1. Introduction
Individual tree crown (ITC) detection and delineation are foundational elements in contemporary forest inventory systems, providing tree-level insights that enable precise forest management and ecological research. In commercial forestry, ITC data facilitates selective timber identification based on specific metrics, such as diameter at breast height (DBH) and crown volume, optimizing resource utilization while supporting sustainable practices [
1]. The detailed inventory not only optimizes resource utilization to maximize economic returns but also aligns with sustainable forest management principles [
2]. Beyond commercial applications, ITC data underpins ecological research by providing critical information on forest structural complexity for biodiversity assessments, carbon sequestration estimation, and ecosystem health monitoring [
3]. The high-resolution remote sensing data enables researchers to quantify crown dimensions and canopy structure while enabling biomass estimation and revealing indicators of forest vigor and disturbance patterns.
Three-dimensional (3D) ITC data offers value for extracting key biophysical parameters like aboveground biomass (AGB), which serves as a critical indicator for carbon sequestration models that predict future carbon fluxes and climate change impacts [
4,
5]. The demand for accurate 3D ITC information has increased significantly in recent years, driven by the increasing complexity of forest management challenges and the pressing need for sustainable resource use. This urgency is compounded by global climate changes and natural disturbances, such as fires, storms, and pest infestations, highlighting the pivotal role of ITC data in adaptive forest management and ecological conservation.
Mixed-wood forests present significant challenges for accurate ITC detection and delineation due to overlapping canopies and variations in tree shapes and sizes, which are primarily influenced by species composition and age structure. The diverse structural characteristics of these forests make it difficult to distinguish individual crowns, especially in dense stands, leading to potential errors in ITC detection and delineation accuracy [
6,
7,
8]. High-resolution Light Detection and Ranging (LiDAR) data have revolutionized this field by providing detailed structural information about forest canopies, enabling precise measurements of tree height, crown diameter, and canopy structures. The integration of deep learning techniques with LiDAR data has significantly enhanced the accuracy and efficiency of ITC detection and delineation [
6,
7,
9,
10].
Current ITC delineation methodologies using LiDAR data are generally divided into two main approaches: (1) 2D Canopy Height Model (CHM)-based methods and (2) 3D LiDAR point-based methods. CHM-based methods are widely adopted due to their simplicity and computational efficiency [
2,
8,
10]. When integrating deep learning techniques with CHM-based methods, two types of networks are commonly applied: semantic segmentation networks and instance segmentation networks. Semantic segmentation networks, such as U-Net [
11], FCN [
12], and DeepLabV3+ [
13,
14] have demonstrated impressive capabilities in semantic segmentation from raster images. While these networks are effective in identifying general individual tree crown areas, they often struggle to distinguish individual crowns in dense or overlapping canopies. Instance segmentation networks, including Mask R-CNN [
15,
16] and its variants, like Cascade R-CNN [
17], offer enhanced ability to distinguish individual crowns, even in dense canopies. Traditional approaches include watershed segmentation [
18,
19], region growing [
8], graph-based methods [
20], and point cloud clustering [
21]. The watershed algorithm, particularly its marker-controlled variants, remains widely used due to its computational efficiency and interpretability. Similarly, Dalponte’s algorithm [
22] utilizes a region growing approach from local maxima seed points on Canopy Height Models to delineate individual tree crowns, demonstrating robust performance across diverse forest structures while maintaining computational efficiency. While these methods perform well on 2D representations of LiDAR data (e.g., CHMs), they do not fully leverage the rich 3D structural information available in LiDAR point clouds. As a result, these methods can face challenges, particularly in heterogeneous forests with intersecting canopies, leading to reduced performance in areas with overlapping crowns.
In parallel, 3D LiDAR point-based methods offer a more detailed structural characterization, making them particularly well-suited to complex forest environments. Algorithms, such as K-means clustering [
23], voxel space projection [
24], and PointNet++ [
25,
26,
27], process raw point clouds directly and have been successfully applied to ITC delineation. Similarly, algorithms operate directly on 3D point clouds rather than rasterized surfaces, leveraging both horizontal and vertical structural characteristics to segment individual trees [
28]. The integration of advanced machine learning techniques, including deep learning models, like PointNet++, significantly improves ITC delineation accuracy by identifying intricate patterns within the data that traditional methods may miss. PointNet was combined with voxelization and height gradient information from LiDAR data to achieve effective tree crown detection rates, ranging from 0.80 to 0.90, and accurate crown breadth estimations (R
2 > 0.79) across various forest environments [
27]. Point-based 3D LiDAR methods better preserve 3D structural details, enabling more accurate crown delineations [
12,
19]. However, these methods are computationally intensive and can be difficult to scale to large forested areas. Similarly, 3D Convolutional Networks, including 3D U-Net and VoxelNet, have been adapted to process LiDAR data volumetrically, maintaining 3D structural integrity [
29]. These approaches, however, often rely on voxelization, which can lead to the loss of fine-grained structural information.
In summary, despite advancements in integrating deep learning with LiDAR data, several challenges remain. Many CHM-based deep learning methods (e.g., FCN and U-Net) excel in either detection or delineation but rarely achieve both simultaneously. Advanced networks, such as Mask R-CNN, can perform treetop detection, bounding box regression, and ITC mask prediction in an integrated manner. However, they face limitations in fully utilizing the 3D structural richness of LiDAR data. Furthermore, 3D LiDAR point-cloud-based methods often suffer from computational inefficiency and a lack of comprehensive 3D training references, especially when applied to extensive forest environments. This highlights a critical research gap: the need for a deep learning framework that can fully exploit 3D LiDAR data while addressing computational constraints and improving the preparation of training datasets.
