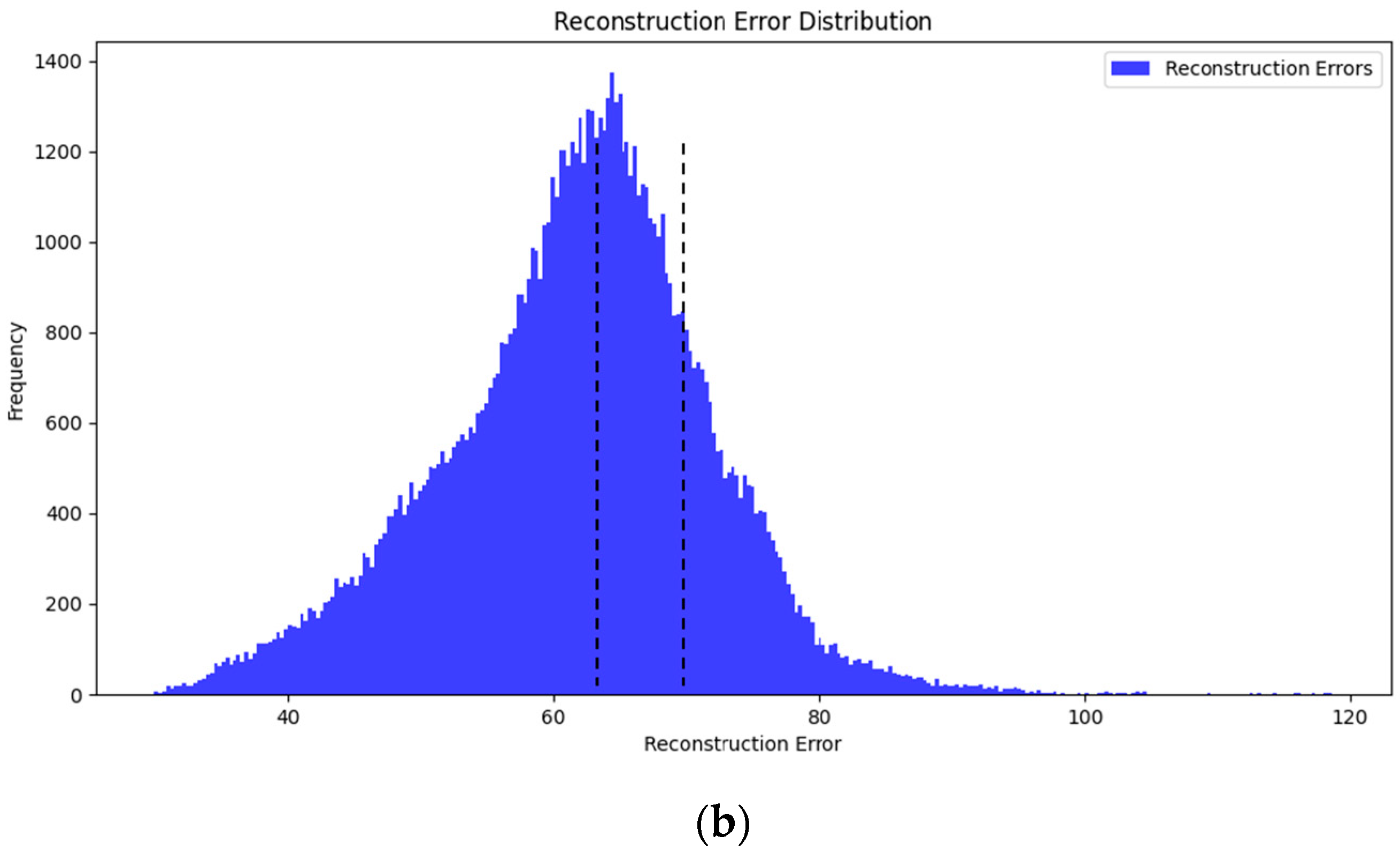
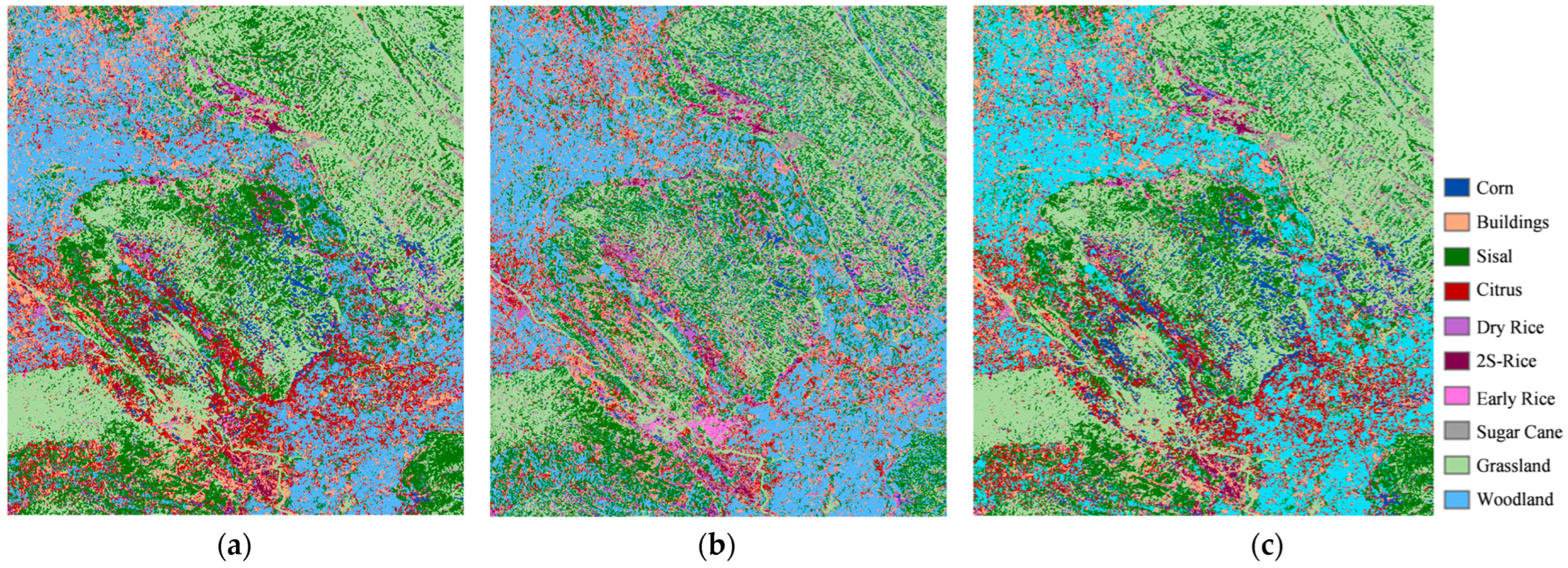
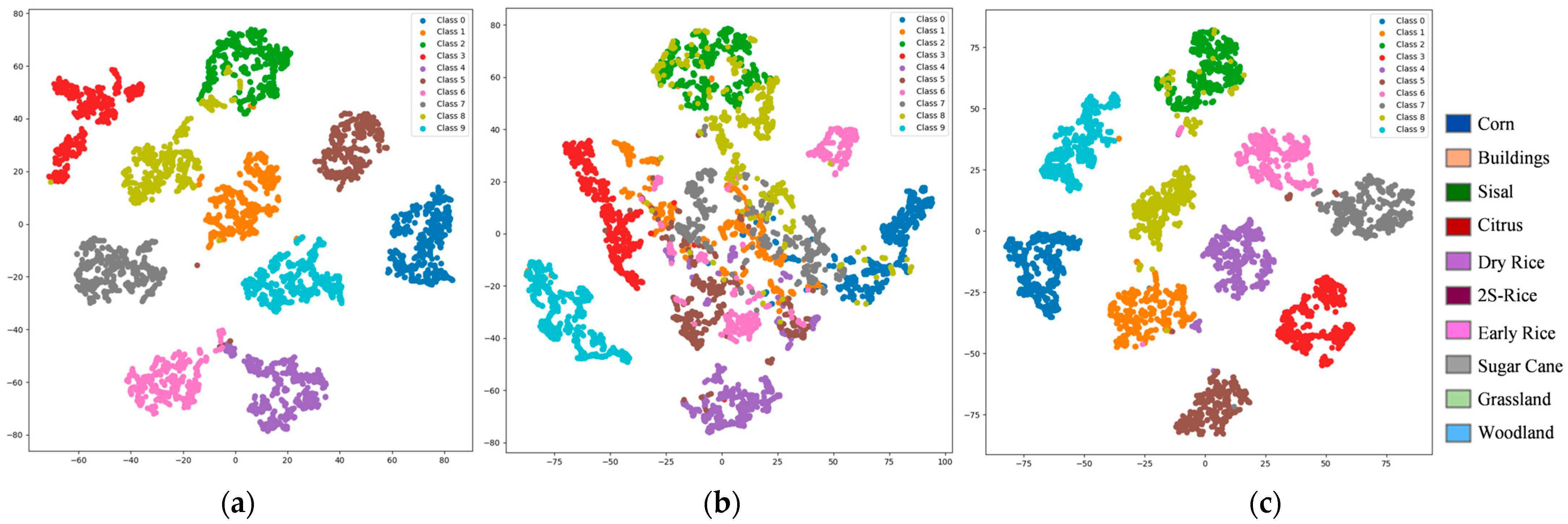
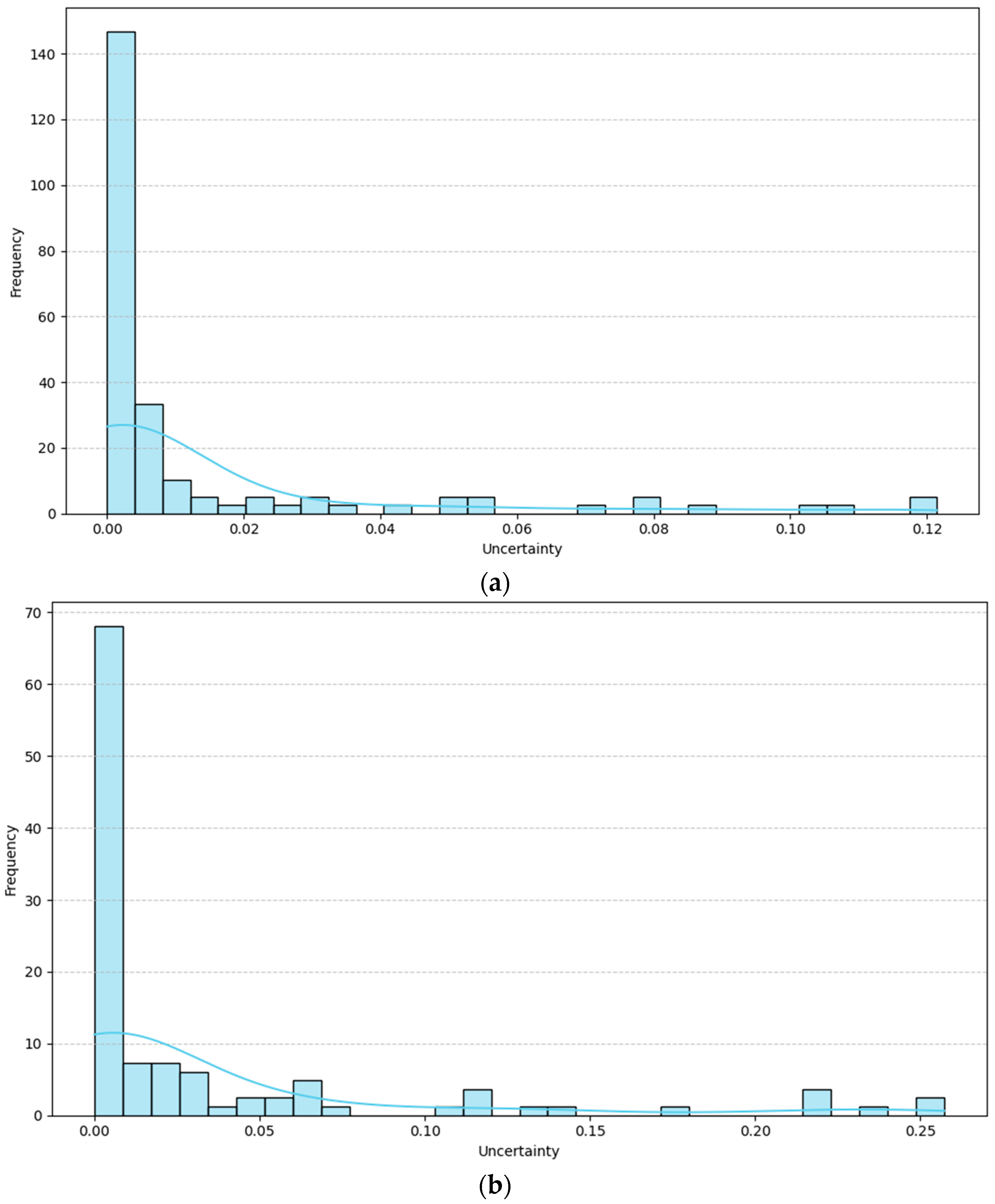
1. Introduction
Agriculture is essential for sustaining human existence [
1]. Humanity must urgently confront the majority of issues associated with food production [
2,
3,
4]. Prompt and precise access to agricultural data can enhance crop management and address challenges related to food production and food security. Remote sensing supplants field surveys to acquire data on crop dynamics, offering an efficient method for mapping crop types on a large scale [
5]. Specifically, due to the enhanced spatial and temporal resolution of remotely sensed imagery, it has emerged as the predominant technique for classifying crop types [
6]. The efficacy of remote sensing technology for crop type identification is frequently limited by intraspecific variability due to uneven training samples, resulting in a long-tailed distribution of intraspecific data. This ultimately hinders the efficacy of classification methods and undermines the precision of crop mapping.
