Leveraging Open-Source Tools to Analyse Ground-Based Forest LiDAR Data in South Australian Forests
Abstract
1. Introduction
2. Survey of Open Source Tools
2.1. Functional Requirements
2.1.1. Primary Tool Functionality
- Stand-level metrics: Stem count, canopy, cover, and biomass estimation.
- Individual tree metrics: DBH, height, volume, stem angle, stem taper, and creation of quantitative structure models (QSMs).
- Branch and crown attributes: Branch volume, length, angle, count, and crown shape, volume and diameter.
- Feature detection: Identification of specific tree features, such as whorls, double leaders, and knots.
2.1.2. Secondary Tool Functionality
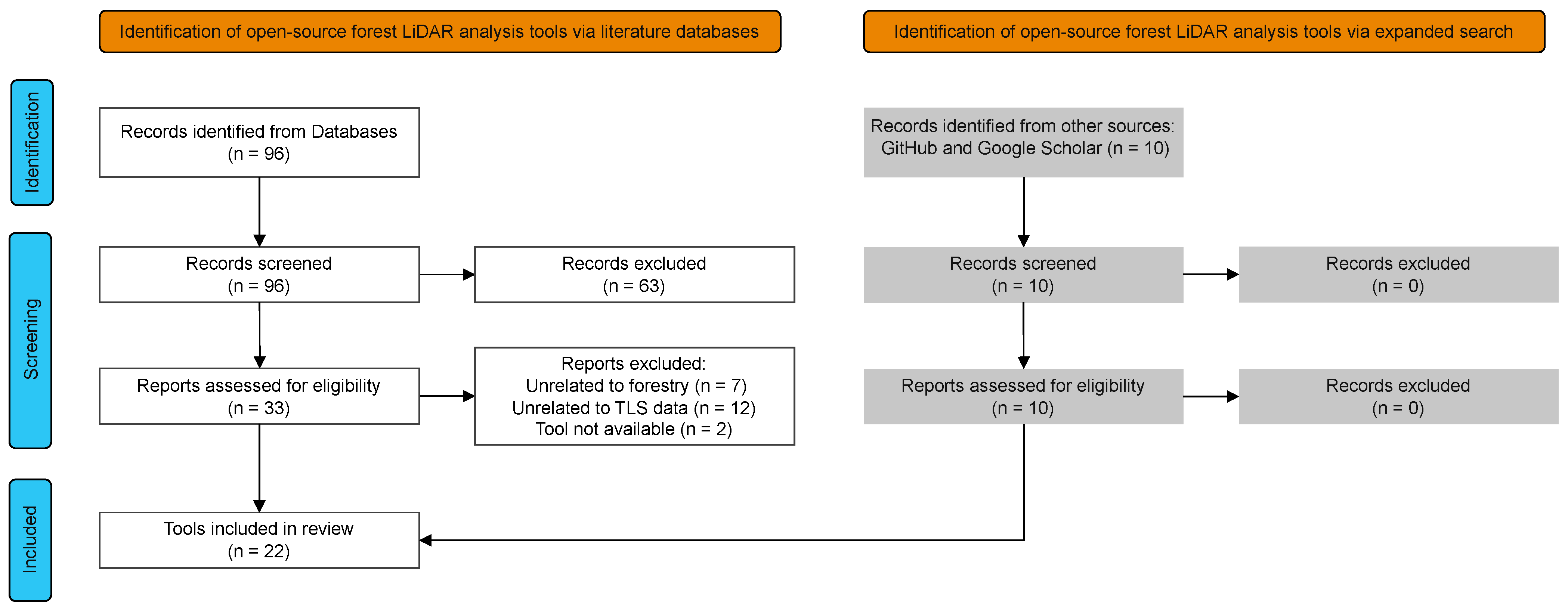
2.2. Semi-Systematic Review
2.2.1. Literature Search
