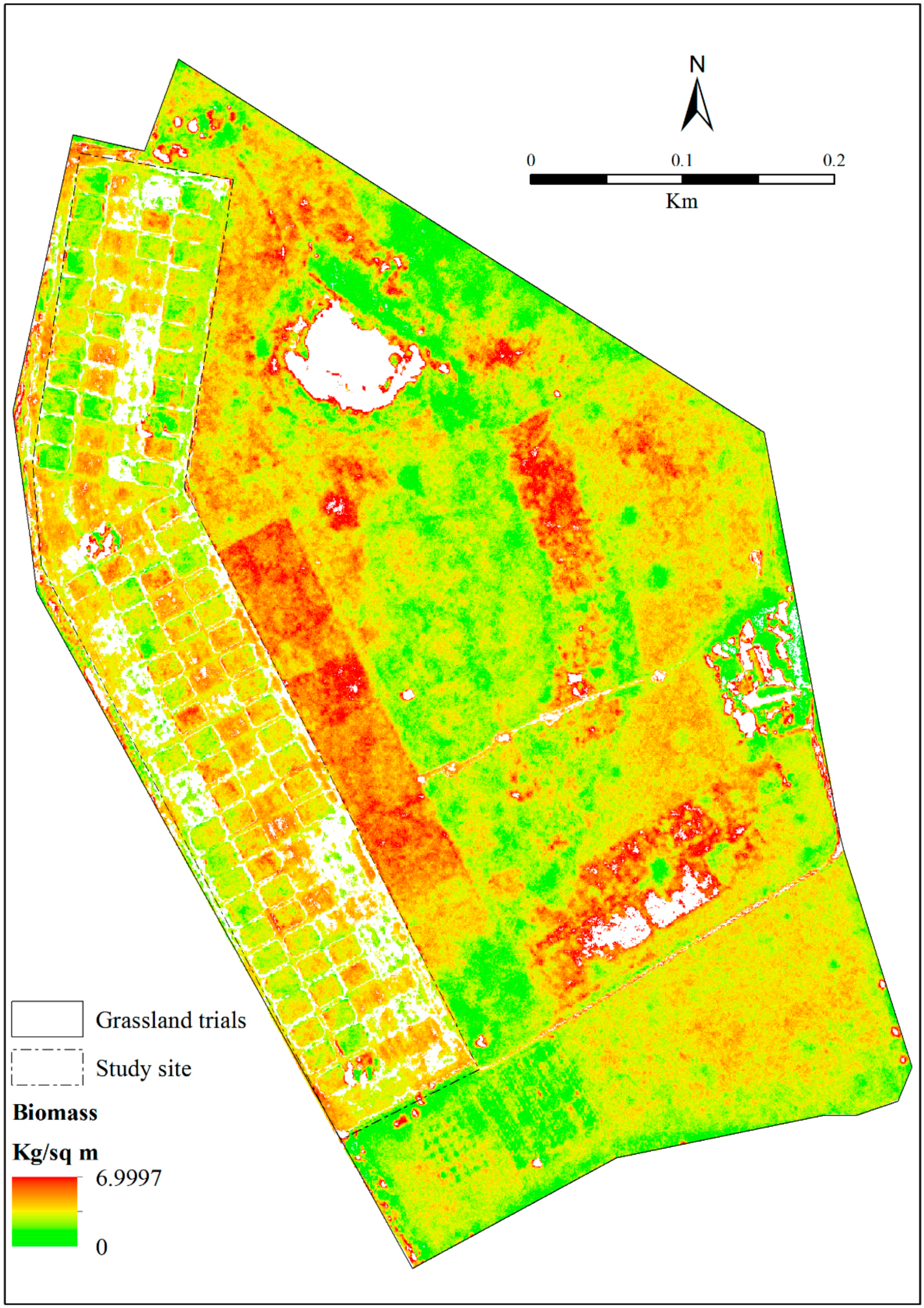
3.5. Estimating Above-Ground Grass Biomass across Different Levels of Grassland Management Treatments Using WV-3-Derived Texture Models Combined with Optimal Vegetation Indices Selected by the SPLSR Algorithm
4. Discussion
4.1. Combining Texture Models with Red-Edge in Predicting above-Ground Grass Biomass
4.2. Biological Behavior of Grasses at Ukulinga Research Farm Based on Literature Review
5. Conclusions
- combining texture models with red-edge derivatives provides a more accurate approach in estimating the above-ground biomass of grass grown under complex grassland management treatments. To the best of our knowledge, this is the first study to evaluate the utility of texture models and red-edge in estimating above-ground grass biomass, across a multitude of grassland management treatment levels,
- the best predictor in estimating above-ground biomass (ABGB) grown under complex grassland management treatments was derived using all data combined,
- texture models perform better than the red-edge vegetation indices in estimating grass above-ground biomass, and
- as expected, the red-edge spectrum-derived vegetation indices outperformed the broadband indices.