In recent years, significant progress has been made in crop classification [
7]. Traditional methods, such as Random Forest [
8] and Support Vector Machine (SVM) [
9,
10], rely on mono-temporal static spectral features, which make it challenging to adapt to dynamic changes during crop growth. With the development of deep learning techniques, particularly the application of Convolutional Neural Networks [
11], classification accuracy has increased dramatically, and these models can effectively extract spectral-spatial feature representations for various crops. However, mono-temporal models overlook the temporal features of crop growth and cannot fully account for long-term trends and seasonal fluctuations [
12,
13,
14]. To address this limitation, time-series-based deep learning models, such as LSTM [
15] and Transformer [
16], along with their variants [
17,
18], have emerged. These models utilise remote sensing time series that capture dynamic changes in the crop growth cycle, enabling the accurate analysis of both short-term fluctuations and long-term trends [
19,
20]. This analysis enhances classification accuracy, particularly when handling complex temporal dependencies [
21]. In recent years, the application of the Transformer in remote sensing crop classification has expanded rapidly. It is not only used for sequence modelling but also for multimodal fusion and multi-scale feature extraction [
17,
18], significantly enhancing the representation ability of complex agricultural scenes. However, the improvement of agrarian classification accuracy depends not only on the classification method but also on the quality and quantity of training samples [
22,
23,
24]. The boundaries of the training dataset are predominantly influenced by intra-class variability [
25,
26], arising from diverse growth conditions attributable to environmental factors (e.g., maize exhibiting varying phenological patterns due to topographical differences) [
27]. These factors lead to significant differences in the spectral and temporal domains, even for the same crop type [
25,
28,
29]. On the other hand, deep learning models tend to focus more on easily distinguishable samples while ignoring confusing or hard samples [
30,
31]. This leads to an overfitting phenomenon on easy samples during the training process, while the model may perform poorly on difficult ones [
32]. The long tail distribution issue caused by unbalanced intra-class variation leads to overfitting, significantly reducing the model’s classification performance (i.e., increasing prediction uncertainty), particularly when dealing with complex crop environments in the real world. Therefore, we require a robust crop classification method that accounts for the long tail distribution of samples within classes to generate reliable crop mapping information. Recent studies have begun to focus on uncertainty modeling to alleviate the above problems. For instance, Ref. [
33] proposed an uncertainty-aware dynamic fusion network (UDFNet), which quantifies the confidence level of multimodal remote sensing data using an energy function to achieve adaptive feature fusion and performs well in land cover classification with significant intra-class variation. Ref. [
34] introduces dual-uncertainty (randomness and cognition) modelling for detecting architectural changes, significantly enhancing the Transformer’s robustness in scenarios with high intra-class variation. A systematic assessment of the uncertainty of the Earth observation segmentation model was conducted, verifying the effectiveness of the integrated and random reasoning methods in the semantic segmentation of satellite images, which can be transferred to the crop classification task [
35].
To date, researchers have paid little attention to intra-class variation or unbalanced sampling, which leads to long tail distribution issues during model training and prediction for crop mapping tasks [
27]. To address the problem of intra-class variation, Sun et al. classified winter wheat by leveraging a time series of vegetation indices to measure the similarity between crops [
36]. Although this method can effectively capture crop growth characteristics, it still cannot adequately address the diversity of crop growth conditions and intra-class variability through time curves. Another strategy is to utilise biorhythmic features (quantified by measuring various biorhythms (e.g., start, peak, and end times) in the vegetation index time series) to improve crop classification performance. For example, Qiu et al. identified winter wheat by analysing the variations that occurred before and after the heading date [
29]. However, the multivariate nature of biorhythmic features has not effectively solved the problem of intra-class sample variability. As intra-class variability increases, the feature space of crop samples becomes more complex, and the boundaries between samples gradually become blurred, resulting in unbalanced samples with varying levels of difficulty. Consequently, this variability is evident not only in the feature disparities within the spectral-temporal domain but also in the quantity of within-class samples across varying levels of difficulty. For instance, some samples may be more challenging to identify due to different growing conditions, such as on higher elevations, or even missing data in time series. To address these challenges, Zeng et al. optimised the sampling process by introducing an adaptive weight function that adjusts the sampling weights based on the degree of data imbalance, thereby significantly improving classification accuracy [
37]. This enables the model to focus more on highly confusing and difficult-to-recognise samples while minimising its reliance on easily classifiable ones. This approach effectively mitigates the overfitting of easy-to-categorise samples while improving the model’s adaptability to handle “more difficult” samples. By incorporating an adaptive learning mechanism, the model can dynamically adjust its learning strategy in response to the crop’s growth conditions and environmental characteristics, thereby improving its classification ability for complex samples and enhancing the model’s robustness and generalisation ability. Although adaptive learning can improve crop mapping performance by focusing on complex samples, the imbalance between easy and hard sample instances still poses a challenge for single-branch deep learning frameworks. This is because single models typically rely on one-time, fixed decision boundaries. When confronted with imbalanced samples, those boundaries often fail to accommodate the diversity and complexity of sample feature representations [
38,
39].
Recent research has employed the Mixture-of-Experts (MoE) framework to tackle this issue. The MoE establishes a network of various specialists, each concentrating on a particular task or data subset, and employs a gating mechanism to select the appropriate experts for classification dynamically. This method can effectively manage sample heterogeneity and improve classification accuracy. In this respect, Lei et al. introduced a multi-level fine-grained crop classification method utilising multi-expert knowledge distillation to tackle classification issues arising from the similarity of crop biological parameters [
40]. Nevertheless, conventional Mixture of Experts (MoE) encounters certain constraints when addressing extremely ambiguous or challenging data, particularly when intra-class difficulty differences are inadequately acknowledged. This may result in the incorrect assignment of hard samples to experts who are unsuitable for them. To overcome this problem, the MoE structure incorporates intra-class difficulty partitioning to ensure that multiple experts train complex samples with varying levels of difficulty. The more challenging examples are allocated to different experts for iterative learning and optimisation, enabling each expert to concentrate on samples of variable difficulty levels. This improvement effectively addresses the shortcomings of traditional MoE in dealing with intra-class sample variations, thereby enhancing the model’s performance in handling highly complex and confusing samples [
41,
42,
43]. Despite these improvements, the MoE still faces the challenge of slow convergence, resulting from the conflicting predictions of multiple experts. Moreover, the introduction of numerous experts significantly expands the model’s parameter space, resulting in longer training times and increased computational resource consumption. To mitigate these problems, we incorporate an uncertainty measurement mechanism that dynamically adjusts the weighting factor by evaluating the prediction confidence of each sample. This innovation combines the strengths of the MoE with uncertainty quantification to provide a more efficient and accurate solution, especially in complex agricultural classification tasks.