To address these challenges and research gaps in ITC detection and delineation within mixed-wood forests, this study proposes a novel two-stage deep learning strategy that integrates CHM-based treetop region segmentation using Mask R-CNN with LiDAR point cloud clustering using the 3D U-Net model. In the first stage, Mask R-CNN is applied to a CHM, leveraging its robust instance segmentation capabilities while maintaining computational efficiency by avoiding direct processing of LiDAR point clouds. The second stage refines crown boundaries in 3D space through a specialized 3D U-Net architecture that integrates spatial and structural information from LiDAR point clouds. The main objectives of this study are as follows:
- i.
To propose a two-stage deep learning framework for improving ITC detection and delineation by fully exploiting LiDAR data.
- ii.
To create a benchmark 2D and 3D reference dataset for airborne LiDAR (LAS) data in mixed-wood forests.
- iii.
To produce 3D and 2D ITC products for practical forestry applications.
3. Methodology
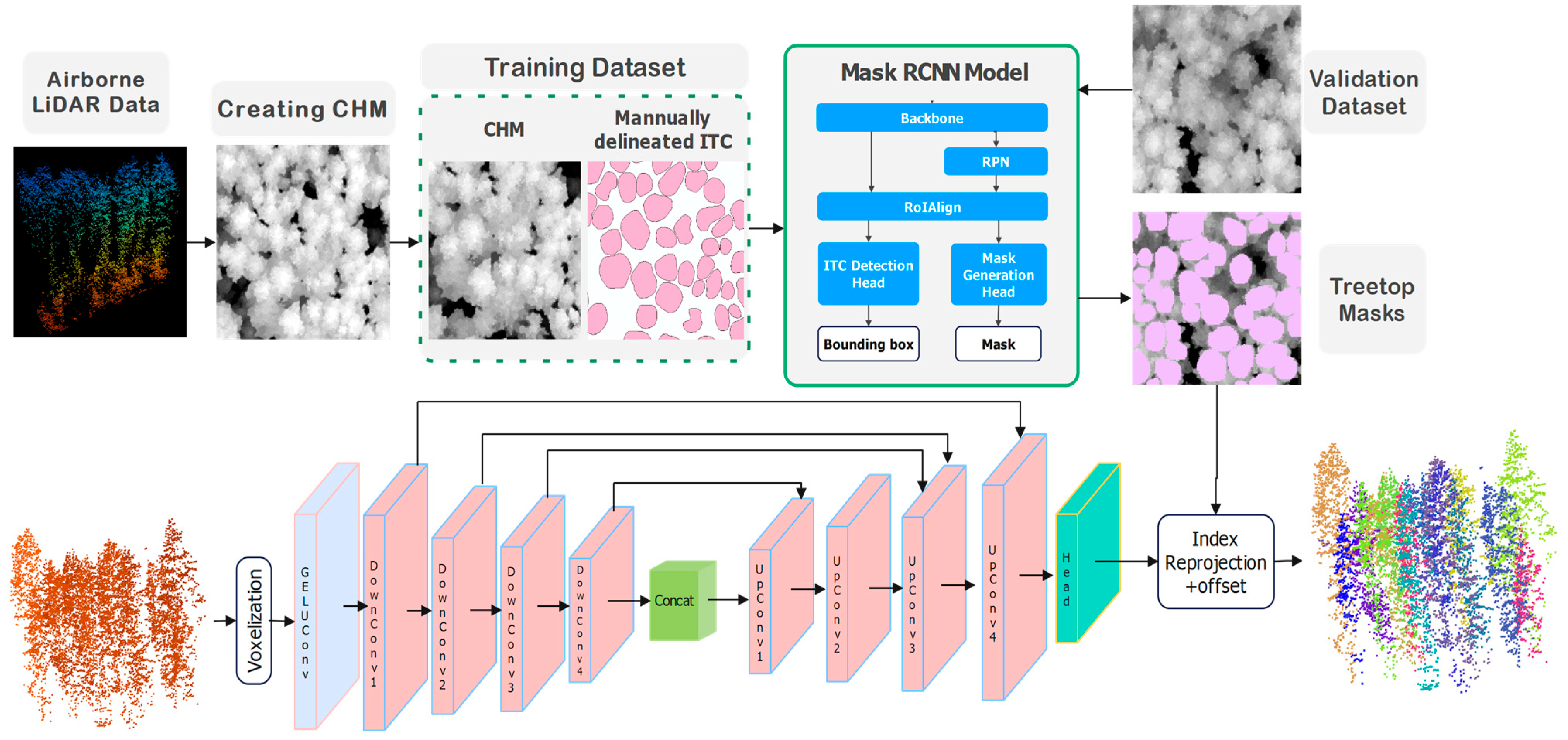
This paper proposes a two-stage framework for individual tree crown (ITC) detection and delineation by integrating CHM-based Mask R-CNN detection and delineation with U-Net model-based LiDAR point cloud clustering.
In the first stage, Mask R-CNN serves as the foundation for initial treetop regions’ identification. This deep learning architecture employs a ResNet50 backbone augmented with Feature Pyramid Networks (FPNs) to process multi-resolution remote sensing imagery. The model systematically generates region proposals across varying scales, enabling it to detect trees of different sizes. The network’s architecture consists of four main components: (1) a backbone network for feature extraction, (2) a Region Proposal Network (RPN) that identifies potential crown locations, (3) a Region of Interest (RoI) alignment layer that precisely maps features to proposed regions, and (4) parallel branches for bounding box regression, classification, and mask generation. Training incorporates overlapping patch sampling with carefully tuned overlap ratios to ensure detection continuity across patch boundaries, while non-maximum suppression algorithms resolve duplicate detections in overlapping areas. The model effectively handles the spectral variability of tree crowns through data augmentation techniques including rotation, scaling, and intensity adjustments.