“Terrestrial laser scanning” OR “LiDAR” AND “Forest Inventory” OR “Forest Measurement” OR “Segmentation” AND “Open Source”
2.2.2. Attribute Testing
2.3. Tool Survey Analysis
3. Functional Assessment of Tools
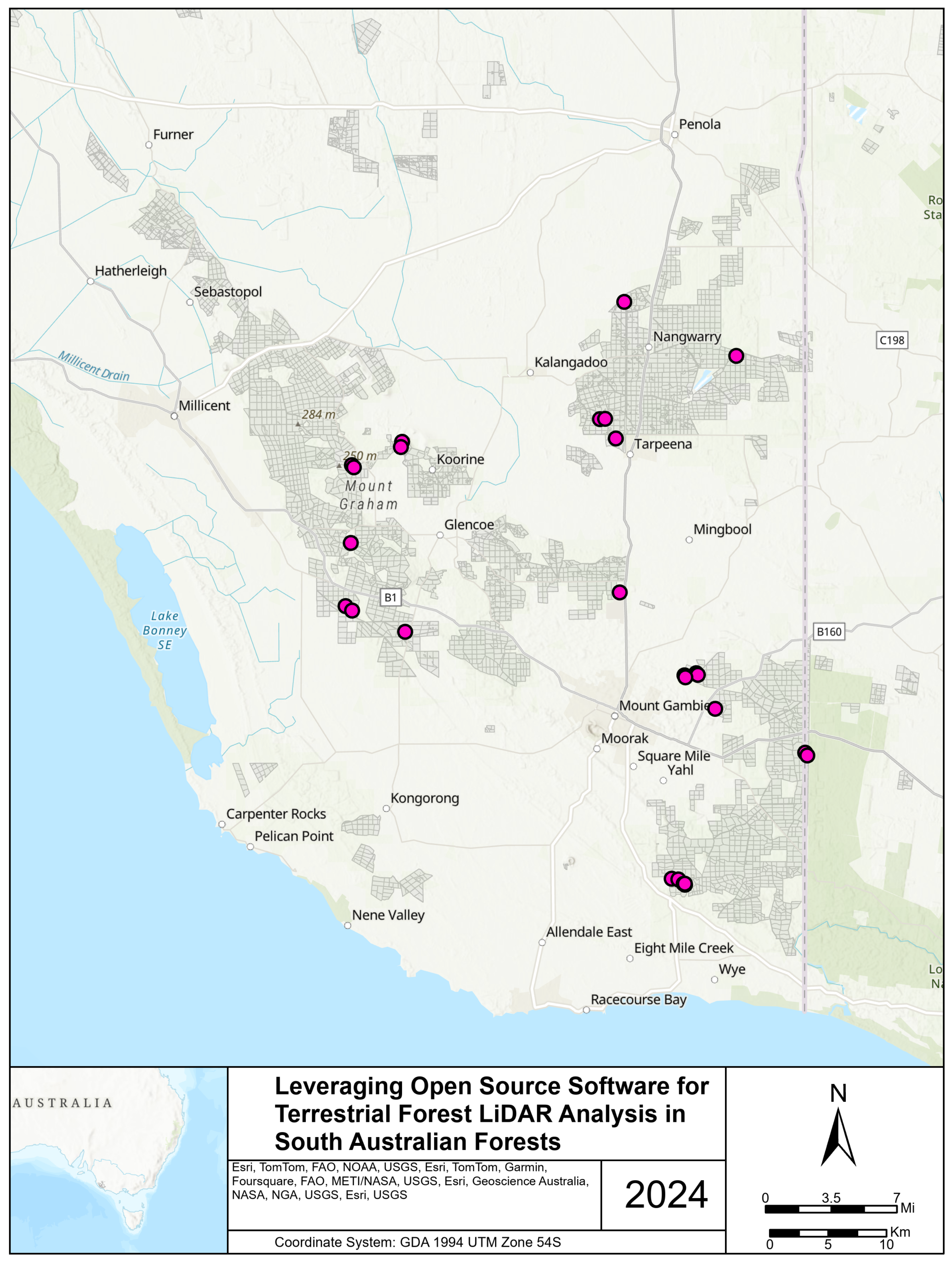
3.1. Materials: Validation Dataset
LiDAR Data Preprocessing
3.2. Methodology
3.2.1. Initial Tool Selection
3.2.2. Tool Calibration
3.2.3. Candidate Tools
3.2.4. Tool Assessment
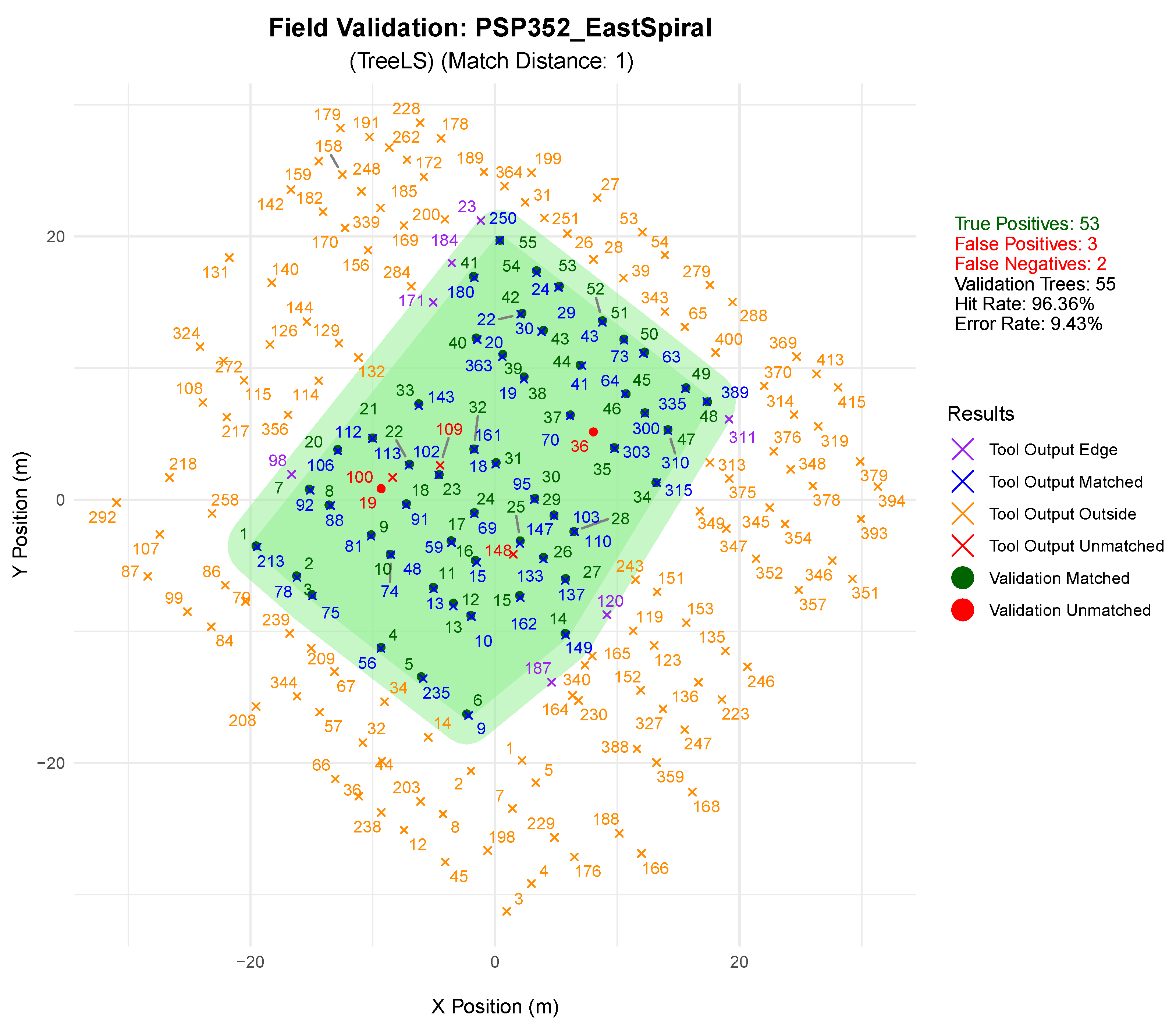
Stem Detection Tool Assessment
Stem Metric Extraction Tool Assessment
4. Results
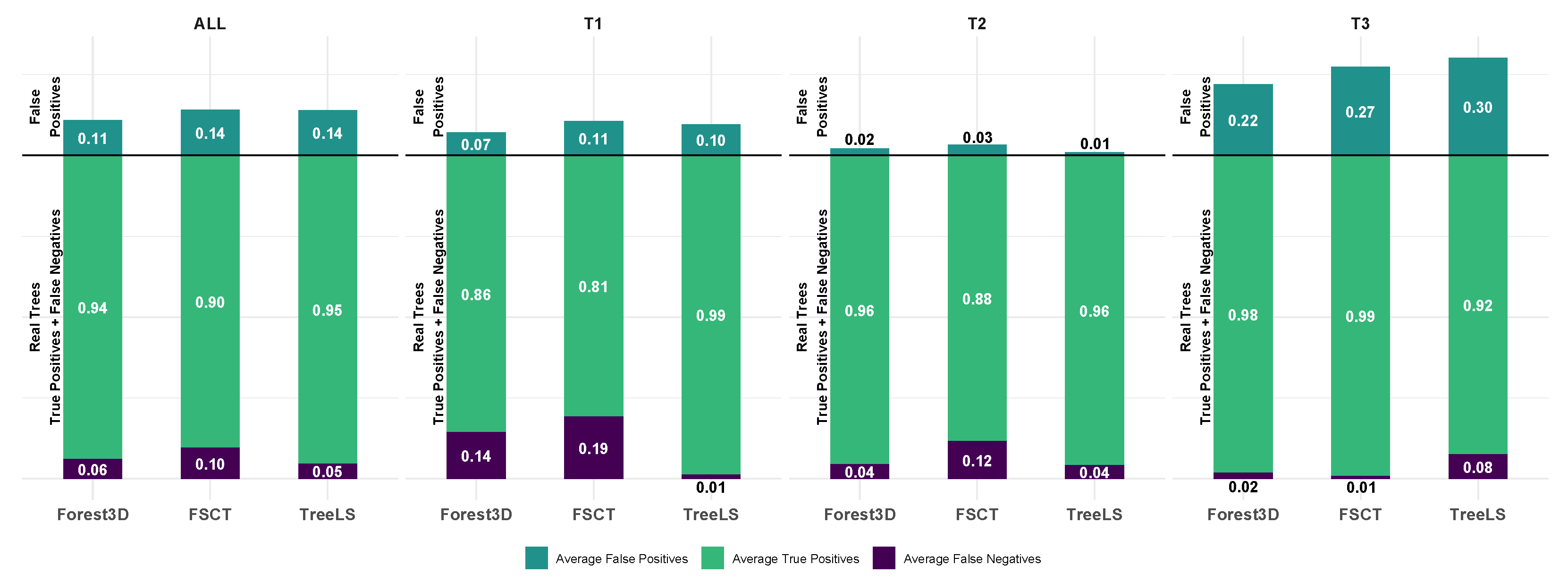
4.1. Stem Segmentation
4.2. Stem Metric Extraction
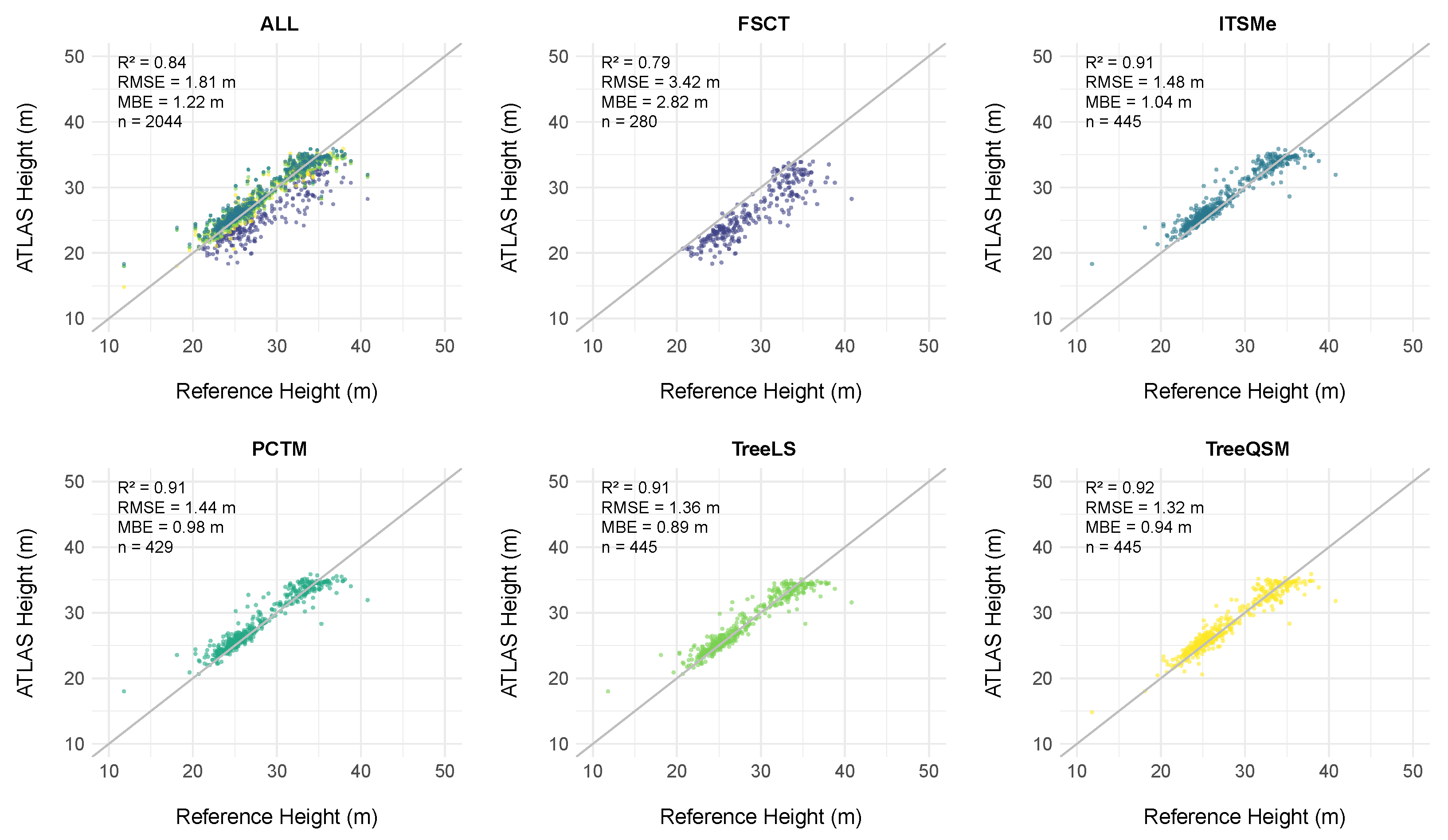
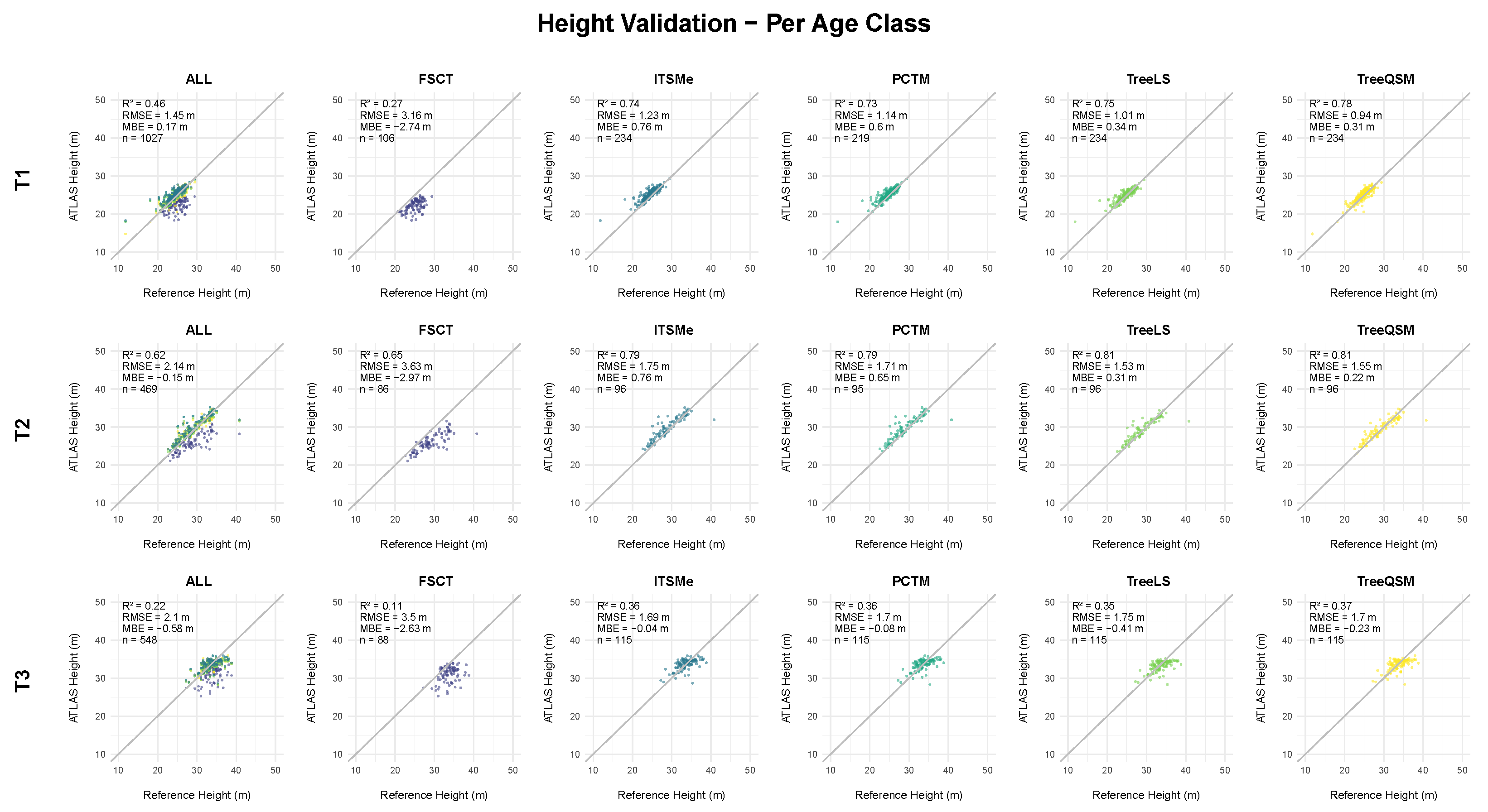
4.2.1. Height Assessment
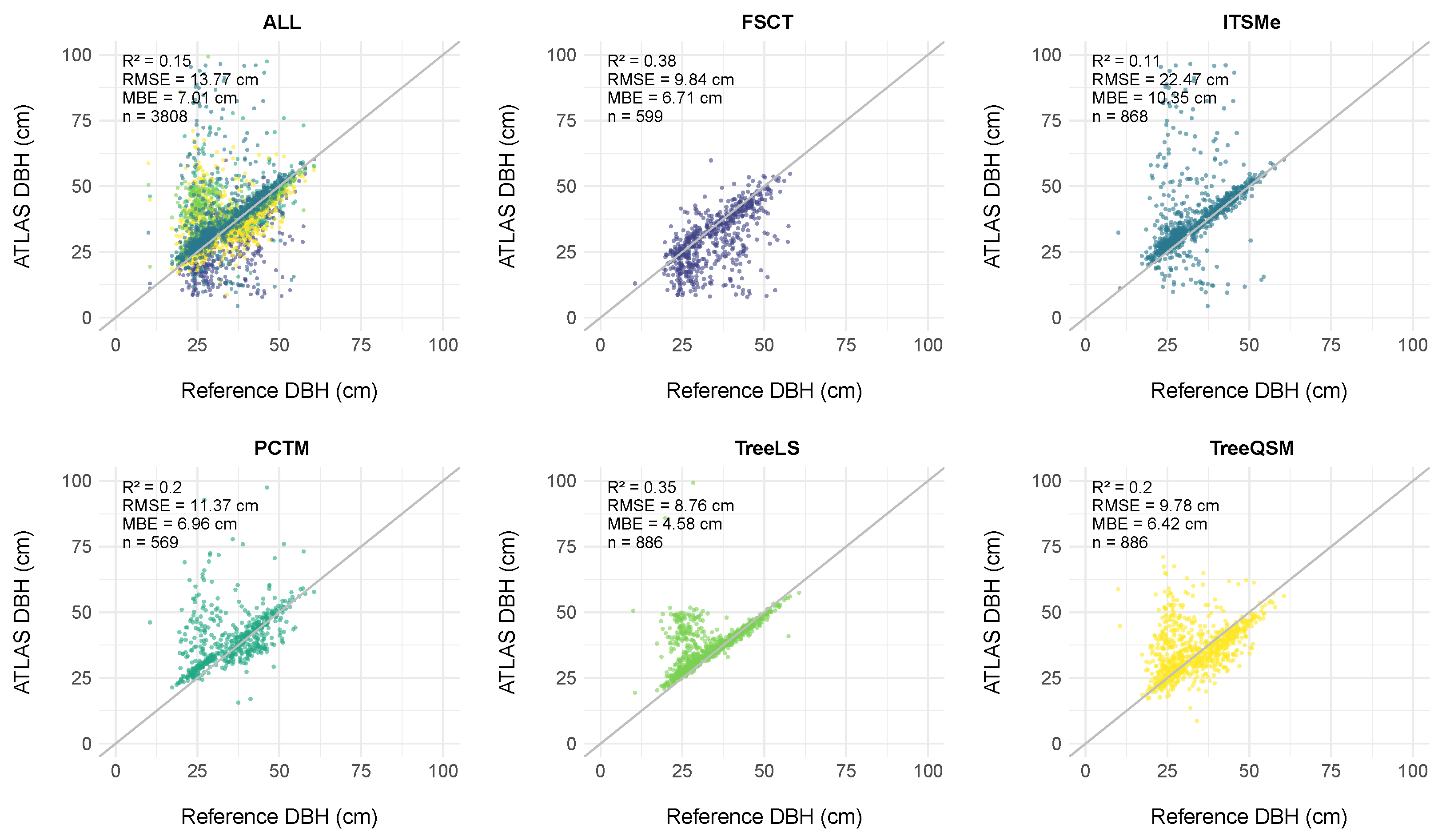