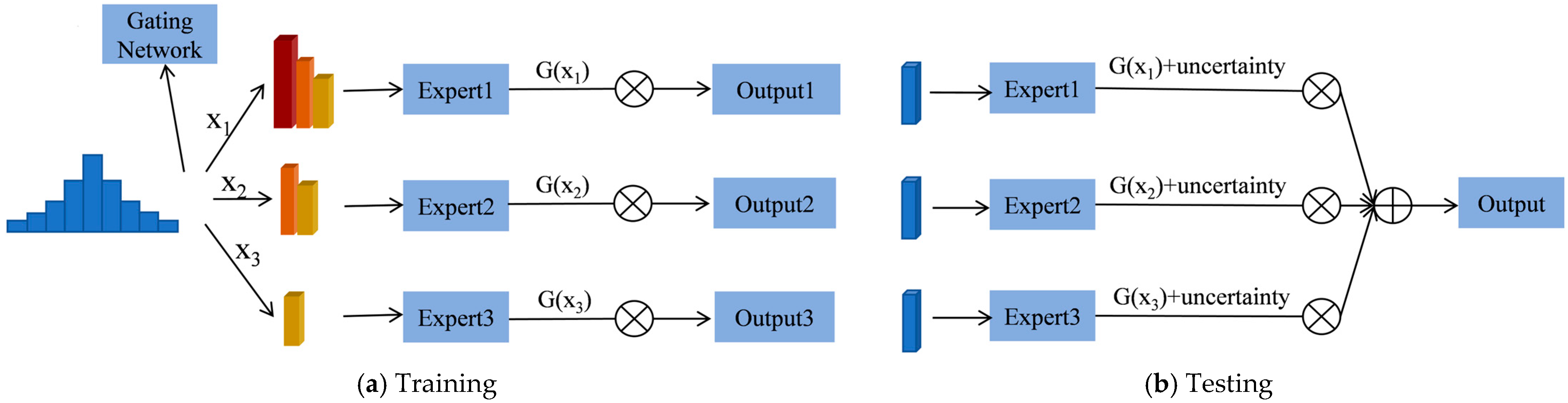
We propose a Difficulty-based Mixture of Experts Vision Transformer framework (DMoE-ViT), which assigns intra-class samples of varying difficulty levels to different expert networks, enabling precise crop classification. The proposed framework consists of three main components: first, a stratified partitioning of samples based on their difficulty levels; second, a multi-expert mechanism to solve the long tail distribution problem caused by the imbalance in sample difficulty distribution; and finally, the incorporation of uncertainty analysis to further enhance classification accuracy. The main contributions of the research are as follows:
- (1)
We propose a difficulty-induced sample quantification strategy to address the long tail distribution of intra-class sample difficulty, thereby improving the training performance of crop classifiers.
- (2)
The multi-expert mechanism dynamically learns weights by focusing on samples at different difficulty levels, thereby improving crop mapping.
The rest of this paper is organized as follows:
Section 2 introduces the proposed DMoE-ViT model.
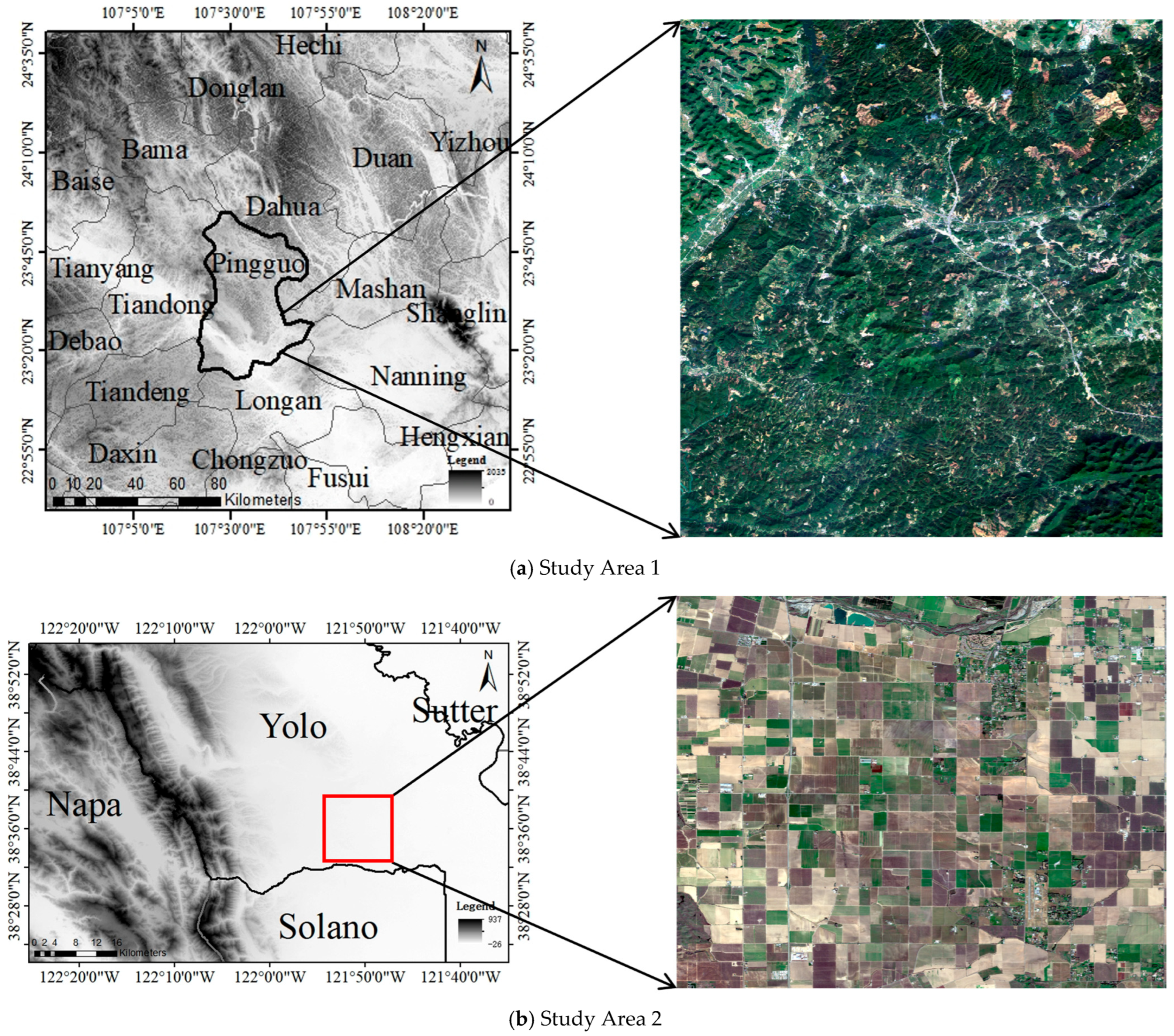
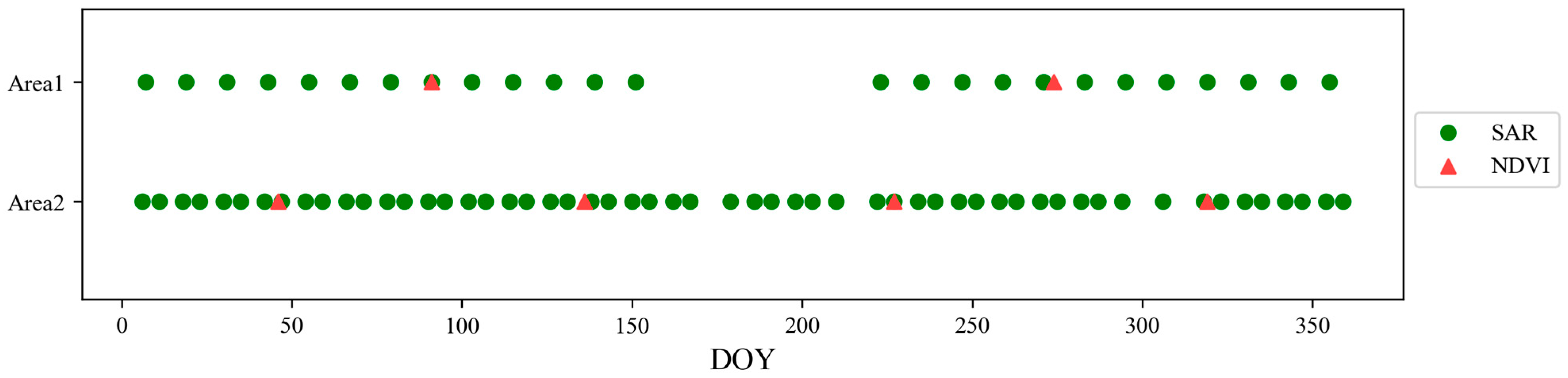
Section 3 describes the study area, along with the data collection and pre-processing methods for optical and SAR satellite time-series data.