The second stage employs 3D U-Net to refine the initial treetop regions from the first stage by incorporating structural information from LiDAR point clouds. This specialized 3D neural network processes point-wise features through a series of operations; initial point cloud data are processed through MLP (Multi-Layer Perceptron) convolution layers, followed by four successive down-sampling blocks that reduce spatial resolution while increasing feature depth. The encoder-decoder architecture with skip connections preserves fine-grained spatial details while capturing broader contextual information. The network’s key innovation lies in its offset prediction mechanism, which calculates displacement vectors from each crown point to its corresponding stem location. This offset-based approach enables precise clustering of points belonging to individual crowns, effectively handling complex crown structures and occlusions. The integration of Mask R-CNN and 3D U-Net creates a powerful complementary framework that utilizes the precise treetop region detection capabilities of CHM imagery with the rich structural information from 3D point clouds, resulting in more accurate 3D ITC detection and delineation.
For the 3D point cloud clustering stage, 3D U-Net was selected after comparing it with PointNet++ [
34] and traditional K-means clustering [
35] algorithms. 3D U-Net demonstrated superior performance in capturing the complex spatial relationships within the point cloud while maintaining computational efficiency. PointNet++ showed promise but required significantly more computational resources when applied to our high-density LiDAR data. The hierarchical feature extraction capabilities of 3D U-Net proved particularly effective for discriminating between adjacent crowns in 3D space, especially in mixed-wood environments where deciduous and coniferous trees exhibit vastly different structural characteristics.
The model was implemented using Python version 3.11.4 within the PyCharm (Version 2023.2) [
36], leveraging packages for data processing, machine learning, and visualization. In the first stage, Mask R-CNN was applied to CHM to accurately detect treetop masks, providing precise initial localizations of individual trees. In the second stage, a 3D U-Net architecture was employed to cluster LiDAR points based on treetop masks, delineating ITC boundaries in three-dimensional space. This refinement process utilizes both spatial and height information from the LiDAR point cloud data. By combining the high-resolution segmentation capability of Mask R-CNN with the spatial and structural refinement provided by the 3D U-Net model, this two-stage approach bridges the gap between CHM-based and LiDAR point-cloud-based delineation. It offers a robust framework for capturing both the horizontal extent and vertical structure of tree crowns. The proposed approach was evaluated using accuracy metrics and detailed result analysis.
Figure 4 illustrates the workflow of the two-stage framework proposed in this study.
3.1. First Stage: 2D Treetop Region Segmentation Using Mask R-CNN
Mask R-CNN represents a state-of-the-art deep learning architecture that integrates object detection and segmentation, enabling end-to-end training for treetop region segmentation tasks [
37]. The selection of Mask R-CNN for CHM processing and 3D U-Net for point cloud clustering was based on their complementary strengths and experimentation. Mask R-CNN was chosen for the first stage due to its superior instance segmentation capabilities, which are essential for distinguishing individual tree crowns in the 2D CHM representation. Unlike semantic segmentation networks (e.g., Fully Connected Network (FCN) and SegNet), which excel at pixel-level classification but struggle with separating adjacent instances, Mask R-CNN’s RPN effectively identifies discrete tree crowns, even in dense canopies. We experimented with several alternative approaches, including Faster R-CNN and YOLO for detection, but found that they lacked the precise mask generation needed for accurate crown delineation. Similarly, semantic segmentation networks, FCN and SegNet, were tested but proved less effective at separating adjacent trees in the CHM.
Figure 4 illustrates the workflow of 2D treetop region segmentation using Mask R-CNN, which consists of the following steps: (i) the pre-processed CHM image is put into the pre-trained ResNet-50 neural network to extract feature maps, (ii) these feature maps are processed by RPN to identify Regions of Interest (ROIs), (iii) the ROI Align operation standardizes the ROIs into a uniform shape, (iv) a FCN classifies ITCs and refines the bounding box positions and sizes, and (v) fully Convolutional Networks generate masks by segmenting pixels corresponding to each tree crown.
The Mask R-CNN model was trained using a dataset of approximately 1400 tree crowns. Random patches were generated using a sliding window approach, ensuring the model was exposed to diverse portions of the image during training. These patches included overlapping regions, allowing the model to learn from both the central and boundary areas of tree crowns. To improve generalization across various tree crown shapes and orientations, data augmentation techniques, random rotations and flipping, were applied. Mask R-CNN model was trained to predict both bounding boxes and segmentation masks for each tree crown. During training, the model minimized a combined loss function that included classification, bounding box regression, and mask prediction. Key hyperparameters, including the learning rate, batch size, and IoU threshold, were optimized to enhance performance and ensure accurate tree crown delineation after conducting a series of methodical tests. The optimal learning rate was determined to be 1e−5 using the AdamW optimizer. This conservative learning rate allowed the model to converge effectively to the optimal solution while avoiding local minima. For batch size, it was found that using 2 images per batch provided the best balance between training stability and computational efficiency given our hardware constraints. Smaller batch sizes resulted in noisy gradient updates, while larger batches required excessive memory without proportional improvements in accuracy. The IoU threshold for non-maximum suppression was set at 0.5 after testing values ranging from 0.3 to 0.7. This threshold value optimally balanced the trade-off between detecting closely positioned trees (which requires a lower threshold) and avoiding duplicate detections (which demands a higher threshold). The model utilized the default anchor scales provided by the Mask R-CNN implementation to accommodate the variable sizes of tree crowns in our dataset. The model was trained for up to 20,000 epochs with 100 iterations per epoch and early stopping based on mean IoU (mIoU) validation metrics with a small improvement threshold of 0.001. The best model was saved whenever the validation mIoU improved beyond this threshold. The training implementation used an overlap ratio of 0.2 for patch creation during inference to ensure complete coverage of the study area. This approach was particularly important given our dataset consisting of high-resolution (0.15 m) Canopy Height Model (CHM) imagery.