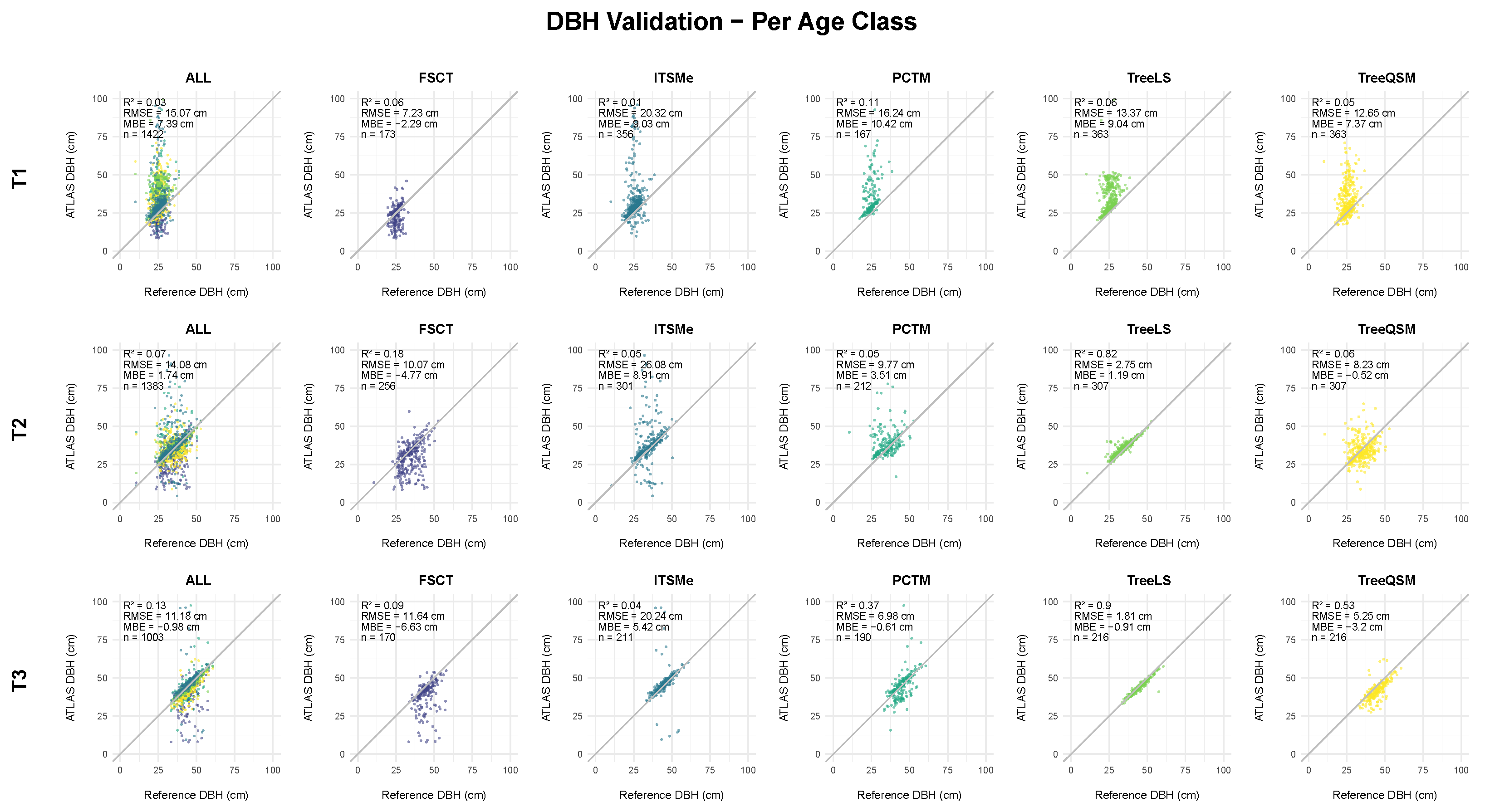
4.2.2. DBH Assessment
5. Discussion
5.1. Stem Segmentation
5.2. Individual Tree Metrics
5.2.1. Height Assessment
5.2.2. DBH Assessment
5.2.3. Comparison to Related Work
5.3. Suggested Implementation
5.4. Limitations
5.5. Future Work
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. Tool Descriptions
Appendix B. Supplementary Statistics
| Thin | Contrast | Mean Difference | SE | df | p Value | 95% CI |
|---|---|---|---|---|---|---|
| T1 | FSCT-ITSMe | |||||
| FSCT-PCTM | ||||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM | ||||||
| T2 | FSCT-ITSMe | |||||
| FSCT-PCTM | ||||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM | ||||||
| T2 | FSCT-ITSMe | |||||
| FSCT-PCTM | ||||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM |
| Thin | Contrast | Mean Difference | SE | df | p Value | 95% CI |
|---|---|---|---|---|---|---|
| T1 | FSCT-ITSMe | |||||
| FSCT-PCTM | 0.024 | |||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM | ||||||
| T2 | FSCT-ITSMe | |||||
| FSCT-PCTM | ||||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM | ||||||
| T2 | FSCT-ITSMe | |||||
| FSCT-PCTM | ||||||
| FSCT-TreeLS | ||||||
| FSCT-TreeQSM | ||||||
| ITSMe-PCTM | ||||||
| ITSMe-TreeLS | ||||||
| ITSMe-TreeQSM | ||||||
| PCTM-TreeLS | ||||||
| PCTM-TreeQSM | ||||||
| TreeLS-TreeQSM |
References
- Shiver, B.D. Sampling Techniques for Forest Resource Inventory; Wiley: New York, NY, USA, 1996. [Google Scholar]
- De Vries, P.G. Sampling Theory for Forest Inventory; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest Inventory with Terrestrial LiDAR: A Comparison of Static and Hand-Held Mobile Laser Scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppä, J.; Kaartinen, H.; Lehtomäki, M.; Pyörälä, J.; Pfeifer, N.; Holopainen, M.; Brolly, G.; Francesco, P.; Hackenberg, J.; et al. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Hartley, R.J.L.; Jayathunga, S.; Massam, P.D.; De Silva, D.; Estarija, H.J.; Davidson, S.J.; Wuraola, A.; Pearse, G.D. Assessing the Potential of Backpack-Mounted Mobile Laser Scanning Systems for Tree Phenotyping. Remote Sens. 2022, 14, 3344. [Google Scholar] [CrossRef]
- CloudCompare. CloudCompare (Version 2.13.1) [GPL Software]. 2024. Available online: http://www.cloudcompare.org/ (accessed on 20 August 2024).