Section 4 details the experimental setup and model parameters.
Section 5 presents the experimental results and their accuracy.
Section 6 analyzes the labeling of the same sample by multiple experts and the effect of sample difficulty classification on the results. Finally,
Section 7 concludes the paper.
4. Experimental Design
To better evaluate the proposed method’s classification performance, we compared it with five currently used deep learning networks: CNN, LSTM, CNN-Attention (CNN-Att), UNet, and ViT. Finally, we give a method for evaluating the model’s classification results.
4.1. Experiment Parameter Setting and Configuration
We first set the sample crop size of the proposed model to 18. Each expert uses 2D convolution to extract spatial information from the input samples. For the temporal feature extraction of crops, based on experimental findings and existing data, the encoder depth is set at 2, the number of MSA heads is fixed at 12, and the FFN input nodes are preset to 1296.
All models mentioned above are trained using the PyTorch 2.0 framework, with Adam as the optimiser, an initial learning rate of 0.00001, a batch size of 96, and a training duration of 35 epochs. Model training is conducted on a server utilising the CentOS 7.6 operating system, which is equipped with two NVIDIA Tesla V100 GPUs (16GB) and an Intel Xeon® Gold 5118 CPU. Utilisation of Python version 3.6 is indicated.
4.2. Model Evaluation Methods
To better analyze the classification results and model capabilities, we used five commonly used accuracy assessment metrics: overall classification accuracy (OA), kappa coefficient, confusion matrix, F1-score, and Recall. OA indicates the proportion of correctly classified samples among all samples and is used to assess the classifier’s performance. The kappa coefficient reflects the consistency of the classification results, while the confusion matrix shows the classification performance for each crop type. The F1-score combines precision and Recall and is defined as their reconciled mean. Recall is a standard metric for assessing the performance of classification models.
6. Discussion
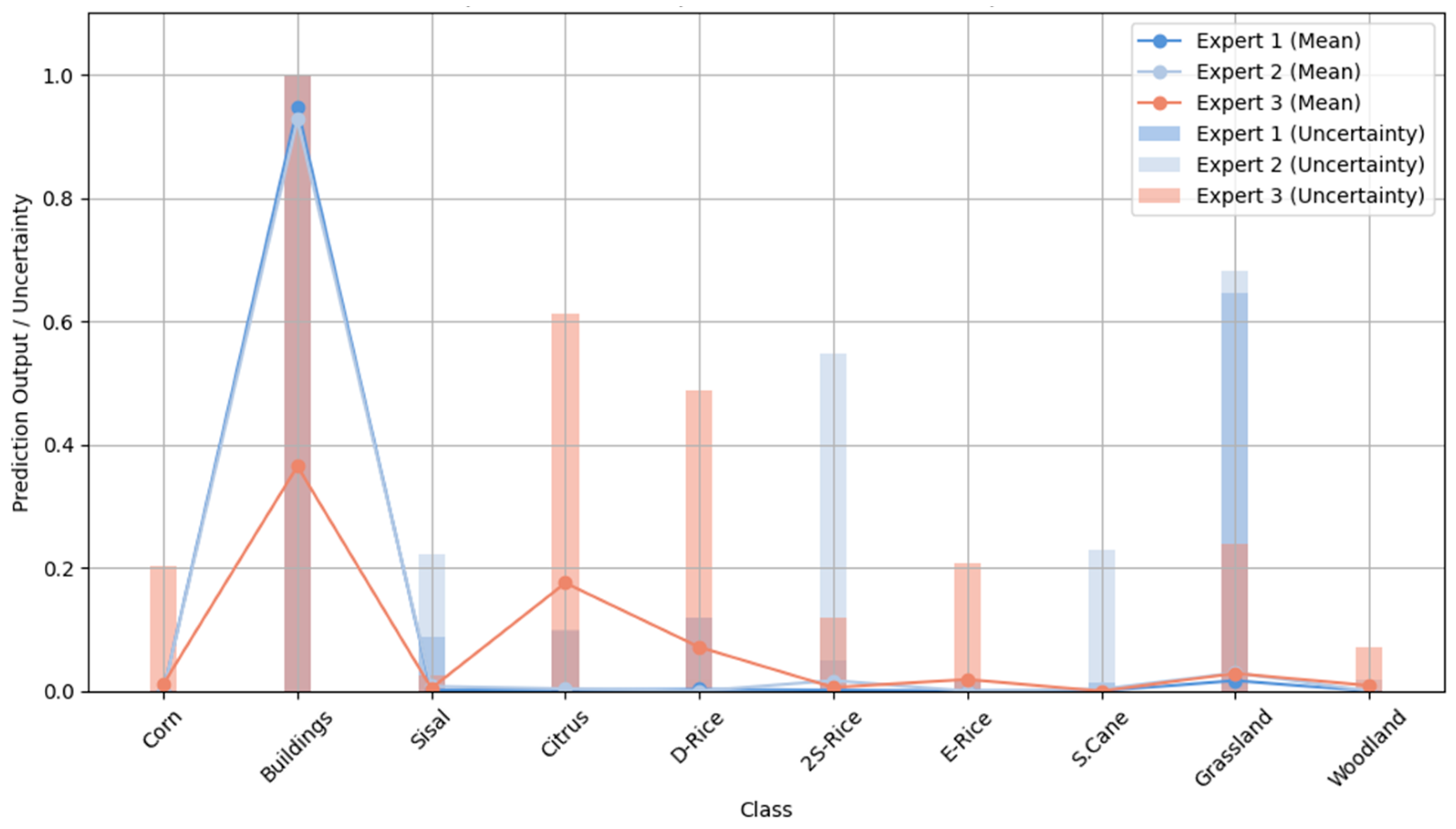
6.1. Multiple Experts’ Output Results for a Single Sample
In
Figure 13, we can intuitively observe the performance differences of the Mixture of Experts model on a single sample. Specifically, simple samples usually exhibit lower uncertainty because the model’s predictions on these samples are more consistent, and their features are typically more straightforward to distinguish, enabling the model to make rapid and accurate decisions. For example, in a simple image classification task, all expert networks tend to provide similar outputs when processing these samples, resulting in more credible final weighted predictions, thereby reducing uncertainty values. In contrast, hard samples exhibit higher uncertainty, suggesting that the model’s predictions for these samples are less credible. These samples typically exhibit more complex features, blurred boundaries, contain noise, and may be influenced by factors such as varying backgrounds, occlusions, or similar categories. Therefore, different expert networks will produce different predictions when processing these samples.