Post-processing included an optimized polygon merging algorithm that utilized spatial indexing through cKDTree to efficiently identify and merge overlapping tree crown detections based on centroid proximity (within a search radius of 8) and an overlap ratio threshold of 0.5, thereby preventing duplicate detections while preserving unique crown morphologies. For prediction filtering, we applied a distance transformation to each Mask R-CNN generated treetop region, retaining only pixels with confidence values above 0.7, which effectively preserves the core structure of each crown ready for the second stage while reducing edge localization errors. For validation, the best-trained model was evaluated on a separate validation set of about 400 trees with three plots.
3.2. Second Stage: 3D ITC Delineation Using 3D U-Net Model
In the second stage, ITC points were clustered based on treetop masks generated in the first stage using a 3D U-Net regression model. This model predicted 3D offset vectors (ΔX, ΔY, ΔZ) for each voxel, representing the displacement from the voxel’s current position to the nearest ITC point or crown centroid. The 3D U-Net architecture was designed to extract and encode features essential for ITC classification. It employed a series of convolutional layers in the encoder to capture features at progressively coarser spatial scales, followed by decoder layers that reintroduced finer spatial details through concatenation and interpolation. The predicted offsets were then used to adjust the position of each voxel, and the resulting positions were mapped back to the original point cloud by assigning voxel labels to the corresponding points.
The 3D U-Net model is built on a U-Net backbone for point cloud processing. The network begins with an input layer that processes the initial point cloud data through an MLP convolution layer. The encoder path consists of four successive down-sampling blocks (marked as DownConv with channel dimensions [32, 64, 128, 256]), each followed by 3D convolution operations to capture hierarchical features at different scales. The decoder path mirrors the encoder with four up-sampling blocks (UpConv with channel dimensions [32, 64, 128, 256]), each followed by fusion blocks that combine features from the corresponding encoder level through skip connections. These skip connections help preserve fine-grained spatial information that might be lost during down-sampling. The fusion blocks integrate information from both the encoder and decoder paths, enabling the network to effectively combine both local and global features. After each fusion operation, 3D convolution layers further process the combined features. The network culminates in a head layer that processes the final features, ultimately producing offset predictions that help identify and cluster crown points to their corresponding treetop regions. The network employs LeakyReLU (α = 0.2) for intermediate layers and tanh for the output layer, with batch normalization after each convolution and 0.2 dropout in the bottleneck. The model was trained using Adam optimizer (learning rate = 0.0005, β1 = 0.9, β2 = 0.999) with a batch size of 8 voxel blocks (64 × 64 × 32) for 50 epochs (with early stopping after 10 epochs of no improvement), minimizing mean squared error loss for offset prediction to cluster crown points to their corresponding treetop regions. The systematic use of down-sampling and up-sampling operations allows the network to capture both fine-grained local details and broader contextual information necessary for accurate crown point clustering.
To improve performance, a binary voxel block was introduced as a secondary input, indicating the initial crown of treetops identified in the previous stage. This modification was essential for guiding the 3D U-Net to recognize treetops and computing offsets relative to these anchor points. The model was trained using the mean squared error (MSE) loss function to ensure high precision in offset predictions. By applying the computed offsets, ITC points were clustered around their corresponding treetops, streamlining the isolation process. Each treetop’s point was then assigned an ID to enable accurate clustering and delineation of tree crowns.
3.3. Evaluation and Accuracy Assessment
To evaluate the proposed framework, we performed a comparative analysis against the itcSegment algorithm (an established direct 3D point cloud segmentation approach) and Mask R-CNN, using manually delineated reference data. As the benchmark method, we selected the algorithm implemented in the itcLiDAR function from the R package itcSegment [
22]. Unlike methods relying on 2D CHM-based segmentation, this technique preserves the complete vertical structure and architectural complexity of forest canopies. The process begins by analyzing the spatial distribution of points within the cloud, using the provided EPSG code (32617) for spatial context and operating at a specified resolution of 0.15 m. Potential initial points or “seeds” within crowns are identified using a multi-scale search strategy (ranging from 3 to 9) combined with a relative height threshold (0.55). Following seed identification, the algorithm effectively partitions the point cloud by assigning spatially proximate points to individual crown segments. This assignment is governed by distance constraints (1 m to10 m) and criteria evaluating 3D structure and height similarity between points, influenced by parameters like TRESHCrown (0.6) and, potentially, cell weighting (1). Points falling below an absolute Height Threshold (1 m) above ground are excluded from the crown segments. Ultimately, the algorithm assigns a unique numerical identifier (itcSegmentTreeID) to each point belonging to a delineated tree, partitioning the point cloud into discrete 3D tree entities and enabling comparison analysis with the proposed method.
Two accuracy assessments were conducted to evaluate the performance of the proposed two-stage network method [
10]. The first assessment focused on the accuracy of Mask R-CNN delineation from the CHM image using manually delineated ground truth data. The second assessment evaluated the final accuracy of the combined two-stage network through a fully automated approach without manual interventions between stages. In this scenario, errors from each network could propagate to subsequent steps, potentially compounding inaccuracies.
