Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation
Abstract
Highlights
- A unified noise detection–correction framework that learns spectral representations with a Transformer, detects noisy samples via Isolation Forest, and corrects reflectance with a cGAN for SOC estimation.
- Outperforms existing noise-handling methods on benchmark satellite datasets.
- Correcting (rather than discarding) noisy samples improves accuracy and coverage of SOC maps at scale.
- Enables reliable remote sensing–based SOC monitoring to support precision agriculture and soil management.
Abstract
1. Introduction
- Proposing a novel framework that integrates Transformer-based feature extraction, Isolation Forest for noise detection, and cGAN for noise correction.
- Addressing the limitations of traditional exclusion-based noise-handling methods by correcting noisy samples to enhance data utility.
- Demonstrating the effectiveness and scalability of the proposed methodology across multiple remote sensing platforms, including datasets with complex noise patterns.
- Enhancing SOC estimation accuracy by leveraging advanced feature representation and reconstruction techniques.
2. Dataset Preparation
2.1. Study Area and Soil Data Collection
2.2. Landsat 8 Data Acquisition and Pre-Processing
2.3. Landsat 8 Image Transformation and Vegetation Indices
3. Methodology
3.1. Overview of the Proposed Framework
3.2. Proposed Noise Detection and Correction Modules
3.2.1. Transformer-Based Feature Extraction
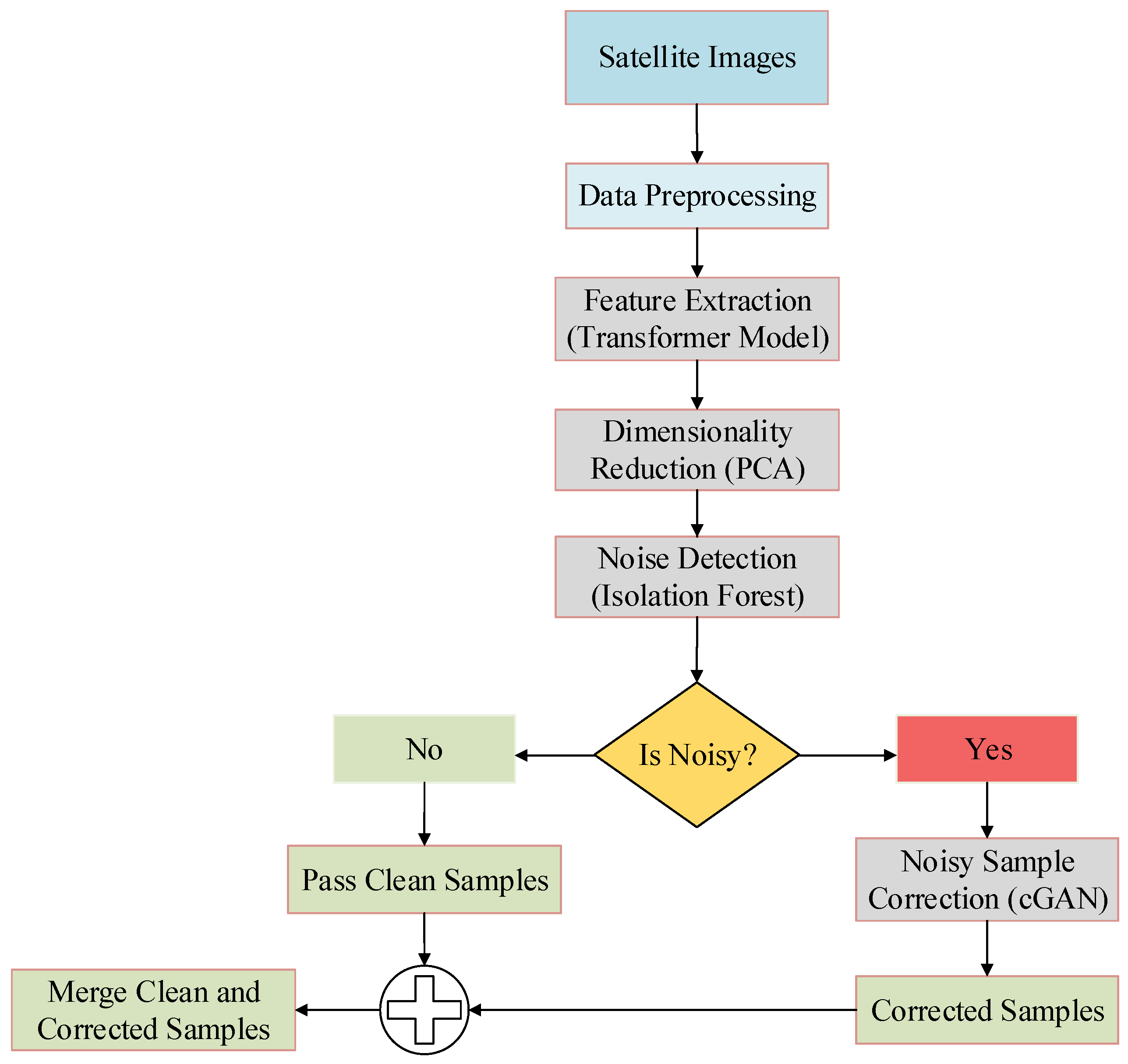
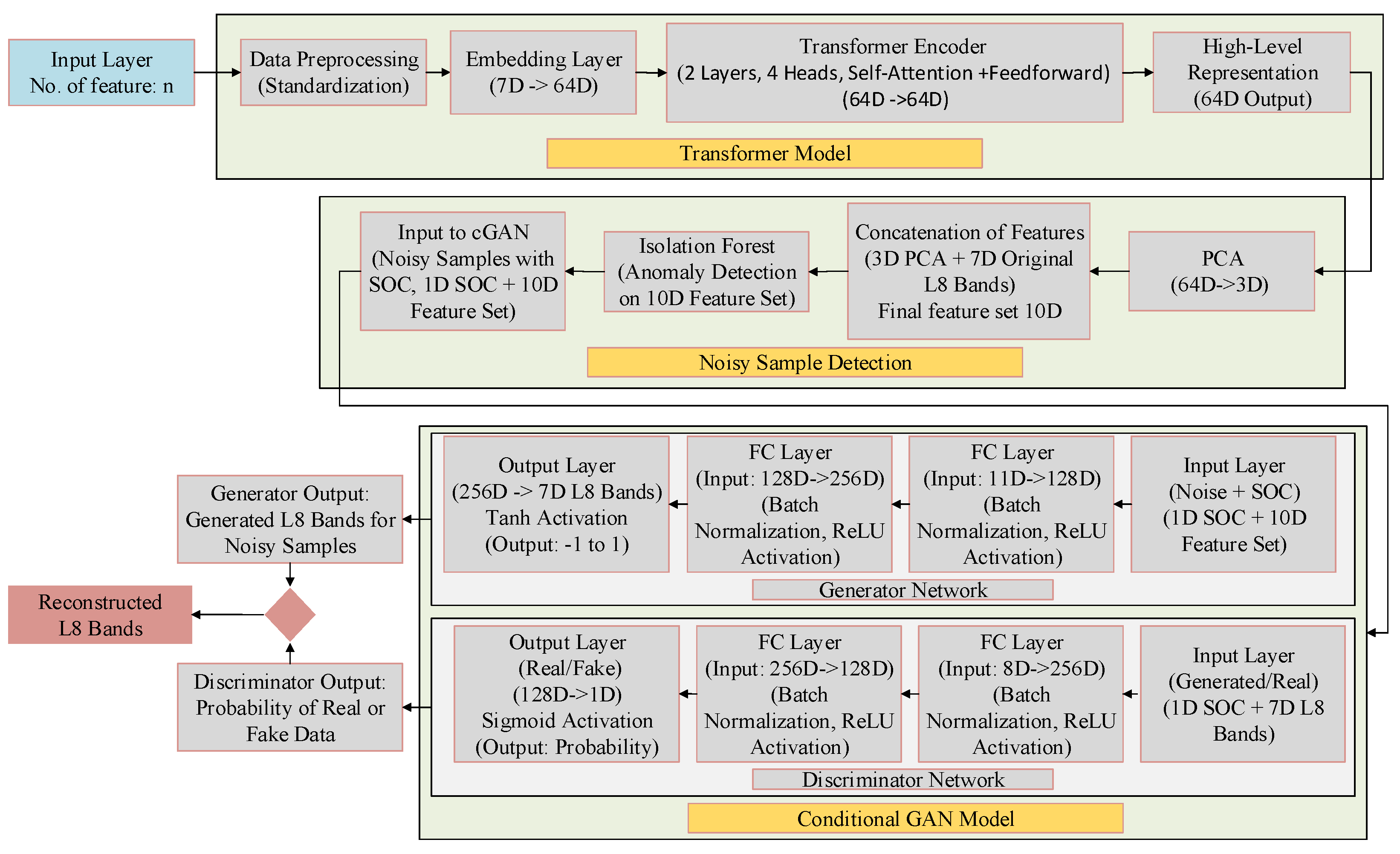
3.2.2. Dimensionality Reduction Using Principal Component Analysis
3.2.3. Noise Detection Using Isolation Forest
3.2.4. Noise Correction Using Conditional GAN
- Conditional guidance: The generator is explicitly conditioned on nearby non-noisy samples and their average SOC, introducing contextual and semantic constraints.
- Direct reconstruction penalty: The term enforces closeness to true reflectance, reducing large deviations.
- Activation and clipping: Tanh activation followed by inverse normalization ensures outputs fall within a valid physical range (e.g., [0, 1]).
- Standardization: Inputs and outputs were standardized during training and de-standardized post-generation to maintain consistency with the original data scale.
3.2.5. Post-Reconstruction Dataset
3.3. Experimental Setup
4. Results
4.1. SOC Estimation Using Raw Landsat 8 Reflectance Bands
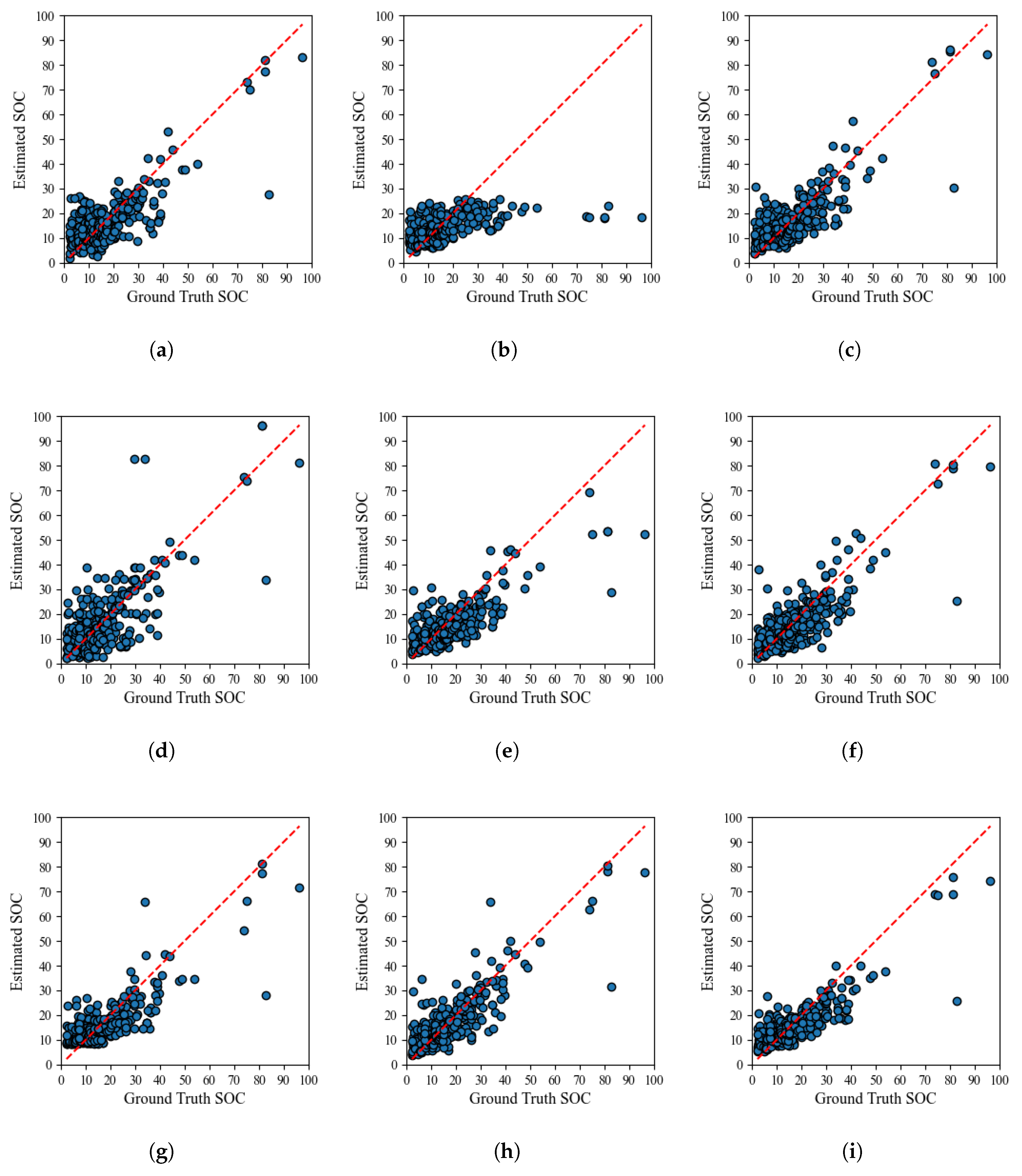
4.2. SOC Estimation Using Landsat 8 Bands with Transformed Features: VIs, SIs, and TCT
4.3. Comparison of Band Selection Techniques for Optimized SOC Estimation

5. Discussion
5.1. Comparison with State-of-the-Art Methods
- Threshold-Based Methods: NDVI-based exclusion is a widely used filtering technique in SOC estimation tasks [19]. These methods rely on predefined thresholds to reduce vegetation-related noise. While effective in datasets dominated by vegetation noise, they struggle with more complex noise sources, such as atmospheric effects or sensor anomalies, leading to lower accuracy in mixed-noise conditions.
- Statistical Detection and Reconstruction: Statistical techniques such as Z-Score [26,27] and MAD [28,36] identify noise based on deviations from statistical norms. When combined with kriging for reconstruction [27,36], these methods improve SOC estimation by leveraging spatial correlations. However, their reliance on assumptions like normal data distribution makes them less effective for heterogeneous noise patterns.