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kumar, L.; Sinha, P.; Taylor, S.; Alqurashi, A.F. Review of the use of remote sensing for biomass estimation to support renewable energy generation. J. Appl. Remote Sens. 2015, 9, 097696. [Google Scholar] [CrossRef]
- Wilson, J.B.; Peet, R.K.; Dengler, J.; Pärtel, M. Plant species richness: The world records. J. Veg. Sci. 2012, 23, 796–802. [Google Scholar] [CrossRef]
- O’Mara, F.P. The role of grasslands in food security and climate change. Ann. Bot. 2012, 110, 1263–1270. [Google Scholar] [CrossRef] [PubMed]
- Andrade, B.O.; Koch, C.; Boldrini, I.I.; Vélez-Martin, E.; Hasenack, H.; Hermann, J.-M.; Kollmann, J.; Pillar, V.D.; Overbeck, G.E. Grassland degradation and restoration: A conceptual framework of stages and thresholds illustrated by southern Brazilian grasslands. Nat. Conserv. 2015, 13, 95–104. [Google Scholar] [CrossRef]
- Conant, R.T.; Paustian, K.; Elliott, E.T. Grassland management and conversion into grassland: Effects on soil carbon. Ecol. Appl. 2001, 11, 343–355. [Google Scholar] [CrossRef]
- Bastin, J.-F.; Barbier, N.; Couteron, P.; Adams, B.; Shapiro, A.; Bogaert, J.; De Cannière, C. Aboveground biomass mapping of African forest mosaics using canopy texture analysis: Toward a regional approach. Ecol. Appl. 2014, 24, 1984–2001. [Google Scholar] [CrossRef]
- Sarker, L.R.; Nichol, J.E. Improved forest biomass estimates using ALOS AVNIR-2 texture indices. Remote Sens. Environ. 2011, 115, 968–977. [Google Scholar] [CrossRef]
- Rosenqvist, Å.; Milne, A.; Lucas, R.; Imhoff, M.; Dobson, C. A review of remote sensing technology in support of the Kyoto Protocol. Environ. Sci. Policy 2003, 6, 441–455. [Google Scholar] [CrossRef]
- Lu, D. The potential and challenge of remote sensing-based biomass estimation. Int. J. Remote Sens. 2006, 27, 1297–1328. [Google Scholar] [CrossRef]
- Meng, S.; Pang, Y.; Zhang, Z.; Jia, W.; Li, Z. Mapping Aboveground Biomass using Texture Indices from Aerial Photos in a Temperate Forest of Northeastern China. Remote Sens. 2016, 8, 230. [Google Scholar] [CrossRef]
- Timothy, D.; Onisimo, M.; Riyad, I. Quantifying aboveground biomass in African environments: A review of the trade-offs between sensor estimation accuracy and costs. Trop. Ecol. 2016, 57, 393–405. [Google Scholar]
- Schino, G.; Borfecchia, F.; De Cecco, L.; Dibari, C.; Iannetta, M.; Martini, S.; Pedrotti, F. Satellite estimate of grass biomass in a mountainous range in central Italy. Agrofor. Syst. 2003, 59, 157–162. [Google Scholar] [CrossRef]
- Griffith, J.A.; Price, K.P.; Martinko, E.A. A multivariate analysis of biophysical parameters of tallgrass prairie among land management practices and years. Environ. Monit. Assess. 2001, 68, 249–271. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Sha, Z.; Yu, M.; Bai, Y.; Zhang, L. A comparison of two models with Landsat data for estimating above ground grassland biomass in Inner Mongolia, China. Ecol. Model. 2009, 220, 1810–1818. [Google Scholar] [CrossRef]
- Huete, A.R. Separation of soil-plant spectral mixtures by factor analysis. Remote Sens. Environ. 1986, 19, 237–251. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Nichol, J.E.; Sarker, M.L.R. Improved biomass estimation using the texture parameters of two high-resolution optical sensors. IEEE Trans. Geosci. Remote Sens. 2011, 49, 930–948. [Google Scholar] [CrossRef]
- Ngubane, Z.; Odindi, J.; Mutanga, O.; Slotow, R. Assessment of the Contribution of WorldView-2 Strategically Positioned Bands in Bracken fern (Pteridium aquilinum (L.) Kuhn) Mapping. S. Afr. J. Geomat. 2014, 3, 210–223. [Google Scholar] [CrossRef]