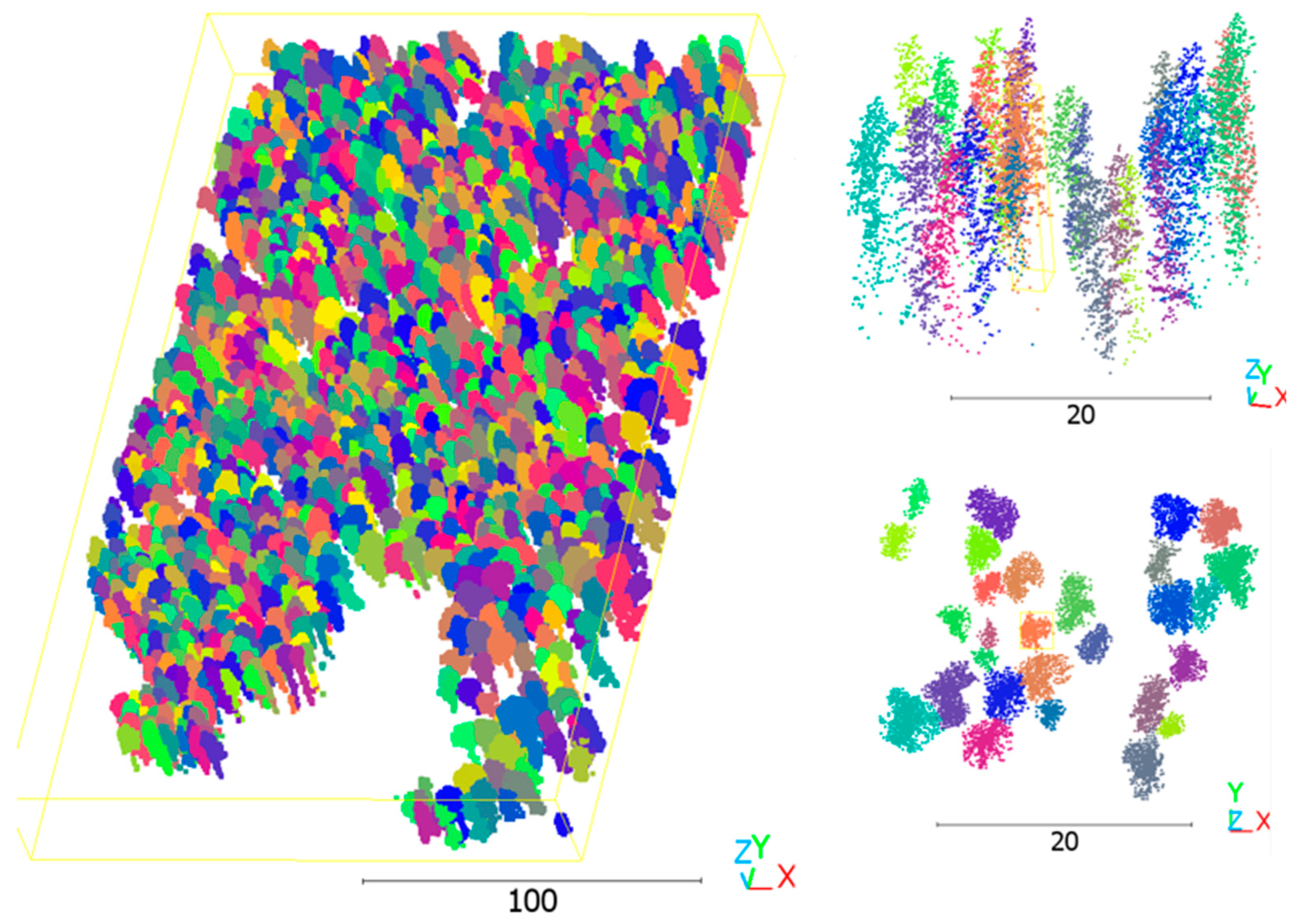
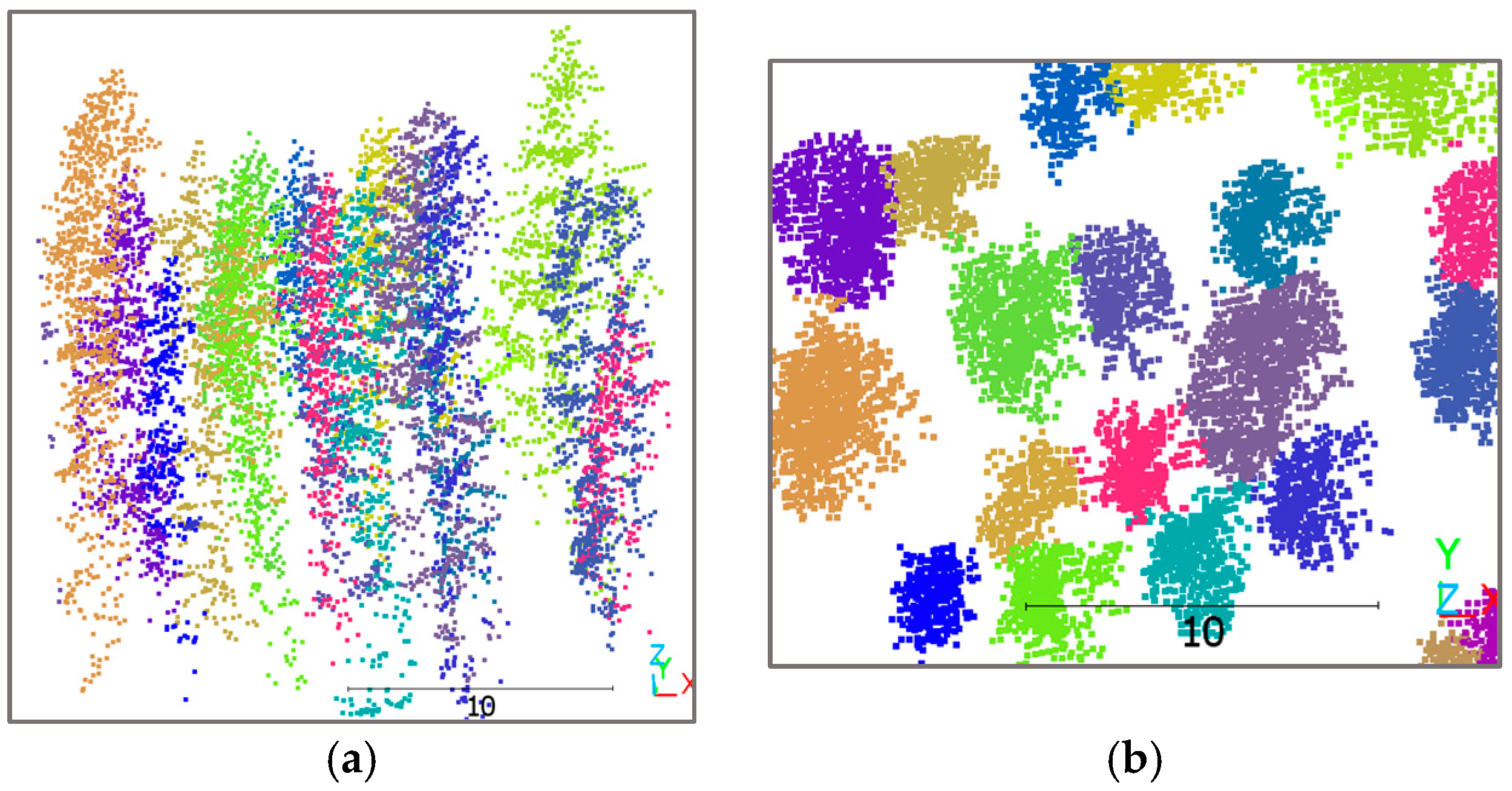
For both assessments, reference data for accuracy evaluation were generated stepwise, tailored to the specific requirements of each network. The 3D LiDAR points of the ITC dataset were derived from manually delineated reference trees on the CHM (
Figure 3). This structured approach ensured that each network’s performance was assessed against precisely relevant criteria, providing detailed insights into both the efficacy of individual networks and the system’s overall performance under both manual and automated conditions.