- Roussel, J.R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Meador, A.S.; Bourdon, J.F.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of airborne laser scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- Jan, H.; Kim, C.; Demol, M.; Raumonen, P.; Piboule, A.; Mathias, D. SimpleForest—A comprehensive tool for 3d reconstruction of trees from forest plot point clouds. bioRxiv 2021. [Google Scholar] [CrossRef]
- de Conto, T.; Olofsson, K.; Görgens, E.B.; Rodriguez, L.C.E.; Almeida, G. Performance of stem denoising and stem modeling algorithms on single tree point clouds from terrestrial laser scanning. Comput. Electron. Agric. 2017, 143, 165–176. [Google Scholar] [CrossRef]
- Rocha, K.D.; Schlickmann, M.B.; Xia, J.; Leite, R.V.; Klauberg, C.; Sharma, A.; Silva, C.A. Characterizing Even and Uneven-Aged Southern Pine Forest Using Terrestrial Laser Scanning. In Proceedings of the IGARSS 2023—2023 IEEE International Geoscience and Remote Sensing Symposium, Pasadena, CA, USA, 16–21 July 2023; pp. 4258–4261. [Google Scholar] [CrossRef]
- Wilkes, P.; Disney, M.; Armston, J.; Bartholomeus, H.; Bentley, L.; Brede, B.; Burt, A.; Calders, K.; Chavana-Bryant, C.; Clewley, D.; et al. TLS2trees: A scalable tree segmentation pipeline for TLS data. Methods Ecol. Evol. 2023, 14, 3083–3099. [Google Scholar] [CrossRef]
- Krisanski, S.; Taskhiri, M.S.; Gonzalez Aracil, S.; Herries, D.; Muneri, A.; Gurung, M.B.; Montgomery, J.; Turner, P. Forest Structural Complexity Tool—An Open Source, Fully-Automated Tool for Measuring Forest Point Clouds. Remote Sens. 2021, 13, 4677. [Google Scholar] [CrossRef]
- Donager, J.J.; Sánchez Meador, A.J.; Blackburn, R.C. Adjudicating Perspectives on Forest Structure: How Do Airborne, Terrestrial, and Mobile Lidar-Derived Estimates Compare? Remote Sens. 2021, 13, 2297. [Google Scholar] [CrossRef]
- Atkins, J.W.; Stovall, A.E.; Silva, C.A. Open-source tools in R for forestry and forest ecology. For. Ecol. Manag. 2022, 503, 119813. [Google Scholar] [CrossRef]
- Nambiar, S.; O’Hehir, J. Increasing production from pine plantations in the Green Triangle. Aust. For. Grow. 2010, 33, 35–37. [Google Scholar]
- Snyder, H. Literature review as a research methodology: An overview and guidelines. J. Bus. Res. 2019, 104, 333–339. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R. 2020. Available online: http://www.rstudio.com/ (accessed on 14 August 2024).
- Ogle, D.H. FSA: Simple Fisheries Stock Assessment Methods. 2023. R Package, Version 0.7.2. Available online: https://fishr-core-team.github.io/FSA/ (accessed on 11 March 2025).
- Kassambara, A. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2023. R Package Version 0.7.2. Available online: https://github.com/kassambara/rstatix (accessed on 11 March 2025).
- Laaksonen, J.; Oja, E. Classification with learning k-nearest neighbors. In Proceedings of the International Conference on Neural Networks (ICNN’96), Washington, DC, USA, 3–6 June 1996; Volume 3, pp. 1480–1483. [Google Scholar] [CrossRef]
- Illingworth, J.; Kittler, J. The Adaptive Hough Transform. IEEE Trans. Pattern Anal. Mach. Intell. 1987, PAMI-9, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Litkey, P.; Hyyppa, J.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Automatic Stem Mapping Using Single-Scan Terrestrial Laser Scanning. IEEE Trans. Geosci. Remote Sens. 2012, 50, 661–670. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. PointNet++: Deep Hierarchical Feature Learning on Point Sets in a Metric Space. arXiv 2017. [Google Scholar] [CrossRef]
- Kükenbrink, D.; Marty, M.; Bösch, R.; Ginzler, C. Benchmarking laser scanning and terrestrial photogrammetry to extract forest inventory parameters in a complex temperate forest. Int. J. Appl. Earth Obs. Geoinf. 2022, 113, 102999. [Google Scholar] [CrossRef]
- Sparks, A.M.; Smith, A.M. Accuracy of a LiDAR-Based Individual Tree Detection and Attribute Measurement Algorithm Developed to Inform Forest Products Supply Chain and Resource Management. Forests 2022, 13, 3. [Google Scholar] [CrossRef]
- MathWorks Inc. Call MATLAB from Python, MATLAB Help Center. 2024. Available online: https://au.mathworks.com/help/matlab/matlab-engine-for-python.html (accessed on 14 August 2024).
- Gautier, L. rpy2-R in Python, GitHub. 2024. Available online: https://github.com/rpy2/rpy2 (accessed on 14 August 2024).
- Rascle, P. CloudComPy, GitHub. 2024. Available online: https://github.com/CloudCompare/CloudComPy (accessed on 14 August 2024).
- Weiser, H.; Schäfer, J.; Winiwarter, L.; Krašovec, N.; Fassnacht, F.E.; Höfle, B. Individual tree point clouds and tree measurements from multi-platform laser scanning in German forests. Earth Syst. Sci. Data 2022, 14, 2989–3012. [Google Scholar] [CrossRef]
- Bryson, M.; Wang, F.; Allworth, J. Using synthetic tree data in deep learning-based tree segmentation using LiDAR point clouds. Remote Sens. 2023, 15, 2380. [Google Scholar] [CrossRef]
- Trochta, J.; Krůček, M.; Vrška, T.; Král, K. 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE 2017, 12, e0176871. [Google Scholar] [CrossRef]
- Fan, G.; Nan, L.; Dong, Y.; Su, X.; Chen, F. AdQSM: A New Method for Estimating Above-Ground Biomass from TLS Point Clouds. Remote Sens. 2020, 12, 3089. [Google Scholar] [CrossRef]
- Bellock, K.E. alphashape: Toolbox for Generating Alpha Shapes, GitHub. 2024. Available online: https://github.com/bellockk/alphashape (accessed on 19 July 2024).
- Boskaljon, F.; Bloembergen, D. Urban Tree Analysis Using 3D Point Clouds. 2024. Available online: http://amsterdamintelligence.com/posts/urban-tree-analysis-using-3d-point-clouds (accessed on 19 December 2024).
- Du, S.; Lindenbergh, R.; Ledoux, H.; Stoter, J.; Nan, L. AdTree: Accurate, Detailed, and Automatic modeling of Laser-Scanned Trees. Remote Sens. 2019, 11, 2074. [Google Scholar] [CrossRef]
- Zhang, W.; Qi, J.; Wan, P.; Wang, H.; Xie, D.; Wang, X.; Yan, G. An Easy-to-Use Airborne LiDAR Data Filtering Method Based on Cloth Simulation. Remote Sens. 2016, 8, 501. [Google Scholar] [CrossRef]
- Letard, M.; Lague, D.; Le Guennec, A.; Lefèvre, S.; Feldmann, B.; Leroy, P.; Girardeau-Montaut, D.; Corpetti, T. 3DMASC: Accessible, explainable 3D point clouds classification. Application to bi-spectral topo-bathymetric lidar data. ISPRS J. Photogramm. Remote Sens. 2024, 207, 175–197. [Google Scholar] [CrossRef]
- Brodu, N.; Lague, D. 3D Terrestrial LiDAR data classification of complex natural scenes using a multi-scale dimensionality criterion: Applications in geomorphology. CoRR 2011. Available online: http://arxiv.org/abs/1107.0550 (accessed on 26 August 2024).