- Ramoelo, A.; Skidmore, A.K.; Cho, M.A.; Schlerf, M.; Mathieu, R.; Heitkönig, I.M. Regional estimation of savanna grass nitrogen using the red-edge band of the spaceborne RapidEye sensor. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 151–162. [Google Scholar] [CrossRef]
- Zhao, P.; Lu, D.; Wang, G.; Wu, C.; Huang, Y.; Yu, S. Examining Spectral Reflectance Saturation in Landsat Imagery and Corresponding Solutions to Improve Forest Aboveground Biomass Estimation. Remote Sens. 2016, 8, 469. [Google Scholar] [CrossRef]
- Broge, N.H.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Pu, R.; Gong, P.; Biging, G.S.; Larrieu, M.R. Extraction of red edge optical parameters from Hyperion data for estimation of forest leaf area index. IEEE Trans. Geosci. Remote Sens. 2003, 41, 916–921. [Google Scholar]
- Delegido, J.; Verrelst, J.; Rivera, J.P.; Ruiz-Verdú, A.; Moreno, J. Brown and green LAI mapping through spectral indices. Int. J. Appl. Earth Obs. Geoinf. 2015, 35, 350–358. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Gholz, H.L. Exploring the relationship between reflectance red edge and chlorophyll content in slash pine. Tree Physiol. 1990, 7, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P. Biophysical and biochemical sources of variability in canopy reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Lu, D. Aboveground biomass estimation using Landsat TM data in the Brazilian Amazon. Int. J. Remote Sens. 2005, 26, 2509–2525. [Google Scholar] [CrossRef]
- Cutler, M.; Boyd, D.; Foody, G.; Vetrivel, A. Estimating tropical forest biomass with a combination of SAR image texture and Landsat TM data: An assessment of predictions between regions. ISPRS J. Photogramm. Remote Sens. 2012, 70, 66–77. [Google Scholar] [CrossRef]
- Eckert, S. Improved forest biomass and carbon estimations using texture measures from WorldView-2 satellite data. Remote Sens. 2012, 4, 810–829. [Google Scholar] [CrossRef]
- Dube, T.; Mutanga, O. Investigating the robustness of the new Landsat-8 Operational Land Imager derived texture metrics in estimating plantation forest aboveground biomass in resource constrained areas. ISPRS J. Photogramm. Remote Sens. 2015, 108, 12–32. [Google Scholar] [CrossRef]
- Wallis, C.I.; Paulsch, D.; Zeilinger, J.; Silva, B.; Fernández, G.F.C.; Brandl, R.; Farwig, N.; Bendix, J. Contrasting performance of Lidar and optical texture models in predicting avian diversity in a tropical mountain forest. Remote Sens. Environ. 2016, 174, 223–232. [Google Scholar] [CrossRef]
- Ozdemir, I.; Karnieli, A. Predicting forest structural parameters using the image texture derived from WorldView-2 multispectral imagery in a dryland forest, Israel. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 701–710. [Google Scholar] [CrossRef]
- Salas, E.A.L.; Boykin, K.G.; Valdez, R. Multispectral and Texture Feature Application in Image-Object Analysis of Summer Vegetation in Eastern Tajikistan Pamirs. Remote Sens. 2016, 8, 78. [Google Scholar] [CrossRef]
- Fynn, R.W.; O’Connor, T.G. Determinants of community organization of a South African mesic grassland. J. Veg. Sci. 2005, 16, 93–102. [Google Scholar] [CrossRef]
- Morris, C.; Fynn, R. The Ukulinga long-term grassland trials: Reaping the fruits of meticulous, patient research. Bull. Grassl. Soc. South. Afr. 2001, 11, 7–22. [Google Scholar]
- De Wit, M.; Blignaut, J.; Nazare, F. Monetary Valuation of the Grasslands in South Africa. 2006. Available online: http://biodiversityadvisor.sanbi.org/wp-content/uploads/2014/07/2006deWit_Background-InfoRep5_Strategic-Monetary-valuation.pdf (accessed on 6 January 2017).