For ITC detection, precision (Equation (1)) and recall (Equation (2)) metrics were utilized to assess the accuracy of detected trees compared with the reference data. The F1 score, calculated from these metrics (Equation (3)), represents the overall detection accuracy. True positive (TP), false positive (FP), and false negative values were used to quantify detection performance: TP represents the number of correctly detected trees, FP denotes incorrectly detected trees, and FN corresponds to ground truth trees that are overlooked in the detection results.
Accuracy indices were calculated based on these three metrics. Precision evaluates the algorithm’s ability to correctly identify trees, while recall assesses its capability to detect all ground truth trees. A detection was considered correct if a single predicted ITC mask was located within the boundaries of a reference tree. If no ITC mask was found or multiple ITC masks were assigned to a single reference tree, the detection was deemed incorrect. Delineation accuracy is defined as the ratio of correctly delineated tree crowns to the total number of reference tree crowns.
In ITC delineation, which involved ITC mask and crown clustering, an alternative delineation definition was used. The delineation accuracy percentage represents the proportion of trees that were correctly identified and delineated by the algorithm relative to the reference dataset. A tree crown is considered correctly delineated if the Intersection over Union (IoU) between the predicted crown and the reference crown exceeds a predetermined threshold of 0.5. The mean Intersection over Union (mIoU) across all IoUs was adopted to measure the overall agreement between model-generated (Aseg) and reference (Aref) tree boundaries, providing a comprehensive view of the model’s performance in delineating ITC. mIoU can be calculated using the following equation.
5. Discussion
5.1. Performance on Coniferous, Deciduous, and Mixed Plots
The relationship between crown area and delineation accuracy varies across different forest types, as observed in the three validation plots. In the coniferous-dominated plot (Plot-1), where trees typically have smaller and more uniform crowns, the proposed two-stage deep learning method achieved high accuracy, reaching delineation accuracy of 91.48% and an F1 score of 0.88. This outperformed both Mask R-CNN (90.03% accuracy, 0.87 F1) and lidR itcSegment (85.58% accuracy, 0.82 F1) within this study. The compact, well-defined crown shapes of coniferous trees enhance the model’s ability to accurately distinguish individual trees, minimizing segmentation errors. The high precision (0.92) and recall (0.89) values indicate that the model effectively detects and delineates coniferous trees with minimal over-segmentation or omission errors. This superior performance in coniferous plots aligns with findings from previous studies utilizing high-resolution LiDAR data [
38,
39]. The proposed method shows a remarkable improvement, likely due to the enhanced ability of the two-stage approach to capture the typically regular crown shapes of coniferous trees. For instance, studies using fused terrestrial and UAV LiDAR reported F scores around 0.78 for coniferous forests, while approaches using local maxima algorithms on UAV-derived Canopy Height Models in mixed conifer forests have shown F scores around 0.86 [
38]. Some deep learning studies, like those using Mask R-CNN, have reported F1 scores up to 0.91, but often in specific contexts like plantations, placing the results among the higher performers, especially for coniferous stands, where accuracy is often higher due to distinct tree shapes [
4].
In contrast, delineation accuracy declined in mixed-wood (Plot-2) and deciduous plots (Plot-3), reaching 84.18% and 82.38%, respectively. This was slightly lower than its performance in the coniferous plot, a common trend observed in ITC delineation due to the complex and often overlapping crowns of deciduous trees. In the deciduous plot, Mask R-CNN achieved a comparable F1 score of 0.83, while itcSegment scored 0.80. The larger and more irregular crown areas in these plots complicate the segmentation process, as the model struggles to differentiate overlapping or closely spaced tree crowns. The deciduous plot exhibited the lowest precision (0.84), indicating a higher likelihood of false positives, likely due to the broader, less-defined tree crowns. Additionally, the fluctuating structure of deciduous trees, influenced by factors like seasonal foliage changes, introduces further uncertainty in delineation. These findings suggest that crown area is a critical factor affecting segmentation accuracy, with smaller, more defined crowns yielding better results [
18]. To improve accuracy in mixed and deciduous forests, integrating additional data sources, such as spectral information or multi-temporal imagery, could help refine crown boundary detection and enhance model performance across diverse forest structures. Research using fused LiDAR reported F scores around 0.8 for broadleaf forests [
40], while other deep learning applications, like Mask R-CNN, have shown variable results in delineating unseen test trees, with an F1 score of 0.64and a score of 0.74 for the tallest trees based on aerial RGB imagery in complex tropical forests [
4]. These findings suggest that the deep learning methods evaluated in the present study perform well relative to established benchmarks for challenging deciduous environments. The mixed-wood plot (Plot-2) presented intermediate results, with the two-stage approach achieving 84.18% accuracy, outperforming Mask R-CNN (80.04%) and itcSegment (76.15%) by approximately 5.2% and 10.5%, respectively. These results were similar to those reported in a comparable study using UAV-LiDAR data in mixed forests [
41]. The improvement is most pronounced in this challenging mixed-forest environment, where the two-stage method’s ability to leverage both 2D and 3D information proves particularly advantageous for distinguishing adjacent trees of different species and structures, making it a promising approach for operational forest inventory applications across diverse forest compositions.