- Machine Learning-Based Methods: ML-based approaches such as LOF [30,31] and OC-SVM [32,33] can adapt to complex noise structures by modeling local and global anomalies. However, their performance depends on proper parameter tuning and dataset characteristics. While robust PCA [34,35] and kriging improve reconstruction, these methods remain computationally intensive and less effective for highly variable noise distributions.
- Deep Learning Approaches: DL methods, including VAEs [37,38] and cVAEs [39], leverage neural networks to model and correct noise. However, they require large, high-quality training datasets to generalize effectively. In real-world remote sensing applications, their tendency to overfit and struggle with highly variable noise distributions limits their effectiveness.
- Proposed Method: Our approach integrates a Transformer-based Isolation Forest for noise detection and a cGAN for noise correction. Unlike traditional threshold-based exclusion methods, it preserves critical spectral information by reconstructing noisy samples rather than discarding them. By leveraging deep learning for both feature extraction and anomaly detection, combined with generative models for reconstruction, this framework ensures adaptability across different remote sensing environments, significantly improving SOC estimation accuracy in complex datasets.
5.2. Impact of Noise Ratio on Model Performance
5.3. Evaluation Under High Vegetation Cover Using Sentinel-2
5.4. Practical Implications
5.5. Limitations and Future Work
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1D-CNN | One-dimensional convolutional neural network |
| ANN | Artificial neural network |
| B | Band |
| BI | Brightness index |
| CBR | CatBoost regressor |
| cGAN | Conditional generative adversarial network |
| CI | Color index |
| cVAE | Conditional variational autoencoder |
| DL | Deep learning |
| DSI | Dry soil index |
| DT | Decision tree |
| EC | Electrical conductivity |
| ESDAC | European Soil Data Centre |
| EVI | Enhanced vegetation index |
| FLAASH | Fast Line-of-sight Atmospheric Analysis of Spectral Hypercubes |
| GB | Gradient boosting |
| GNDVI | Green normalized difference vegetation index |
| HSI | Hyperspectral imaging |
| K | Potassium |
| KNN | k-Nearest neighbors |
| L8 | Landsat 8 |
| LOF | Local outlier factor |
| LR | Linear regression |
| LUCAS | Land Use/Cover Area frame Survey |
| MAD | Median absolute deviation |
| ML | Machine learning |
| MSAVI | Modified soil-adjusted vegetation index |
| N | Nitrogen |
| NDVI | Normalized difference vegetation index |
| OC-SVM | One-class support vector machine |
| P | Phosphorus |
| PCA | Principal component analysis |
| RF | Random forest |
| RVI | Ratio vegetation index |
| S2 | Sentinel-2 |
| SAVI | Soil-adjusted vegetation index |
| SC | Scenario |
| SI | Salinity index |
| SOC | Soil organic carbon |
| SVR | Support vector regression |
| TCT | Tasseled Cap transformation |
| VAE | Variational autoencoder |
| VIs | Vegetation indices |
References
- Fageria, N. Role of soil organic matter in maintaining sustainability of cropping systems. Commun. Soil Sci. Plant Anal. 2012, 43, 2063–2113. [Google Scholar] [CrossRef]
- Bhattacharya, S.S.; Kim, K.H.; Das, S.; Uchimiya, M.; Jeon, B.H.; Kwon, E.; Szulejko, J.E. A review on the role of organic inputs in maintaining the soil carbon pool of the terrestrial ecosystem. J. Environ. Manag. 2016, 167, 214–227. [Google Scholar] [CrossRef]