- Sibanda, M.; Mutanga, O.; Rouget, M. Testing the capabilities of the new WorldView-3 spaceborne sensor’s red-edge spectral band in discriminating and mapping complex grassland management treatments. Int. J. Remote Sens. 2017, 38, 1–22. [Google Scholar] [CrossRef]
- Schumacher, P.; Mislimshoeva, B.; Brenning, A.; Zandler, H.; Brandt, M.; Samimi, C.; Koellner, T. Do Red Edge and Texture Attributes from High-Resolution Satellite Data Improve Wood Volume Estimation in a Semi-Arid Mountainous Region? Remote Sens. 2016, 8, 540. [Google Scholar] [CrossRef]
- Chica-Olmo, M.; Abarca-Hernandez, F. Computing geostatistical image texture for remotely sensed data classification. Comput. Geosci. 2000, 26, 373–383. [Google Scholar] [CrossRef]
- Wang, L.; Sousa, W.P.; Gong, P.; Biging, G.S. Comparison of IKONOS and QuickBird images for mapping mangrove species on the Caribbean coast of Panama. Remote Sens. Environ. 2004, 91, 432–440. [Google Scholar] [CrossRef]
- Safari, A.; Sohrabi, H. Ability of Landsat-8 OLI derived texture metrics in estimating aboveground carbon stocks of coppice Oak Forests. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 751–754. [Google Scholar] [CrossRef]
- Kang, Y.; Özdoğan, M.; Zipper, S.C.; Román, M.O.; Walker, J.; Hong, S.Y.; Marshall, M.; Magliulo, V.; Moreno, J.; Alonso, L. How Universal Is the Relationship between Remotely Sensed Vegetation Indices and Crop Leaf Area Index? A Global Assessment. Remote Sens. 2016, 8, 597. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote estimation of chlorophyll content in higher plant leaves. Int. J. Remote Sens. 1997, 18, 2691–2697. [Google Scholar] [CrossRef]
- Fernández-Manso, A.; Fernández-Manso, O.; Quintano, C. SENTINEL-2A red-edge spectral indices suitability for discriminating burn severity. Int. J. Appl. Earth Obs. Geoinf. 2016, 50, 170–175. [Google Scholar] [CrossRef]
- Santoso, H.; Gunawan, T.; Jatmiko, R.H.; Darmosarkoro, W.; Minasny, B. Mapping and identifying basal stem rot disease in oil palms in North Sumatra with QuickBird imagery. Precis. Agric. 2011, 12, 233–248. [Google Scholar] [CrossRef]
- Tucker, C.J. A critical review of remote sensing and other methods for non-destructive estimation of standing crop biomass. Grass Forage Sci. 1980, 35, 177–182. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Cabezas, J.; Galleguillos, M.; Perez-Quezada, J.F. Predicting Vascular Plant Richness in a Heterogeneous Wetland Using Spectral and Textural Features and a Random Forest Algorithm. IEEE Geosci. Remote Sens. Lett. 2016, 13, 646–650. [Google Scholar] [CrossRef]
- Merzlyak, M.; Gitelson, A.A.; Chivkunova, O.; Solovchenko, A.; Pogosyan, S. Application of reflectance spectroscopy for analysis of higher plant pigments. Russ. J. Plant Physiol. 2003, 50, 704–710. [Google Scholar] [CrossRef]