- Xi, Z.; Hopkinson, C. 3D Graph-Based Individual-Tree Isolation (Treeiso) from Terrestrial Laser Scanning Point Clouds. Remote Sens. 2022, 14, 6116. [Google Scholar] [CrossRef]
- Laino, D.; Cabo, C.; Prendes, C.; Janvier, R.; Ordonez, C.; Nikonovas, T.; Doerr, S.; Santin, C. 3DFin: A software for automated 3D forest inventories from terrestrial point clouds. For. Int. J. For. Res. 2024, 97, 479–496. [Google Scholar] [CrossRef]
- Molina-Valero, J.A.; Martínez-Calvo, A.; Ginzo Villamayor, M.J.; Novo Pérez, M.A.; Álvarez González, J.G.; Montes, F.; Pérez-Cruzado, C. Operationalizing the use of TLS in forest inventories: The R package FORTLS. Environ. Model. Softw. 2022, 150, 105337. [Google Scholar] [CrossRef]
- Windrim, L.; Bryson, M. Detection, Segmentation, and Model Fitting of Individual Tree Stems from Airborne Laser Scanning of Forests Using Deep Learning. Remote Sens. 2020, 12, 1469. [Google Scholar] [CrossRef]
- Tian, Z.; Li, S. Graph-Based Leaf–Wood Separation Method for Individual Trees Using Terrestrial Lidar Point Clouds. IEEE Trans. Geosci. Remote Sens. 2022, 60, 5705111. [Google Scholar] [CrossRef]
- Louise, T. ITSMe: Submission Release, GitHub. 2022. Available online: https://github.com/lmterryn/ITSMe (accessed on 26 August 2024).
- rapidlasso GmbH. LAStools—Efficient LiDAR Processing Software. 2023. Available online: https://rapidlasso.com/lastools/ (accessed on 21 August 2024).
- Roussel, J.R. lasR: Fast and Pipeable Airborne LiDAR Data Tools. R Package Version 0.10.0. GitHub. 2024. Available online: https://github.com/r-lidar/lasR (accessed on 26 August 2024).
- Ayrey, E.; Fraver, S.; Kershaw, J.A.; Kenefic, L.S.; Hayes, D.; Weiskittel, A.R.; Roth, B.E. Layer stacking: A novel algorithm for individual forest tree segmentation from LiDAR point clouds. Can. J. Remote Sens. 2017, 43, 16–27. [Google Scholar] [CrossRef]
- Wang, D.; Momo Takoudjou, S.; Casella, E. LeWoS: A universal leaf-wood classification method to facilitate the 3D modeling of large tropical trees using terrestrial LiDAR. Methods Ecol. Evol. 2020, 11, 376–389. [Google Scholar] [CrossRef]
- Roussel, J.R.; Auty, D. Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. Manual, CRAN. 2024. Available online: https://cran.r-project.org/package=lidR (accessed on 19 July 2024).
- Hackenberg, J.; Bontemps, J.D. Improving quantitative structure models with filters based on allometric scaling theory. Appl. Geomat. 2023, 15, 1019–1029. [Google Scholar] [CrossRef]
- Wang, D. Unsupervised semantic and instance segmentation of forest point clouds. ISPRS J. Photogramm. Remote Sens. 2020, 165, 86–97. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Burt, A.; Disney, M.; Calders, K. Extracting individual trees from lidar point clouds using treeseg. Methods Ecol. Evol. 2019, 10, 438–445. [Google Scholar] [CrossRef]
- Puliti, S.; McLean, J.P.; Cattaneo, N.; Fischer, C.; Astrup, R. Tree height-growth trajectory estimation using uni-temporal UAV laser scanning data and deep learning. For. Int. J. For. Res. 2023, 96, 37–48. [Google Scholar] [CrossRef]
| Overview | Tool Details | Data Features | Stand Metrics | Stem Metrics | Branch Metrics | Crown Metrics | Detection | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Platform | Test Version | Licence | Publisher | Automation | Use Case | File Operations | Point Cloud Subsampling | Scan Cropping | Ground Extraction | Tree Segmentation | Leaf/Wood Segmentation | Outputted Data Format | Stem Count | Canopy Cover | Biomass | DBH | Height | Volume | Angle | Taper | QSM Construction | Volume | Length | Angle | Count | Statistics | Shape | Volume | Diameter | Whorl | Double Leader | Knot |
| AdQSM | GLSL | 1.7 | “Open Tool” | Fan et al. | No | Individual Trees | .csv/.txt | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||||
| alphashape3d | R | 1.3.1 | GNU GPL-2 | Lafarge, Pateiro-Lopez | Yes | Crown Data | R Dataframe | ✓ | ✓ | ✓ | |||||||||||||||||||||||
| CloudCompare | C++ | 2.13.1 | GNU GPL-2 | Independent software | Yes | All Data | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | Most Filetypes | ✓ | ✓ | ✓ | |||||||||||||||||
| FORTLS | R | 1.4.0 | GNU GPL -3 | Molina-Valero et al. | Yes | All Scans | ✓ | ✓ | R Dataframe /.txt/.csv | ✓ | ✓ | ✓ | ✓ | ||||||||||||||||||||
| Forest3D | Python | 2021 | MIT License | Windrim and Bryson | Yes | Uniform Stands | ✓ | ✓ | ✓ | .csv/.ply | ✓ | ✓ | ✓ | ||||||||||||||||||||
| FSCT | Python | 2022 | GNU GPL -3 | Sean Krisanski | Yes | All Scans | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | .las/.csv /.html/.png | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||
| ITSMe | R | 1.0.0 | MIT License | Terryn, L | Yes | Individual Trees | R Dataframe | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||||||||
| LAStools | C++ | 2.0.2 | Partially Open Source | rapidlasso | Yes | All Data | ✓ | ✓ | ✓ | Most Filetypes | |||||||||||||||||||||||
| lidR | R | 4.1.1 | GNU GPL -3 | Roussel et al. | Yes | All Scans | ✓ | ✓ | ✓ | ✓ | ✓ | Most Filetypes | ✓ | ✓ | ✓ | ✓ | |||||||||||||||||
| PointCloud Tree Modelling (PCTM) | Python | 0.2 | GNU GPL -3 | Amsterdam Intellegence | Yes | Individual Trees | .csv | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||||||||