- Weil, R.R.; Islam, K.R.; Stine, M.A.; Gruver, J.B.; Samson-Liebig, S.E. Estimating active carbon for soil quality assessment: A simplified method for laboratory and field use. Am. J. Altern. Agric. 2003, 18, 3–17. [Google Scholar] [CrossRef]
- Loria, N.; Lal, R.; Chandra, R. Handheld In Situ Methods for Soil Organic Carbon Assessment. Sustainability 2024, 16, 5592. [Google Scholar] [CrossRef]
- Liu, S.; Chen, J.; Guo, L.; Wang, J.; Zhou, Z.; Luo, J.; Yang, R. Prediction of soil organic carbon in soil profiles based on visible–near-infrared hyperspectral imaging spectroscopy. Soil Tillage Res. 2023, 232, 105736. [Google Scholar] [CrossRef]
- Li, Y.; Chang, C.; Wang, Z.; Zhao, G. Remote sensing prediction and characteristic analysis of cultivated land salinization in different seasons and multiple soil layers in the coastal area. Int. J. Appl. Earth Obs. Geoinf. 2022, 111, 102838. [Google Scholar] [CrossRef]
- Ge, X.; Ding, J.; Jin, X.; Wang, J.; Chen, X.; Li, X.; Liu, J.; Xie, B. Estimating agricultural soil moisture content through UAV-based hyperspectral images in the arid region. Remote Sens. 2021, 13, 1562. [Google Scholar] [CrossRef]
- Datta, D.; Paul, M.; Murshed, M.; Teng, S.W.; Schmidtke, L. Soil Moisture, Organic Carbon, and Nitrogen Content Prediction with Hyperspectral Data Using Regression Models. Sensors 2022, 22, 7998. [Google Scholar] [CrossRef] [PubMed]
- Sargeant, J.; Teng, S.W.; Murshed, M.; Paul, M.; Brennan, D. Estimating Soil Organic Carbon from Multispectral Images Using Physics-Informed Neural Networks. In Proceedings of the Asian Conference on Computer Vision, Hanoi, Vietnam, 8–12 December 2024; pp. 2632–2649. [Google Scholar]
- Rahman, M.; Teng, S.W.; Murshed, M.; Paul, M.; Brennan, D. Deep Learning-based Adaptive Downsampling of Hyperspectral Bands for Soil Organic Carbon Estimation. IEEE Access 2025, 13, 95392–95409. [Google Scholar] [CrossRef]
- Rahman, M.; Teng, S.W.; Murshed, M.; Paul, M.; Brennan, D. Addressing Limitations of Common Methods in Attention-Based Hyperspectral Band Selection Algorithms. In Proceedings of the 2024 IEEE International Conference on Digital Image Computing: Techniques and Applications (DICTA), Perth, Australia, 27–29 November 2024; pp. 640–647. [Google Scholar]
- Rahman, M.; Teng, S.W.; Murshed, M.; Paul, M.; Brennan, D. BSDR: A data-efficient deep learning-based hyperspectral band selection algorithm using discrete relaxation. Sensors 2024, 24, 7771. [Google Scholar] [CrossRef]
- Datta, D.; Paul, M.; Murshed, M.; Teng, S.W.; Schmidtke, L. Comparative Analysis of Machine and Deep Learning Models for Soil Properties Prediction from Hyperspectral Visual Band. Environments 2023, 10, 77. [Google Scholar] [CrossRef]
- Stiglitz, R.; Mikhailova, E.; Post, C.; Schlautman, M.; Sharp, J. Using an inexpensive color sensor for rapid assessment of soil organic carbon. Geoderma 2017, 286, 98–103. [Google Scholar] [CrossRef]
- Nodi, S.S.; Paul, M.; Robinson, N.; Wang, L.; Rehman, S.U. Determination of Munsell Soil Colour Using Smartphones. Sensors 2023, 23, 3181. [Google Scholar] [CrossRef] [PubMed]
- Nodi, S.S.; Paul, M.; Robinson, N.; Wang, L.; Rehman, S.U.; Kabir, M.A. Munsell soil colour prediction from the soil and soil colour book using patching method and deep learning techniques. Sensors 2025, 25, 287. [Google Scholar] [CrossRef] [PubMed]
- Demattê, J.A.; Poppiel, R.R.; Novais, J.J.M.; Rosin, N.A.; Minasny, B.; Savin, I.Y.; Grunwald, S.; Chen, S.; Hong, Y.; Huang, J.; et al. Frontiers in earth observation for global soil properties assessment linked to environmental and socio-economic factors. Innovation 2025, 6, 100985. [Google Scholar] [CrossRef]