- El-Shikha, D.M.; Barnes, E.M.; Clarke, T.R.; Hunsaker, D.J.; Haberland, J.A.; Pinter, P., Jr.; Waller, P.M.; Thompson, T.L. Remote sensing of cotton nitrogen status using the Canopy Chlorophyll Content Index (CCCI). Trans. ASABE 2008, 51, 73–82. [Google Scholar] [CrossRef]
- Fitzgerald, G.; Rodriguez, D.; O’Leary, G. Measuring and predicting canopy nitrogen nutrition in wheat using a spectral index—The canopy chlorophyll content index (CCCI). Field Crop. Res. 2010, 116, 318–324. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.; Zur, Y.; Stark, R.; Gritz, U. Non-destructive and remote sensing techniques for estimation of vegetation status. Pap. Nat. Resour. 2001, 273, 205–210. [Google Scholar]
- Kelsey, K.C.; Neff, J.C. Estimates of aboveground biomass from texture analysis of Landsat imagery. Remote Sens. 2014, 6, 6407–6422. [Google Scholar] [CrossRef]
- Ouma, Y.O.; Tetuko, J.; Tateishi, R. Analysis of co-occurrence and discrete wavelet transform textures for differentiation of forest and non-forest vegetation in very-high-resolution optical-sensor imagery. Int. J. Remote Sens. 2008, 29, 3417–3456. [Google Scholar] [CrossRef]
- Chun, H.; Keleş, S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2010, 72, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahman, E.M.; Mutanga, O.; Odindi, J.; Adam, E.; Odindo, A.; Ismail, R. A comparison of partial least squares (PLS) and sparse PLS regressions for predicting yield of Swiss chard grown under different irrigation water sources using hyperspectral data. Comput. Electron. Agric. 2014, 106, 11–19. [Google Scholar] [CrossRef]
- Frazer, G.W.; Magnussen, S.; Wulder, M.A.; Niemann, K.O. Simulated impact of sample plot size and co-registration error on the accuracy and uncertainty of LiDAR-derived estimates of forest stand biomass. Remote Sens. Environ. 2011, 115, 636–649. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, X.; Schull, M.A.; Román-Colón, M.O.; Yao, T.; Wang, Z.; Zhang, Q.; Jupp, D.L.; Lovell, J.L.; Culvenor, D.S. Measuring effective leaf area index, foliage profile, and stand height in New England forest stands using a full-waveform ground-based lidar. Remote Sens. Environ. 2011, 115, 2954–2964. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.; Corsi, F.; van Wieren, S.E.; Sobhan, I. Estimation of green grass/herb biomass from airborne hyperspectral imagery using spectral indices and partial least squares regression. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 414–424. [Google Scholar] [CrossRef]
- Skidmore, A.K.; Ferwerda, J.G.; Mutanga, O.; Van Wieren, S.E.; Peel, M.; Grant, R.C.; Prins, H.H.; Balcik, F.B.; Venus, V. Forage quality of savannas—simultaneously mapping foliar protein and polyphenols for trees and grass using hyperspectral imagery. Remote Sens. Environ. 2010, 114, 64–72. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 610–621. [Google Scholar] [CrossRef]