5.2. Strengths and Limitations of the Proposed Method
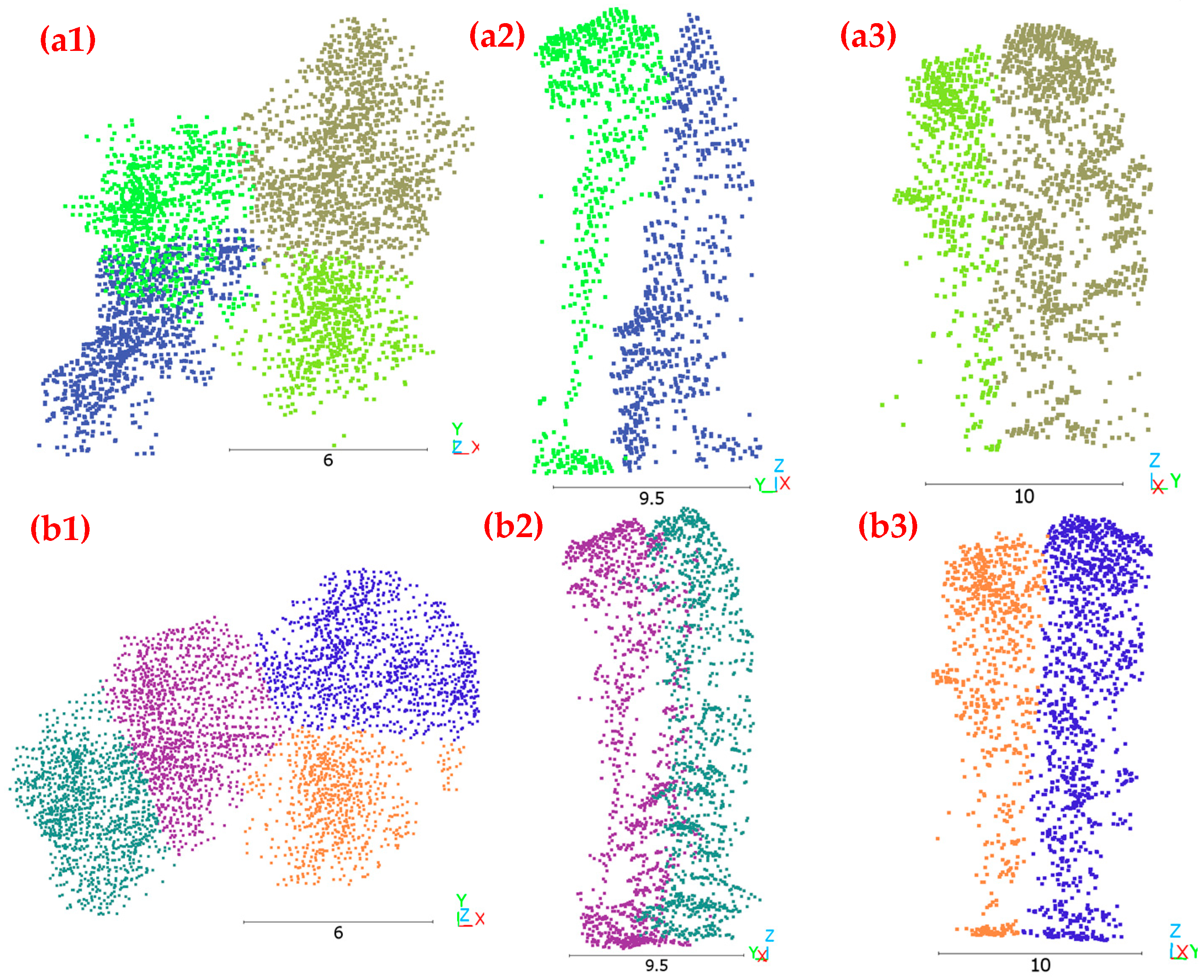
The proposed method demonstrates strengths in 3D ITC delineation compared to the traditional itcSegment approach. As illustrated in
Figure 7, the two-stage deep learning method delineates tree crowns in both horizontal and vertical profiles, addressing a critical limitation of conventional CHM-based approaches. The method’s ability to distinguish individual trees in complex vertical arrangements is evident where the blue-colored tree is positioned beneath the green-colored tree in
Figure 7(a1) yet segmented as a distinct entity. This capability represents a substantial advancement over itcSegment, which struggles with points at adjacent boundaries and under-canopy positions, as shown in
Figure 7(b2). Furthermore, the proposed approach preserves the natural irregularity of crown boundaries, which is especially important for deciduous trees with large volumes and irregular extensions, contrasting with itcSegment’s tendency to create artificially smooth crown boundaries, as shown in
Figure 7(b1,b3). By using treetop regions as clustering references, the two-stage approach achieves more realistic and biologically relevant delineation results, aligning with actual forest structures rather than applying arbitrary geometric constraints.