- Pande, C.B.; Kadam, S.A.; Jayaraman, R.; Gorantiwar, S.; Shinde, M. Prediction of soil chemical properties using multispectral satellite images and wavelet transforms methods. J. Saudi Soc. Agric. Sci. 2022, 21, 21–28. [Google Scholar] [CrossRef]
- Datta, D.; Paul, M.; Murshed, M.; Teng, S.W.; Schmidtke, L.M. Novel Dry Soil and Vegetation Indices to Predict Soil Contents from Landsat 8 Satellite Data. In Proceedings of the 2023 IEEE International Conference on Digital Image Computing: Techniques and Applications (DICTA), Port Macquarie, Australia, 28 November–1 December 2023; pp. 152–159. [Google Scholar]
- Yuzugullu, O.; Fajraoui, N.; Don, A.; Liebisch, F. Satellite-based soil organic carbon mapping on European soils using available datasets and support sampling. Sci. Remote Sens. 2024, 9, 100118. [Google Scholar] [CrossRef]
- Pettorelli, N. The Normalized Difference Vegetation Index; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Liao, Z.; He, B.; Quan, X. Modified enhanced vegetation index for reducing topographic effects. J. Appl. Remote Sens. 2015, 9, 096068. [Google Scholar] [CrossRef]
- Muzhoffar, D.A.F.; Sakuno, Y.; Taniguchi, N.; Hamada, K.; Shimabukuro, H.; Hori, M. Automatic Detection of Floating Macroalgae via Adaptive Thresholding Using Sentinel-2 Satellite Data with 10 m Spatial Resolution. Remote Sens. 2023, 15, 2039. [Google Scholar] [CrossRef]
- Lee, J.K.; Acharya, T.D.; Lee, D.H. Exploring Land Cover Classification Accuracy of Landsat 8 Image Using Spectral Index Layer Stacking in Hilly Region of South Korea. Sens. Mater. 2018, 30, 2927–2941. [Google Scholar] [CrossRef]
- Shiffler, R.E. Maximum Z scores and outliers. Am. Stat. 1988, 42, 79–80. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, Y. Geostatistical estimation methods: Kriging. In Quantitative Geosciences: Data Analytics, Geostatistics, Reservoir Characterization and Modeling; Springer: Berlin/Heidelberg, 2019; pp. 373–401. [Google Scholar]
- Voloh, B.; Watson, M.R.; König, S.; Womelsdorf, T. MAD saccade: Statistically robust saccade threshold estimation via the median absolute deviation. J. Eye Mov. Res. 2020, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Wei, K.; Xiong, W.; Yu, J.; Qi, B. Noise estimation for image sensor based on local entropy and median absolute deviation. Sensors 2019, 19, 339. [Google Scholar] [CrossRef]
- Breunig, M.M.; Kriegel, H.P.; Ng, R.T.; Sander, J. LOF: Identifying density-based local outliers. In Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data, Dallas, TX, USA, 16–18 May 2000; pp. 93–104. [Google Scholar]
- Campos, G.O.; Zimek, A.; Sander, J.; Campello, R.J.; Micenková, B.; Schubert, E.; Assent, I.; Houle, M.E. On the evaluation of unsupervised outlier detection: Measures, datasets, and an empirical study. Data Min. Knowl. Discov. 2016, 30, 891–927. [Google Scholar] [CrossRef]
- Ananias, P.H.M.; Negri, R.G. Anomalous behaviour detection using one-class support vector machine and remote sensing images: A case study of algal bloom occurrence in inland waters. Int. J. Digit. Earth 2021, 14, 921–942. [Google Scholar] [CrossRef]
- Alam, S.; Sonbhadra, S.K.; Agarwal, S.; Nagabhushan, P. One-class support vector classifiers: A survey. Knowl.-Based Syst. 2020, 196, 105754. [Google Scholar] [CrossRef]
- Candès, E.J.; Li, X.; Ma, Y.; Wright, J. Robust principal component analysis? J. ACM 2011, 58, 1–37. [Google Scholar] [CrossRef]