- Zhang, L.; Cheng, Q.; Li, C. Improved model for estimating the biomass of Populus euphratica forest using the integration of spectral and textural features from the Chinese high-resolution remote sensing satellite GaoFen-1. J. Appl. Remote Sens. 2015, 9, 096010. [Google Scholar] [CrossRef]
- Takayama, T.; Iwasaki, A. Optimal Wavelength Selection on Hyperspectral Data with Fused Lasso for Biomass Estimation of Tropical Rain Forest. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, III-8, 101–108. [Google Scholar] [CrossRef]
- Fujiki, S.; Okada, K.-I.; Nishio, S.; Kitayama, K. Estimation of the stand ages of tropical secondary forests after shifting cultivation based on the combination of WorldView-2 and time-series Landsat images. ISPRS J. Photogramm. Remote Sens. 2016, 119, 280–293. [Google Scholar] [CrossRef]
- Shen, W.; Li, M.; Huang, C.; Wei, A. Quantifying Live Aboveground Biomass and Forest Disturbance of Mountainous Natural and Plantation Forests in Northern Guangdong, China, Based on Multi-Temporal Landsat, PALSAR and Field Plot Data. Remote Sens. 2016, 8, 595. [Google Scholar] [CrossRef]
- Kuplich, T.; Curran, P.J.; Atkinson, P.M. Relating SAR image texture to the biomass of regenerating tropical forests. Int. J. Remote Sens. 2005, 26, 4829–4854. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The red edge position and shape as indicators of plant chlorophyll content, biomass and hydric status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Sibanda, M.; Mutanga, O.; Rouget, M. Examining the potential of Sentinel-2 MSI spectral resolution in quantifying above ground biomass across different fertilizer treatments. ISPRS J. Photogramm. Remote Sens. 2015, 110, 55–65. [Google Scholar] [CrossRef]
- Sibanda, M.; Mutanga, O.; Rouget, M. Discriminating Rangeland Management Practices Using Simulated HyspIRI, Landsat 8 OLI, Sentinel 2 MSI, and VENμS Spectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 1–13. [Google Scholar] [CrossRef]
- Gara, T.W.; Murwira, A.; Ndaimani, H. Predicting forest carbon stocks from high resolution satellite data in dry forests of Zimbabwe: Exploring the effect of the red-edge band in forest carbon stocks estimation. Geocarto Int. 2016, 31, 176–192. [Google Scholar] [CrossRef]
- Mutanga, O.; Adam, E.; Cho, M.A. High density biomass estimation for wetland vegetation using WorldView-2 imagery and random forest regression algorithm. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 399–406. [Google Scholar] [CrossRef]
- Tainton, N.; Groves, R.; Nash, R. Time of mowing and burning veld: Short term effects on production and tiller development. Proc. Annu. Congr. Grassl. Soc. South. Afr. 1977, 12, 59–64. [Google Scholar] [CrossRef]
- Kirkman, K.P.; Collins, S.L.; Smith, M.D.; Knapp, A.K.; Burkepile, D.E.; Burns, C.E.; Fynn, R.W.; Hagenah, N.; Koerner, S.E.; Matchett, K.J. Responses to fire differ between South African and North American grassland communities. J. Veg. Sci. 2014, 25, 793–804. [Google Scholar] [CrossRef]