| SimpleForest | Computree | 5.0 | GNU GPL | Jan Hackenberg | Yes | All Scans | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | CompuTree Data | ✓ | ✓ | ||||||||||||||||||
| Spanner | R | 1.0.1 | GNU GPL -3 | Sánchez Meador et al. | Yes | All Scans | ✓ | R Dataframes + lidR LAS Object | ✓ | ||||||||||||||||||||||||
| TreeLS | R | 2.0.5 | GNU GPL -3 | De Conto et al. | Yes | Uniform Stands | ✓ | R Dataframes + lidR LAS Objects | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||||||||||
| TreeQSM | MATLAB | 2.4.1 | GNU GPL -3 | Raumonen et al. | Yes | Individual Trees | MATLAB Table | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||||
| WhorlDetector | R / Python | 2023 | GNU GPL -3 | SmartForest | Yes | Individual Trees | .csv | ✓ | |||||||||||||||||||||||||
| 3D Forest | C++ | 0.52 | GNU GPL-3 | 3D Forest Team | Yes | All Scans | ✓ | ✓ | ✓ | .txt/.ply/.pcd /shapefile | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||||||
| GBSeparation | Python | 1.0 | “Open Source” | Tian Zhilin | Yes | Individual Trees | ✓ | .txt | |||||||||||||||||||||||||
| LayerStacking | R | 2017 | MIT License | Elias Ayrey | Yes | All Scan Data | ✓ | R Dataframe | |||||||||||||||||||||||||
| LeWoS | MATLAB | 4.1 | MIT License | Wang, Di | Yes | All Scan Data | ✓ | MATLAB Table | |||||||||||||||||||||||||
| SSSC | MATLAB | 2020 | GNU GPL-3 | Wang, Di | Yes | All Scans | ✓ | ✓ | MATLAB Table | ||||||||||||||||||||||||
| TLS2Trees | Python | 2023 | GNU GPL-3 | Phil Wilkes | Yes | All Scan Data | ✓ | ✓ | ✓ | .las/.csv/.ply | ✓ | ✓ | ✓ | ||||||||||||||||||||
| TREESEG | Ubuntu | 0.2.2 | MIT License | Burt A et al. | Yes | All Scans | ✓ | ✓ | .pcd/.txt | ||||||||||||||||||||||||
| Development Stage | Stocking (stems/ha) | DBH (cm) | Height (m) |
|---|---|---|---|
| T1 | 702 | 25.4 | 24.6 |
| T2 | 449 | 34.7 | 29.1 |
| T3 | 244 | 44.7 | 33.4 |
| Tool Name | Development Stage | True Positive | False Negative | False Positive | |||
|---|---|---|---|---|---|---|---|
| FSCT | |||||||
| All | 0.904 | 0.174 | 0.096 | 0.174 | 0.141 | 0.189 | |
| T1 | 0.808 | 0.079 | 0.192 | 0.079 | 0.107 | 0.105 | |
| T2 | 0.884 | 0.259 | 0.116 | 0.259 | 0.033 | 0.034 | |
| T3 | 0.991 | 0.018 | 0.009 | 0.018 | 0.274 | 0.247 | |
| Forest3D | |||||||
| All | 0.939 | 0.074 | 0.061 | 0.074 | 0.109 | 0.210 | |
| T1 | 0.856 | 0.095 | 0.144 | 0.095 | 0.072 | 0.080 | |
| T2 | 0.955 | 0.038 | 0.045 | 0.038 | 0.022 | 0.027 | |
| T3 | 0.981 | 0.027 | 0.019 | 0.027 | 0.221 | 0.315 | |
| TreeLS | |||||||
| All | 0.953 | 0.081 | 0.047 | 0.081 | 0.141 | 0.307 | |
| T1 | 0.987 | 0.018 | 0.013 | 0.018 | 0.097 | 0.063 | |
| T2 | 0.959 | 0.073 | 0.041 | 0.073 | 0.009 | 0.030 | |
| T3 | 0.925 | 0.109 | 0.075 | 0.109 | 0.302 | 0.469 | |
| Tool | Development Stage | Processing Time (m) | Stocking (per ha) | |
|---|---|---|---|---|
| Field | Tool | |||
| FSCT | ||||
| T1 | 424 | 702 | 644 | |
| T2 | 231 | 449 | 413 | |
| T3 | 182 | 244 | 326 | |
| Forest3D | ||||
| T1 | 110 | 702 | 659 | |
| T2 | 67 | 449 | 439 | |
| T3 | 44 | 244 | 307 | |
| TreeLS | ||||
| T1 | 19 | 702 | 763 | |
| T2 | 11 | 449 | 436 | |
| T3 | 7 | 244 | 314 | |
| Toolkit | Analysis Time (/tree) |
|---|---|
| FSCT | 148 s |
| TreeLS | N/A |
| ITSMe | 2 s |
| PCTM | 86 s |
| TreeQSM | 30 s |
| Contrast | Mean Difference | SE | df | p Value | 95% CI |
|---|---|---|---|---|---|
| FSCT-ITSMe | |||||
| FSCT-PCTM | |||||
| FSCT-TreeLS | |||||
| FSCT-TreeQSM | |||||
| ITSMe-PCTM | |||||
| ITSMe-TreeLS | |||||
| ITSMe-TreeQSM | |||||
| PCTM-TreeLS | |||||
| PCTM-TreeQSM | |||||
| TreeLS-TreeQSM |
| Contrast | Mean Difference | SE | df | p Value | 95% CI [Lower, Upper] |
|---|---|---|---|---|---|
| FSCT-ITSMe | |||||
| FSCT-PCTM | |||||
| FSCT-TreeLS | |||||
| FSCT-TreeQSM | |||||
| ITSMe-PCTM | |||||
| ITSMe-TreeLS | |||||
| ITSMe-TreeQSM | |||||
| PCTM-TreeLS | |||||
| PCTM-TreeQSM | |||||
| TreeLS-TreeQSM |
| Stem Segmentation | DBH Tool | Height Tool | |
|---|---|---|---|
| T1 | TreeLS | FSCT | TreeQSM |
| T2 | TreeLS | TreeLS | TreeQSM |
| T3 | FSCT | TreeLS | ITSMe |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Keeffe, S.; Thomas, B.H.; O’Hehir, J.; Rombouts, J.; Balasso, M.; Cunningham, A. Leveraging Open-Source Tools to Analyse Ground-Based Forest LiDAR Data in South Australian Forests. Remote Sens. 2025, 17, 1934. https://doi.org/10.3390/rs17111934
O’Keeffe S, Thomas BH, O’Hehir J, Rombouts J, Balasso M, Cunningham A. Leveraging Open-Source Tools to Analyse Ground-Based Forest LiDAR Data in South Australian Forests. Remote Sensing. 2025; 17(11):1934. https://doi.org/10.3390/rs17111934
Chicago/Turabian StyleO’Keeffe, Spencer, Bruce H. Thomas, Jim O’Hehir, Jan Rombouts, Michelle Balasso, and Andrew Cunningham. 2025. "Leveraging Open-Source Tools to Analyse Ground-Based Forest LiDAR Data in South Australian Forests" Remote Sensing 17, no. 11: 1934. https://doi.org/10.3390/rs17111934
APA StyleO’Keeffe, S., Thomas, B. H., O’Hehir, J., Rombouts, J., Balasso, M., & Cunningham, A. (2025). Leveraging Open-Source Tools to Analyse Ground-Based Forest LiDAR Data in South Australian Forests. Remote Sensing, 17(11), 1934. https://doi.org/10.3390/rs17111934