- Zhou, T.; Tao, D. Godec: Randomized low-rank & sparse matrix decomposition in noisy case. In Proceedings of the 28th International Conference on Machine Learning, ICML 2011, Bellevue, WA, USA, 28 June 28–2 July 2011. [Google Scholar]
- Lin, Q.; Li, C. Kriging based sequence interpolation and probability distribution correction for gaussian wind field data reconstruction. J. Wind. Eng. Ind. Aerodyn. 2020, 205, 104340. [Google Scholar] [CrossRef]
- Nowroozilarki, Z.; Mortazavi, B.J.; Jafari, R. Variational autoencoders for biomedical signal morphology clustering and noise detection. IEEE J. Biomed. Health Inform. 2023, 28, 169–180. [Google Scholar] [CrossRef]
- Sadeghi, M.; Alameda-Pineda, X. Switching variational auto-encoders for noise-agnostic audio-visual speech enhancement. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 6663–6667. [Google Scholar]
- Zhang, C.; Barbano, R.; Jin, B. Conditional variational autoencoder for learned image reconstruction. Computation 2021, 9, 114. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Soydaner, D. Attention mechanism in neural networks: Where it comes and where it goes. Neural Comput. Appl. 2022, 34, 13371–13385. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Xu, H.; Pang, G.; Wang, Y.; Wang, Y. Deep isolation forest for anomaly detection. IEEE Trans. Knowl. Data Eng. 2023, 35, 12591–12604. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.H. Isolation forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 413–422. [Google Scholar]
- Liu, F.T.; Ting, K.M.; Zhou, Z.H. Isolation-based anomaly detection. ACM Trans. Knowl. Discov. Data (TKDD) 2012, 6, 1–39. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional generative adversarial nets. arXiv 2014, arXiv:1411.1784. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Orgiazzi, A.; Ballabio, C.; Panagos, P.; Jones, A.; Fernández-Ugalde, O. LUCAS Soil, the largest expandable soil dataset for Europe: A review. Eur. J. Soil Sci. 2018, 69, 140–153. [Google Scholar] [CrossRef]
- Loveland, T.R.; Irons, J.R. Landsat 8: The plans, the reality, and the legacy. Remote Sens. Environ. 2016, 185, 1–6. [Google Scholar] [CrossRef]
- Barsi, J.A.; Schott, J.R.; Hook, S.J.; Raqueno, N.G.; Markham, B.L.; Radocinski, R.G. Landsat-8 thermal infrared sensor (TIRS) vicarious radiometric calibration. Remote Sens. 2014, 6, 11607–11626. [Google Scholar] [CrossRef]
- Thorne, K.; Markharn, B.; Barker, J.; Slater, P.; Biggar, S. Radiometric calibration of Landsat. Photogramm. Eng. Remote Sens. 1997, 63, 853–858. [Google Scholar]
- Gao, B.C.; Montes, M.J.; Davis, C.O.; Goetz, A.F. Atmospheric correction algorithms for hyperspectral remote sensing data of land and ocean. Remote Sens. Environ. 2009, 113, S17–S24. [Google Scholar]
- Maurer, T. How to pan-sharpen images using the gram-schmidt pan-sharpen method–A recipe. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 40, 239–244. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Dehni, A.; Lounis, M. Remote sensing techniques for salt affected soil mapping: Application to the Oran region of Algeria. Procedia Eng. 2012, 33, 188–198. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Matsushita, B.; Yang, W.; Chen, J.; Onda, Y.; Qiu, G. Sensitivity of the enhanced vegetation index (EVI) and normalized difference vegetation index (NDVI) to topographic effects: A case study in high-density cypress forest. Sensors 2007, 7, 2636–2651. [Google Scholar] [CrossRef] [PubMed]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Baig, M.H.A.; Zhang, L.; Shuai, T.; Tong, Q. Derivation of a tasselled cap transformation based on Landsat 8 at-satellite reflectance. Remote Sens. Lett. 2014, 5, 423–431. [Google Scholar] [CrossRef]