- Mentis, M.; Tainton, N. The effect of fire on forage production and quality. In Ecological Effects of Fire in South African Ecosystems; Springer: Berlin/Heidelberg, Germany, 1984; pp. 245–254. [Google Scholar]
- Van-Wyk, D. The Effects of Type, Season and Frequency of Defoliation on Species Diversity, Richness, Evenness and Production of the Mowing-Burning Trials at Ukulinga Research Farm in the Southern Tall Grassveld; University of Natal: Pietermaritzburg, South Africa, 1998. [Google Scholar]
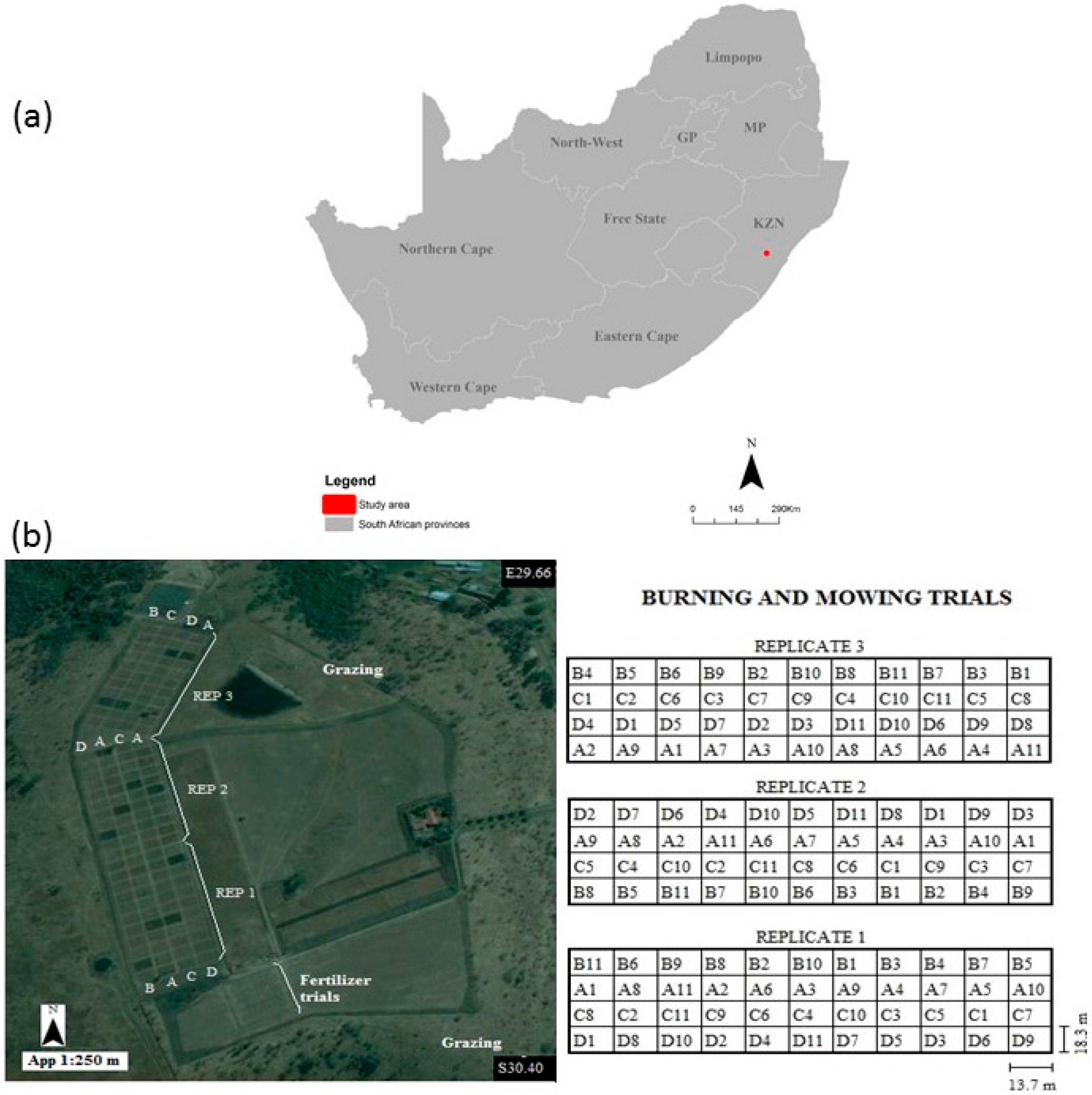
| Treatment Level | Treatment | Samples | Plots |
|---|---|---|---|
| C1 | Control | 60 | 3 |
| C2 | Annual burn (in August) | 60 | 3 |
| C3 | Annual burn (after Spring rain) | 60 | 3 |
| C4 | Biennial burn (in August) | 60 | 3 |
| C5 | Biennial burn (after Spring rain) | 60 | 3 |
| C7 | Triennial burn (in August) | 60 | 3 |
| C8 | Triennial burn (after Spring rain) | 60 | 3 |
| C10 | Mowing (in August) | 60 | 3 |
| C11 | Mowing (after Spring rain) | 60 | 3 |
| D1 | Control | 60 | 3 |
| D2 | Annual burn (in August) | 60 | 3 |
| D3 | Annual burn (after Spring rain) | 60 | 3 |
| D4 | Biennial burn (in August) | 60 | 3 |
| D5 | Biennial burn (after Spring rain) | 60 | 3 |
| D7 | Triennial burn (in August) | 60 | 3 |
| D8 | Triennial burn (after Spring rain) | 60 | 3 |
| D10 | Mowing (in August) | 60 | 3 |
| D11 | Mowing (after Spring rain) | 60 | 3 |
| Total | 1080 | 54 |
| Phase | Analysis | Variable | Description | Reference |
|---|---|---|---|---|
| 1 | Bands | WV-3 B2-B8 | Single-bands—reflectance values | |
| vs. | ||||
| Broadband VIs | Broadband VIs | |||
| Chlorophyll Index Green | Kang et al. [42], Gitelson et al. [43] | |||
| Green normalised difference VI | Fernández-Manso et al. [44] | |||
| Green blue normalised difference VI | Santoso et al. [45] | |||
| Normalised difference VI | Tucker [46] | |||
| Soil adjusted vegetation index | Huete [47] | |||
| Enhanced vegetation index | Cabezas et al. [48] | |||
| 2 | Broadband VIs + bands | Red-Edge Indices | ||
| vs. | Browning reflectance index | Merzlyak et al. [49] | ||
| Red-Edge Vis | Canopy chlorophyll content index | El-Shikha et al. [50] | ||
| Normalised difference near-infrared red-edge index | ||||
| Normalised difference red-edge index | Fitzgerald et al. [51] | |||
| Tasseled cap: Soil brightness Index | Cabezas et al. [48] | |||
| Anthocyanin reflectance Index | Gitelson et al. [52] | |||
| 3 | All VI + Bands | Single Band Textures, windows (3 and 5) | ||
| vs. | Texture type: | |||
| Single-band textures | Mean | Wallis [31] Kelsey et al. [53] Schumacher et al. [38] Ouma et al. [54] Salas et al. [33] Zhao et al. [21] | ||
| Variance | ||||
| Homogeneity | Wallis [31] Kelsey et al. [53] Schumacher et al. [38] Ouma et al. [54] Salas et al. [33] Zhao et al. [21] | |||