While the two-stage method improves delineation accuracy by capturing vertical canopy structures more effectively, it still inherits the limitations of Mask R-CNN, particularly regarding boundary accuracy and small tree detection. Mask R-CNN occasionally produces imprecise crown boundaries and fails to detect smaller trees within dense canopies, errors that subsequently propagate to the second stage of our pipeline. This error accumulation is particularly problematic, as treetops missed in the initial detection phase cannot be recovered in the subsequent 3D segmentation stage, resulting in permanent omissions in the final delineation results. Additionally, CHM-based methods like Mask R-CNN face inherent challenges with deciduous trees, which often lack the pronounced height variations found in coniferous species. The relatively homogeneous crown surfaces of deciduous trees frequently lead to under-segmentation, as the subtle height transitions between adjacent crowns are insufficient for effective boundary delineation.
Although the two-stage approach outperforms both Mask R-CNN and itcSegment across all forest types, the performance gap between coniferous (91.48% accuracy) and deciduous forests (82.38% accuracy) indicates remaining challenges in complex canopy environments. Similar performance patterns have been observed in previous studies using high-resolution LiDAR data [
4,
27,
41], where deciduous tree delineation typically yielded lower accuracy than coniferous trees due to complex crown structures. Additionally, the computational demands of the two-stage deep learning approach are considerably greater than those of traditional methods, potentially limiting its application in time-sensitive or resource-constrained scenarios.
Future work should focus on refining the initial detection stage to improve small tree recognition and enhance boundary definition for deciduous trees, potentially by incorporating texture features or spectral information to supplement the height-based differentiation currently employed. Additionally, post-processing techniques, such as applying a secondary refinement step using point cloud clustering, could help recover missed trees and improve recall in complex forest environments.
5.3. 2D and 3D Reference Datasets for ITC Delineation Using LiDAR Data
The development of both 2D and 3D reference datasets plays a crucial role in evaluating the accuracy of ITC delineation models, with each offering distinct advantages and limitations. However, to the best of our knowledge, these aspects have not been extensively discussed in the literature. The 2D reference dataset, primarily derived from CHM, provides a simplified top-down view of the forest structure, making it useful for efficiently annotating individual tree crowns. However, the limited vertical information in CHM-based datasets often results in fuzzy ITC boundaries, particularly in areas where crowns overlap or trees exhibit irregular growth patterns [
4,
15]. This limitation is especially pronounced in mixed-wood and deciduous forests, where the lack of detailed height differentiation complicates accurate crown delineation. The crown boundaries of coniferous trees, characterized by well-defined treetops, are more easily identified using 2D datasets, whereas deciduous trees, with their broad and interconnected canopies, often exhibit segmentation inconsistencies. Despite these challenges, the 2D dataset remains a practical tool for rapid forest assessment and management, especially in coniferous-dominated areas.
In contrast, the 3D reference dataset, constructed from LiDAR point clouds, offers a more comprehensive representation of the forest’s structure by capturing the full vertical profile of individual trees. The 3D dataset overcomes the limitations of 2D datasets by providing detailed structural information, enabling more precise forest management and analysis. Although creating a 3D dataset is time consuming, it effectively captures the three-dimensional nature of each ITC, closely reflecting the true structure of individual trees in a forest. This level of detail is particularly beneficial for deciduous trees, where stem information is critical for accurately identifying tree numbers and delineating ITCs. The proposed methodology implemented a progressive approach beginning with 2D annotations to generate 3D training samples, followed by manual verification to produce final 3D reference data. This hybrid workflow reduces the burden of training data acquisition while maintaining annotation quality. The progression from 2D to 3D reference datasets establishes an accessible pathway for developing sophisticated 3D delineation algorithms, which is particularly crucial as deep learning applications in forestry continue to advance.
With the growing adoption of deep learning approaches in forestry applications, the importance of benchmark datasets cannot be overstated. While public access to our dataset would enhance research impact and reproducibility, the labor-intensive nature of manual tree crown delineation has limited our current validation to 1600 of the estimated 5500 trees in the study area. We intend to make the complete benchmark dataset, including all trees over the study area, publicly accessible online upon completion of validation work in the future, providing a valuable resource for the advancement of LiDAR-based ITC detection and delineation methods.
6. Conclusions
This study introduced a novel two-stage deep learning framework for improving individual tree crown (ITC) detection and delineation in mixed-wood forests. By integrating CHM-based treetop region segmentation using Mask R-CNN with LiDAR point cloud clustering via a specialized 3D U-Net architecture, our approach effectively addresses the limitations of both 2D and 3D methodologies. The first stage leverages Mask R-CNN’s robust instance segmentation capabilities while maintaining computational efficiency, while the second stage refines crown boundaries in 3D space by incorporating the rich structural information available in LiDAR point clouds. Evaluated against manually delineated reference data, our approach outperforms established methods, including Mask R-CNN alone and the lidR itcSegment algorithm, achieving mean mIoU scores of 0.82 for coniferous plots, 0.81 for mixed-wood plots, and 0.79 for deciduous plots. This study demonstrates the great potential of the two-stage deep learning approach as a robust solution for 3D ITC delineation in mixed-wood forests.
Our work will contribute to the field by establishing a comprehensive benchmark dataset of 2D and 3D references for airborne LiDAR data in mixed-wood forests. This benchmark addresses a critical gap in the literature by providing high-quality training data that capture the complexity of heterogeneous forest environments.
The 3D and 2D ITC products generated through this research have significant practical applications in sustainable forest management, biodiversity assessment, and carbon stock estimation. By providing accurate individual tree-level information, forest managers can make more informed decisions regarding resource allocation, conservation strategies, and climate change mitigation efforts. Future work should focus on refining the initial detection stage to improve small tree recognition and enhance boundary definition for deciduous trees, potentially by incorporating texture features or spectral information to supplement the height-based differentiation currently employed. Additionally, post-processing techniques, such as applying a secondary refinement step using point cloud clustering, could help recover missed trees and improve recall in complex forest environments.