- Mulla, D.; McBratney, A.B. Soil Spatial Variability; Soil physics companion; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Fearn, T. Assessing calibrations: Sep, rpd, rer and r 2. NIR News 2002, 13, 12–13. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Y.; Zhu, X. Soil organic carbon estimation using remote sensing data-driven machine learning. PeerJ 2024, 12, e17836. [Google Scholar] [CrossRef] [PubMed]
- Datta, D.; Paul, M.; Murshed, M.; Teng, S.W.; Schmidtke, L.M. Unveiling Soil-Vegetation Interactions: Reflection Relationships and an Attention-Based Deep Learning Approach for Carbon Estimation. In Proceedings of the 2024 IEEE International Conference on Multimedia and Expo Workshops (ICMEW), Niagara Falls, ON, Canada, 29 August 2024; pp. 1–6. [Google Scholar]
- John, K.; Abraham Isong, I.; Michael Kebonye, N.; Okon Ayito, E.; Chapman Agyeman, P.; Marcus Afu, S. Using machine learning algorithms to estimate soil organic carbon variability with environmental variables and soil nutrient indicators in an alluvial soil. Land 2020, 9, 487. [Google Scholar] [CrossRef]
- Luedtke, J.; Ahmed, S. A sample approximation approach for optimization with probabilistic constraints. SIAM J. Optim. 2008, 19, 674–699. [Google Scholar] [CrossRef]
- Paul, M.; Datta, D.; Murshed, M.; Teng, S.W.; Schmidtke, L.M. Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation. SSRN, 2025; in press. [Google Scholar]
- Datta, D. Transformer-Guided Noise Detection and Correction Framework. 2025. Available online: https://github.com/DristiDatta/Transformer_Guided_Noise_Detection (accessed on 22 August 2025).



| Soil Type | NDVI Range | S. No. | Min | Max | Mean | Median | Std. | CV(%) |
|---|---|---|---|---|---|---|---|---|
| Bare soil | 510 | 2.20 | 96.50 | 15.05 | 12.50 | 11.21 | 74.48 |
| Index Name | Formula | Source |
|---|---|---|
| Ratio Vegetation Index (RVI) | [57] | |
| Normalized Difference Vegetation Index (NDVI) | [58] | |
| Green NDVI (GNDVI) | [54] | |
| Enhanced Vegetation Index (EVI) | [59] | |
| Soil-Adjusted Vegetation Index (SAVI) | [60] | |
| Modified SAVI (MSAVI) | [55] | |
| Brightness Index (BI) | [56] | |
| Salinity Index (SI) | [56] | |
| Color Index (CI) | [56] |
| TCT Component | B2 | B3 | B4 | B5 | B6 | B7 |
|---|---|---|---|---|---|---|
| Brightness | 0.3029 | 0.2786 | 0.4733 | 0.5599 | 0.5080 | 0.1872 |
| Greenness | −0.2941 | −0.2430 | −0.5424 | 0.7276 | 0.0713 | −0.1608 |
| Wetness | 0.1511 | 0.1973 | 0.3283 | 0.3407 | −0.7117 | −0.4559 |
| TCT4 | −0.8239 | 0.0849 | 0.4396 | −0.0580 | 0.2013 | −0.2773 |
| TCT5 | −0.3294 | 0.0557 | 0.1056 | 0.1855 | −0.4349 | 0.8085 |
| TCT6 | 0.1079 | −0.9023 | 0.4119 | 0.0575 | −0.0259 | 0.0252 |
| Model | Hyperparameters and Values |
|---|---|
| LR | Standardization: True; Random state: 42 |
| SVR | Kernel: rbf; C: 1.0; Epsilon: 0.2; Batch size: 32; Epochs: 100; Learning rate: 0.001 |
| ANN | fc1: 128; fc2: 64; fc3: 1; Activation: ReLU; Dropout: 0.3; Batch size: 32; Epochs: 100; Learning rate: 0.001 |
| DT | Random state: 42; Max Depth: None |
| KNN | Neighbors: 5; Metric: Euclidean |
| 1D-CNN | Conv layers: 3; Filters: 16/32/64; Kernel size: 3; Padding: 1; fc1: 128; Dropout: 0.3; Batch size: 32; Epochs: 100; Learning rate: 0.001 |
| GB | Estimators: 100; Random state: 42 |
| RF | Estimators: 100; Random state: 42 |
| CBR | Learning rate: 0.1; Depth: 10; Loss: RMSE; Iterations: 100 |
| Group | Input | LR | SVR | ANN | DT | KNN | 1D CNN | GB | RF | CBR |