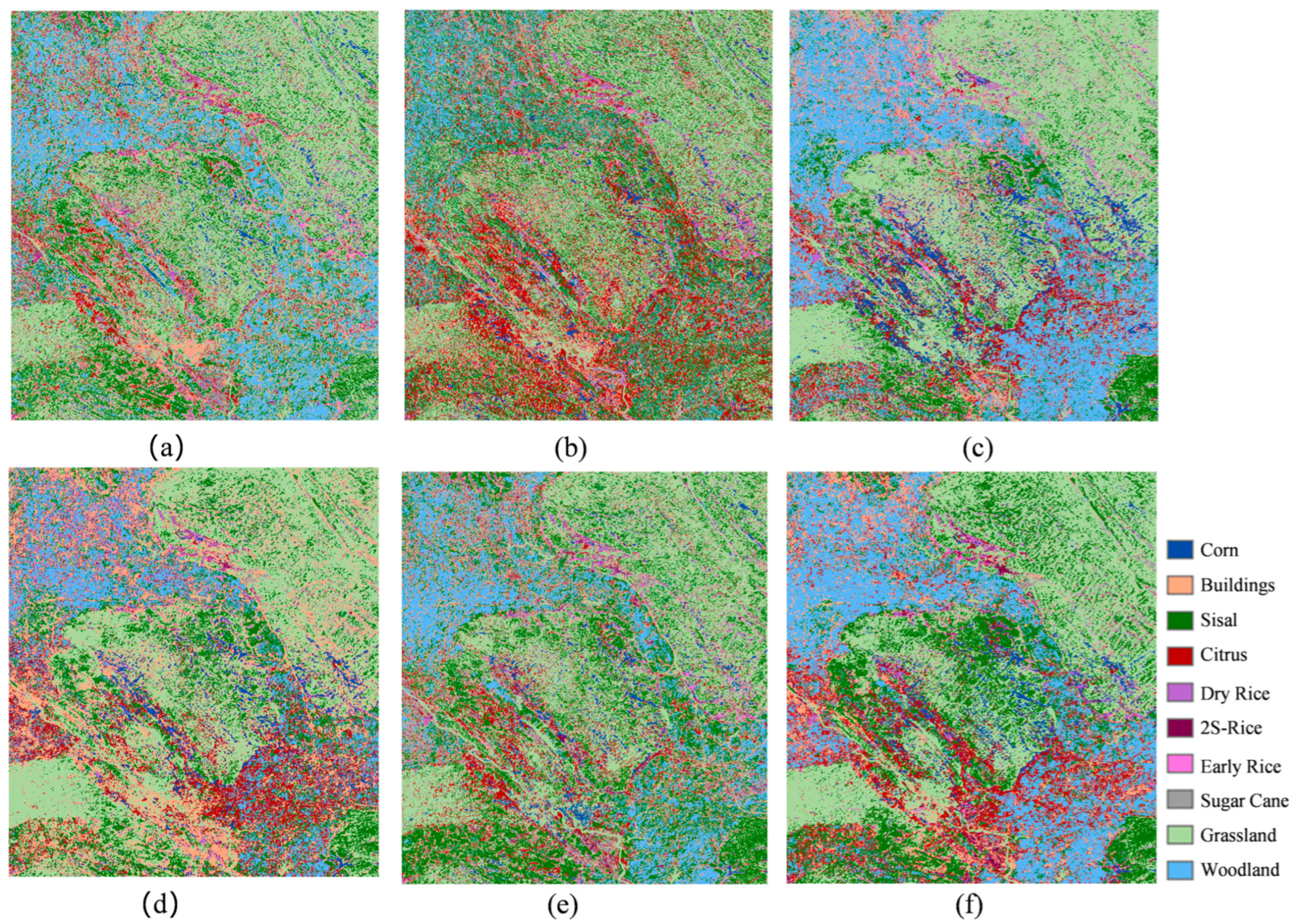
6.2. The Impact of Long Tail Sample Difficulty Classification on Experimental Results
In this study, we compared the effects of distinguishing between long tail sample difficulty and not distinguishing between long tail sample difficulty on crop classification models, as shown in
Figure 14 and
Figure 15 and
Table 7. Through the analysis of the experimental results, we found that the classification accuracy was significantly improved by classifying samples according to their difficulty, especially for high-difficulty samples. First, in an experiment that does not distinguish the difficulty of samples, all types of samples, including easy, moderate, and hard samples, are input to each expert network. Due to the complexity and indistinguishable features of difficult-to-classify samples, this method of inputting samples poses challenges for the model to effectively learn the features of these samples, resulting in low recognition accuracy of difficult-to-classify types such as grassland and double-cropping rice.
In contrast, in the experiment to distinguish the difficulty of samples, a better recognition effect was achieved by inputting samples of different difficulty levels into the expert network. Specifically, Expert 1 handles all samples (including easy and moderate samples), Expert 2 handles moderate and hard samples, and Expert 3 handles only hard samples. This approach enables each expert network to focus on learning the features of a particular hard sample, allowing the model to more precisely capture and optimize the features of these samples, thereby improving the recognition of those samples. Specifically, experiments that distinguish sample difficulty have significantly improved accuracy for high-difficulty categories. For example, the accuracy of the grass category improved from 83.4% to 89.3%, a 5.9% increase. The accuracy rate of double-cropping rice increased from 76.7% to 99.3%, representing a 22.6% improvement. This demonstrates that the model can effectively enhance the recognition performance of challenging samples by accurately assessing their difficulty.
On the other hand, for categories that are relatively easy to classify, such as sisal, woodland, and citrus, there was little change in accuracy, suggesting that the difficulty of distinguishing samples has little effect on these easier categories. At the same time, it also indicates that the difference in sample difficulty has a minimal impact on the classification results for low-difficulty categories. In summary, by comparing the results of the two experiments, we can conclude that the classification accuracy of high-difficulty samples is significantly improved by the difficulty of distinguishing them, especially for categories with high classification difficulty. For simpler classes, the effect of sample difficulty on model performance is relatively small. Therefore, the method of distinguishing sample difficulty has obvious advantages in improving classification accuracy, especially for complex tasks and high-difficulty samples.
6.3. The Relative Contribution and Significance of Optical and Radar Signatures
To quantitatively evaluate the contribution of each modality in the crop classification task, we designed a systematic ablation study. Under the controlled-variable approach, with a fixed model architecture, hyperparameters, and training data, we constructed seven data configurations, ranging from single-modality to full-modality combinations (NDVI-only, VV-only, VH-only, NDVI + VV, NDVI + VH, VV + VH, and NDVI + VV + VH), to compare classification performance across different modal inputs comprehensively. The evaluation framework integrates four dimensions: overall accuracy, marginal gain, category dominance, and relative contribution index. Specifically, we first assessed overall performance using overall accuracy. Subsequently, we calculated the marginal gain from each modality in combined scenarios, counted the frequency with which each modality achieved the highest F1-score across different crop categories, and finally synthesized these metrics through weighted normalization to generate a relative contribution index. This process establishes an evident modality importance ranking, providing a quantitative basis for optimising multimodal remote sensing data fusion strategies.