| Contrast | Wallis [31] Kelsey et al. [53] Schumacher et al. [38] Ouma et al. [54] Salas et al. [33] Zhao et al. [21] | |||
| Dissimilarity | ||||
| Entropy | ||||
| Second moment | ||||
| Correlation | ||||
| 4 | Band texture variables | Band-ratios texture | B2/B3, B2/B5, B2/B7, B2/B8, B3/B5, B3/B7, B3/B8, B5/B7, B5/B8, B2/B6, B3/B6, B6/B7, B6/B8, B6/B8, B8/B7, | |
| vs. | ||||
| All combined data | ||||
| C2 | 0.00 | ||||||||||||||||
| C3 | 0.00 | 0.00 | |||||||||||||||
| C4 | 0.00 | 0.89 | 0.00 | 0.00 | Significant (α = 0.05) | ||||||||||||
| C5 | 0.00 | 1.00 | 0.00 | 1.00 | 1.00 | Non-Significant | |||||||||||
| C7 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||||||
| C8 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | |||||||||||
| C10 | 0.00 | 0.53 | 0.04 | 0.00 | 0.04 | 0.00 | 0.14 | ||||||||||
| C11 | 0.00 | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.14 | |||||||||
| D1 | 0.00 | 0.02 | 0.00 | 0.94 | 0.37 | 0.00 | 0.00 | 0.00 | 0.00 | ||||||||
| D2 | 0.00 | 0.04 | 0.00 | 0.98 | 0.53 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | |||||||
| D3 | 0.00 | 0.00 | 0.00 | 0.73 | 0.14 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | ||||||
| D4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |||||
| D5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.24 | 0.14 | 0.53 | 0.03 | ||||
| D7 | 0.00 | 0.08 | 0.00 | 1.00 | 0.69 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 1.00 | 1.00 | 0.00 | 0.08 | |||
| D8 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.37 | 0.24 | 0.69 | 0.01 | 1.00 | 0.14 | ||
| D10 | 0.00 | 0.00 | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.92 | 0.83 | 0.99 | 0.00 | 1.00 | 0.69 | 1.00 | |
| D11 | 0.00 | 0.97 | 0.00 | 1.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.83 | 0.92 | 0.53 | 0.00 | 0.00 | 0.97 | 0.00 | 0.01 |
| Treatment | C1 | C2 | C3 | C4 | C5 | C7 | C8 | C10 | C11 | D1 | D2 | D3 | D4 | D5 | D7 | D8 | D10 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sibanda, M.; Mutanga, O.; Rouget, M.; Kumar, L. Estimating Biomass of Native Grass Grown under Complex Management Treatments Using WorldView-3 Spectral Derivatives. Remote Sens. 2017, 9, 55. https://doi.org/10.3390/rs9010055
Sibanda M, Mutanga O, Rouget M, Kumar L. Estimating Biomass of Native Grass Grown under Complex Management Treatments Using WorldView-3 Spectral Derivatives. Remote Sensing. 2017; 9(1):55. https://doi.org/10.3390/rs9010055
Chicago/Turabian StyleSibanda, Mbulisi, Onisimo Mutanga, Mathieu Rouget, and Lalit Kumar. 2017. "Estimating Biomass of Native Grass Grown under Complex Management Treatments Using WorldView-3 Spectral Derivatives" Remote Sensing 9, no. 1: 55. https://doi.org/10.3390/rs9010055
APA StyleSibanda, M., Mutanga, O., Rouget, M., & Kumar, L. (2017). Estimating Biomass of Native Grass Grown under Complex Management Treatments Using WorldView-3 Spectral Derivatives. Remote Sensing, 9(1), 55. https://doi.org/10.3390/rs9010055