|---|---|---|---|---|---|---|---|---|---|---|
| L8 7 bands | SC.1 | 0.28 | 0.39 | 0.42 | 0.43 | 0.53 | 0.50 | 0.45 | 0.55 | 0.56 |
| 9.11 | 8.50 | 8.16 | 7.98 | 7.27 | 7.37 | 7.57 | 6.80 | 6.93 | ||
| 1.19 | 1.28 | 1.33 | 1.39 | 1.49 | 1.49 | 1.35 | 1.50 | 1.50 | ||
| SC.2 | 0.40 | 0.40 | 0.50 | 0.37 | 0.48 | 0.55 | 0.59 | 0.60 | 0.62 | |
| 7.36 | 7.59 | 6.57 | 7.29 | 6.79 | 6.05 | 5.95 | 5.63 | 5.75 | ||
| 1.32 | 1.29 | 1.49 | 1.41 | 1.44 | 1.63 | 1.66 | 1.81 | 1.69 | ||
| SC.3 (proposed) | 0.40 | 0.33 | 0.61 | 0.32 | 0.57 | 0.60 | 0.57 | 0.67 | 0.64 | |
| 7.67 | 8.92 | 6.22 | 8.08 | 6.54 | 6.12 | 6.65 | 5.80 | 6.08 | ||
| 1.37 | 1.23 | 1.70 | 1.32 | 1.61 | 1.74 | 1.62 | 1.84 | 1.75 | ||
| L8 7 bands + transformed features | SC.4 | 0.32 | 0.41 | 0.48 | 0.53 | 0.53 | 0.54 | 0.56 | 0.59 | 0.59 |
| 8.84 | 8.40 | 7.34 | 7.24 | 6.97 | 6.78 | 6.93 | 6.86 | 6.82 | ||
| 1.23 | 1.30 | 1.49 | 1.53 | 1.56 | 1.62 | 1.56 | 1.58 | 1.59 | ||
| SC.5 | 0.47 | 0.41 | 0.51 | 0.50 | 0.53 | 0.59 | 0.63 | 0.56 | 0.63 | |
| 6.75 | 7.55 | 6.44 | 6.50 | 6.40 | 5.81 | 5.64 | 6.00 | 5.83 | ||
| 1.45 | 1.30 | 1.53 | 1.55 | 1.52 | 1.70 | 1.74 | 1.68 | 1.67 | ||
| SC.6 (proposed) | 0.52 | 0.30 | 0.59 | 0.31 | 0.55 | 0.56 | 0.62 | 0.66 | 0.68 | |
| 6.81 | 9.09 | 6.28 | 7.62 | 6.76 | 6.40 | 6.23 | 5.83 | 5.77 | ||
| 1.55 | 1.20 | 1.68 | 1.42 | 1.57 | 1.66 | 1.70 | 1.82 | 1.84 | ||
| Band selection with state-of-art methods (from SC.6) | SC.7 | 0.53 | 0.29 | 0.61 | 0.33 | 0.53 | 0.50 | 0.62 | 0.64 | 0.65 |
| 6.79 | 9.14 | 6.16 | 7.78 | 6.88 | 6.69 | 6.25 | 5.96 | 5.96 | ||
| 1.56 | 1.19 | 1.72 | 1.37 | 1.54 | 1.60 | 1.69 | 1.78 | 1.78 | ||
| SC.8 | 0.40 | 0.30 | 0.61 | 0.23 | 0.52 | 0.57 | 0.61 | 0.62 | 0.65 | |
| 7.69 | 9.13 | 6.17 | 8.13 | 6.80 | 6.34 | 6.30 | 6.11 | 5.90 | ||
| 1.37 | 1.20 | 1.72 | 1.33 | 1.56 | 1.68 | 1.68 | 1.73 | 1.79 | ||
| SC.9 | 0.50 | 0.31 | 0.61 | 0.26 | 0.56 | 0.58 | 0.61 | 0.65 | 0.65 | |
| 6.97 | 9.01 | 6.11 | 7.75 | 0.57 | 6.23 | 6.34 | 5.87 | 5.95 | ||
| 1.52 | 1.21 | 1.73 | 1.39 | 1.61 | 1.71 | 1.67 | 1.84 | 1.80 |
| Method | Noise-Detection Approach | Noise-Correction Approach | ↑ | RMSE ↓ | RPD ↑ | (%) | RMSE (%) | RPD (%) |
|---|---|---|---|---|---|---|---|---|
| Threshold-Based Exclusion | NDVI [19] | None | 0.55 | 6.80 | 1.50 | 21.82 | 14.71 | 22.67 |
| Statistical Detection | Z-Score [26] | Kriging [27,36] | 0.61 | 6.68 | 1.67 | 9.84 | 13.17 | 10.18 |
| Statistical Detection | MAD [28,29] | Kriging [27,36] | 0.62 | 5.88 | 1.62 | 8.06 | 1.36 | 13.58 |
| ML Detection | LOF [30,31] | Robust PCA [34,35] | 0.59 | 6.51 | 1.68 | 13.56 | 10.91 | 9.52 |
| Hybrid ML | OC-SVM [32,33] | Kriging [27,36] | 0.66 | 6.07 | 1.75 | 1.52 | 4.45 | 5.14 |
| DL Detection | VAEs [37,38] | cVAEs [39] | 0.54 | 7.28 | 1.53 | 24.07 | 20.33 | 20.26 |
| Proposed Method | Transformer-guided Isolation Forest | cGAN | 0.67 | 5.80 | 1.84 | – | – | – |
| Noise Level | Input | LR | SVR | ANN | DT | KNN | 1dCNN | GB | RF | CBR |
|---|---|---|---|---|---|---|---|---|---|---|
| 100% Noise | Baseline | −0.51 | −0.12 | −0.25 | −0.47 | −0.12 | −0.23 | 0.16 | 0.07 | 0.31 |
| 17.28 | 17.01 | 16.89 | 18.32 | 15.62 | 16.79 | 14.01 | 15.09 | 13.20 | ||
| 0.90 | 0.94 | 0.91 | 0.93 | 1.02 | 0.92 | 1.15 | 1.05 | 1.29 | ||
| Proposed | 0.61 | 0.48 | 0.59 | 0.81 | 0.75 | 0.50 | 0.86 | 0.72 | 0.86 | |
| 8.94 | 12.17 | 9.41 | 6.16 | 8.21 | 10.36 | 5.20 | 6.75 | 5.63 | ||
| 1.73 | 1.49 | 1.63 | 3.03 | 2.01 | 1.51 | 3.01 | 2.47 | 3.00 | ||
| 50% Noise | Baseline | −0.51 | 0.08 | −0.04 | 0.33 | 0.38 | 0.00 | 0.40 | 0.36 | 0.44 |