6.3.1. Heatmap of the Importance of Each Modal to Category
Table 8 utilizes a symbolic heatmap to visually illustrate the local importance of the three modalities—NDVI, VV, and VH—across various land cover categories. The symbolic representation is defined as follows: ▲ (> 30%) denotes very high importance, ☆ (15%–30%) indicates high significance, ○ (5%–15%) represents moderate importance, □ (0%–5%) signifies low relevance, and ▼ (< 0%) designates a negative contribution. The results demonstrate that VV polarisation assumes an absolutely dominant role for several crop types, including early rice (▲), sugarcane (▲), and double-cropping rice (▲). VH polarisation provides substantial textural enhancement for sugarcane (▲), grassland (☆), and dryland-to-paddy conversion (☆). NDVI exhibits specialised utility in categories associated with vegetation health, such as citrus (☆), corn (○), and woodland (○). Simultaneously, the analysis reveals distinct inter-modal interactions: NDVI exhibits a negative impact (▼) on building and early rice identification, while VH contributes minimally (□) for citrus, bordering on redundancy. These patterns highlight the existence of both complementary and conflicting mechanisms among the different modalities.
The local contribution value of each mode to a specific category within the table is defined as:
Among the various accuracy metrics, denotes the classification accuracy for category Y, while represents the combination of the other two modalities, excluding modality X.
6.3.2. Global Modality Contribution Ranking
Through a comprehensive analysis of
Table 8 and
Table 9, it is clear that VV dominates the highest number of land cover categories, followed by VH and NDVI. By further classifying these dominant categories based on their characteristic attributes, a distinct pattern of modality-specific strengths emerges: VV assumes a globally dominant role, VH exhibits texture-discrimination dominance, while NDVI demonstrates a vegetation-oriented advantage. The quantitative results from the Ratio metric in
Table 9 show that VV has the highest proportion, consistent with its top Ranking in the relative contribution index. VH ranks second, and NDVI third. These findings not only confirm the core driving role of VV in the overall classification performance but also further elucidate the complementary nature and hierarchical importance of the three modalities in specific discrimination tasks. This provides a solid quantitative foundation for optimising the design of multimodal fusion strategies.
The Overall Contribution value within each combination in the table is defined as:
Within the Accuracy section, denotes the classification accuracy for category Y, while represents the combination of the other two modalities excluding modality X. The column labelled Ranking indicates the importance ranking of each modality. In contrast, the column labeled “Ratio” shows the contribution ratio per scene—specifically, the proportion of Overall Contribution relative to the total number of all modality images.
6.4. Graded-Difficulty Proportionate Melting Experiment
To systematically validate the effectiveness of the proposed 3:2:1 difficulty stratification strategy, this section was designed to conduct comprehensive ablation experiments targeting the allocation ratios across three difficulty levels (Easy, Moderate, Hard). On the validation set of the first study area, the following four stratification configurations were evaluated:
Uniform Distribution (1:1:1): Each difficulty level accounts for 33.3% as the baseline;
Easy-Focused (4:2:1): Prioritizes foundational feature learning (Easy 57.1%, Moderate 28.6%, Hard 14.3%);
Moderate-Difficulty Emphasis (2:2:1): Prioritizes learning complex patterns (Easy 40%, Moderate 40%, Hard 20%);
Proposed Strategy (3:2:1): Gradual curriculum learning (Easy 50%, Moderate 33.3%, Hard 16.7%).
The experimental results are shown in
Table 10. Key evaluation metrics include overall classification accuracy (OA), Kappa coefficient, F1-score for hard samples, and training convergence speed (the number of iterations required to reach 95% of the final OA).
Table 10 presents comparative experimental results on a typical agricultural remote sensing dataset, demonstrating that the proposed 3:2:1 strategy achieves the best overall accuracy (OA), F1-score, and Kappa coefficient. Specifically, the OA of the proposed strategy is 5.5% higher than that of the 1:1:1 baseline, with improved convergence stability and fewer required training epochs. Further analysis reveals that the 1:1:1 strategy leads to premature and excessive exposure to hard samples, which hinders the stable learning of fundamental discriminative features. The 4:2:1 strategy results in insufficient training of hard samples, causing underfitting in the corresponding expert network. Meanwhile, the 2:2:1 strategy undermines the foundational role of easy samples, resulting in instability in overall feature learning. In contrast, the 3:2:1 strategy strikes an optimal balance between consolidating basic features and tackling challenging cases, thereby validating the rationality of its design.
This study constructs a Mixture-of-Experts (MoE) network that shares a core conceptual resonance with recent advanced multi-scale attention frameworks. For instance, in the task of lithological unit classification in vegetated areas, Hui et al. [
49] introduced Multi-Scale Enhanced Cross-Attention (MSECA) and Wavelet-Enhanced Cross-Attention (WCCA) to strengthen the model’s ability to capture both fine-grained local details and long-range contextual dependencies. The MoE design in this work can be viewed as a corresponding, yet more generalized, “model capacity allocation” mechanism: The expert networks handling simple samples function similarly to MSECA, focusing on extracting robust local features from clear and common patterns. The expert networks handling challenging samples emulate the role of WCCA, aiming to model the broader contextual information required for complex and sparse samples.