| 16.88 | 16.05 | 15.83 | 12.31 | 12.33 | 15.40 | 11.91 | 12.46 | 11.72 | ||
| 0.97 | 1.04 | 1.03 | 1.39 | 1.38 | 1.08 | 1.41 | 1.33 | 1.41 | ||
| Proposed | 0.39 | 0.32 | 0.36 | −0.10 | 0.48 | 0.42 | 0.51 | 0.34 | 0.67 | |
| 11.93 | 13.95 | 11.19 | 13.49 | 10.67 | 11.39 | 10.72 | 11.04 | 8.93 | ||
| 1.36 | 1.23 | 1.48 | 1.40 | 1.61 | 1.45 | 1.54 | 1.53 | 1.91 | ||
| 33% Noise | Baseline | 0.12 | 0.20 | 0.22 | 0.17 | 0.36 | 0.21 | 0.48 | 0.28 | 0.58 |
| 11.45 | 11.45 | 10.86 | 10.35 | 9.69 | 10.59 | 8.15 | 9.40 | 7.95 | ||
| 1.10 | 1.12 | 1.18 | 1.24 | 1.31 | 1.20 | 1.63 | 1.38 | 1.63 | ||
| Proposed | 0.42 | 0.36 | 0.42 | 0.46 | 0.44 | 0.44 | 0.58 | 0.55 | 0.63 | |
| 9.64 | 10.30 | 9.43 | 9.25 | 9.20 | 9.13 | 7.93 | 8.19 | 7.72 | ||
| 1.33 | 1.27 | 1.34 | 1.39 | 1.40 | 1.39 | 1.60 | 1.54 | 1.66 | ||
| 25% Noise | Baseline | 0.12 | 0.28 | 0.21 | 0.00 | 0.33 | 0.30 | 0.42 | 0.49 | 0.57 |
| 11.38 | 10.78 | 10.81 | 10.60 | 9.26 | 10.48 | 8.57 | 8.12 | 7.55 | ||
| 1.07 | 1.18 | 1.13 | 1.20 | 1.34 | 1.21 | 1.42 | 1.51 | 1.68 | ||
| Proposed | 0.47 | 0.36 | 0.53 | 0.29 | 0.52 | 0.43 | 0.56 | 0.50 | 0.64 | |
| 9.68 | 10.72 | 9.10 | 11.01 | 9.03 | 9.82 | 8.39 | 8.90 | 7.66 | ||
| 1.38 | 1.26 | 1.48 | 1.26 | 1.52 | 1.38 | 1.67 | 1.60 | 1.83 | ||
| 20% Noise | Baseline | 0.12 | 0.26 | 0.24 | 0.20 | 0.20 | 0.13 | 0.53 | 0.36 | 0.50 |
| 10.86 | 10.47 | 10.03 | 9.41 | 9.48 | 10.70 | 7.71 | 8.27 | 7.94 | ||
| 1.24 | 1.94 | 1.22 | 1.33 | 1.35 | 1.19 | 1.59 | 1.58 | 1.57 | ||
| Proposed | 0.38 | 0.36 | 0.45 | 0.33 | 0.38 | 0.50 | 0.58 | 0.58 | 0.55 | |
| 9.39 | 9.90 | 8.91 | 8.72 | 9.06 | 8.49 | 7.50 | 7.30 | 7.78 | ||
| 1.31 | 1.26 | 1.36 | 1.45 | 1.36 | 1.43 | 1.62 | 1.69 | 1.61 |
| Soil Type | NDVI Range | S. No. | Min | Max | Mean | Median | Std. | CV(%) |
|---|---|---|---|---|---|---|---|---|
| Mixed | 485 | 2.3 | 172.3 | 17.12 | 13.80 | 16.37 | 95.28 |
| Input | LR | SVR | ANN | DT | KNN | 1D CNN | GB | RF | CBR |
|---|---|---|---|---|---|---|---|---|---|
| Raw Data | 0.04 | 0.02 | 0.07 | −1.21 | −0.02 | 0.02 | 0.00 | −0.07 | −0.02 |
| 15.14 | 15.66 | 15.01 | 20.49 | 15.47 | 15.21 | 15.37 | 15.79 | 15.62 | |
| 1.03 | 1.01 | 1.04 | 0.78 | 1.01 | 1.03 | 1.02 | 0.99 | 1.00 | |
| OC-SVM | 0.10 | 0.03 | 0.05 | 0.04 | 0.04 | 0.14 | 0.12 | 0.13 | 0.15 |
| & Kriging | 11.43 | 15.64 | 15.04 | 19.49 | 15.23 | 15.36 | 14.79 | 14.68 | 14.78 |
| 1.09 | 1.02 | 1.05 | 0.98 | 1.00 | 1.13 | 1.11 | 1.03 | 1.10 | |
| Proposed | 0.20 | 0.12 | 0.21 | 0.21 | 0.26 | 0.39 | 0.32 | 0.37 | 0.37 |
| 11.09 | 15.94 | 15.76 | 19.68 | 13.50 | 9.51 | 12.47 | 11.95 | 12.11 | |
| 1.22 | 1.11 | 1.13 | 1.12 | 1.20 | 1.67 | 1.26 | 1.34 | 1.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, M.; Datta, D.; Murshed, M.; Teng, S.W.; Schmidtke, L.M. Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation. Remote Sens. 2025, 17, 3463. https://doi.org/10.3390/rs17203463
Paul M, Datta D, Murshed M, Teng SW, Schmidtke LM. Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation. Remote Sensing. 2025; 17(20):3463. https://doi.org/10.3390/rs17203463
Chicago/Turabian StylePaul, Manoranjan, Dristi Datta, Manzur Murshed, Shyh Wei Teng, and Leigh M. Schmidtke. 2025. "Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation" Remote Sensing 17, no. 20: 3463. https://doi.org/10.3390/rs17203463
APA StylePaul, M., Datta, D., Murshed, M., Teng, S. W., & Schmidtke, L. M. (2025). Transformer-Guided Noise Detection and Correction in Remote Sensing Data for Enhanced Soil Organic Carbon Estimation. Remote Sensing, 17(20), 3463. https://doi.org/10.3390/rs17203463