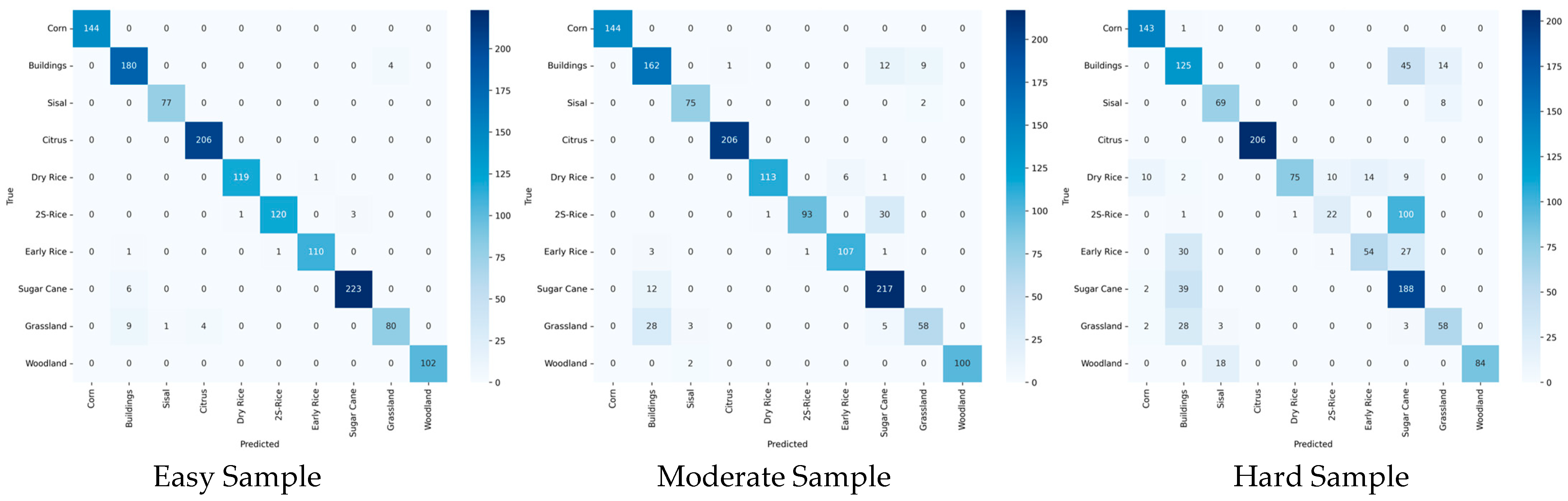
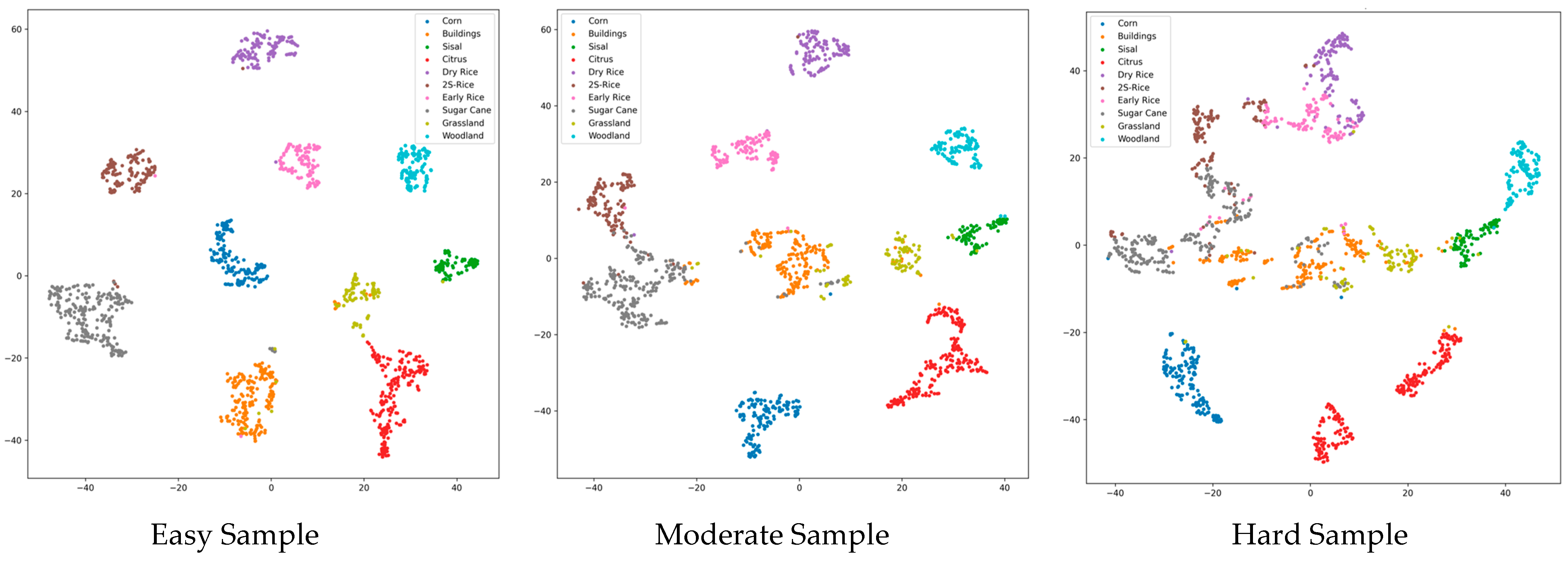
6.5. Stratified Statistical Analysis on Samples of Varying Difficulty Levels
To rigorously validate the efficacy of the difficulty-aware mechanism and to enable fine-grained evaluation within each difficulty stratum, we establish independent assessment frameworks for the three difficulty levels (easy, moderate, and hard), reporting Accuracy, Recall, F1-score, Kappa, and Overall Accuracy (OA) for each subset, along with per-class precision.
Table 11 presents the complete stratified results,
Table 12 details the per-class stratified accuracies,
Figure 16 illustrates the confusion matrices for the different difficulty subsets, and
Figure 17 depicts the t-SNE visualisations of the respective difficulty subsets.
The results in
Table 11 demonstrate a natural performance degradation with increasing difficulty (Easy > Moderate > Hard), fully aligning with the anticipated distribution. Our approach achieves an overall OA of 0.964, attaining or approaching the optimal single-expert performance across all subsets, thereby highlighting the MoE framework’s success in synergistically integrating specialised capabilities for distinct difficulty strata.
Table 12 further reveals that head categories (Corn, Citrus, Sisal, Woodland) approach or reach 100% in the Easy and Moderate subsets, with our method maintaining 100.0% throughout, indicating efficient, lossless processing of simple samples and high inter-crop discriminability in
Figure 16 and
Figure 17. Tail-end challenging categories markedly benefit from expert collaboration; for instance, 2S-Rice surges from 17.74% to 96.77% in the Difficult subset, attributable to Expert-3’s deep modelling of complex spectral features. Similarly, Early Rice (48.21% → 98.21%), Dry Rice (62.50% → 99.17%), Buildings (67.93% → 97.83%), and Grassland (61.70% → 85.11%) achieve dramatic improvements, underscoring the combined contributions of Expert-2 and Expert-3. These categories, characterized by high entropy due to seasonal variations, cropping patterns, or land-cover confusion, represent prototypical long tail complex cases. Although the Difficult subset averages ~70% in isolation, our method—through dynamic weighted fusion via the gating network and uncertainty calibration—transforms potential performance drags into global gains, yielding an ultimate overall OA of 96.4%.
7. Conclusions
In this paper, a Difficulty-based Mixture of Experts Vision Transformer framework (DMoE-ViT) is proposed for accurate crop type mapping. This framework utilizes the Vision Transformer (ViT) architecture and a Mixture of Experts model (MoE) gating mechanism to handle samples of varying difficulty levels in a multi-expert model. Long tail samples were categorized into three difficulty levels—easy, moderate, and hard—and split into three datasets. Each dataset was fed into separate ViT networks, generating distinct output results. The DMoE-ViT model computed appropriate weights for each dataset based on its difficulty level. During testing, the same data samples and their labels were passed through the three networks, and the predictions were weighted according to the model and aggregated to produce the final output.
Additionally, uncertainty was calculated for the final prediction, with an output weight assigned based on this uncertainty, which was then multiplied by the aggregated result to yield the final prediction. Our experimental results demonstrate that this approach enhances the model’s performance by incorporating a sample difficulty classification mechanism, enabling the model to adjust its learning process according to the complexity of the long tail samples. The implementation of the MoE gating mechanism enhances the efficiency of processing diverse sample types and improves the stability and accuracy of models across many categories. This method can provide a better understanding of the relationship between sample complexity and model performance when dealing with hard samples, highlighting the importance of introducing difficulty perception strategies in training deep learning models. Future endeavours may boost the model’s performance through various improvements. The precision of classification can be enhanced by refining the difficulty classification process using methods such as clustering algorithms, deep learning-based feature extraction, or domain-specific expertise.
Additionally, adjusting the sample classification threshold can more accurately distinguish between different difficulty levels. Second, model flexibility and performance can be improved by exploring alternative expert architectures, such as CNNS, RNNS, or hybrid models. Finally, the robustness of the model to different sample complexities can also be improved by integrating uncertainty quantification or data enhancement strategies.