Tree Type Classification from ALS Data: A Comparative Analysis of 1D, 2D, and 3D Representations Using ML and DL Models
Abstract
1. Introduction
1.1. Related Work
- 3D voxel grids, encoding local point densities [42].
1.2. Research Gap and Contribution
1.3. Outline
2. Materials
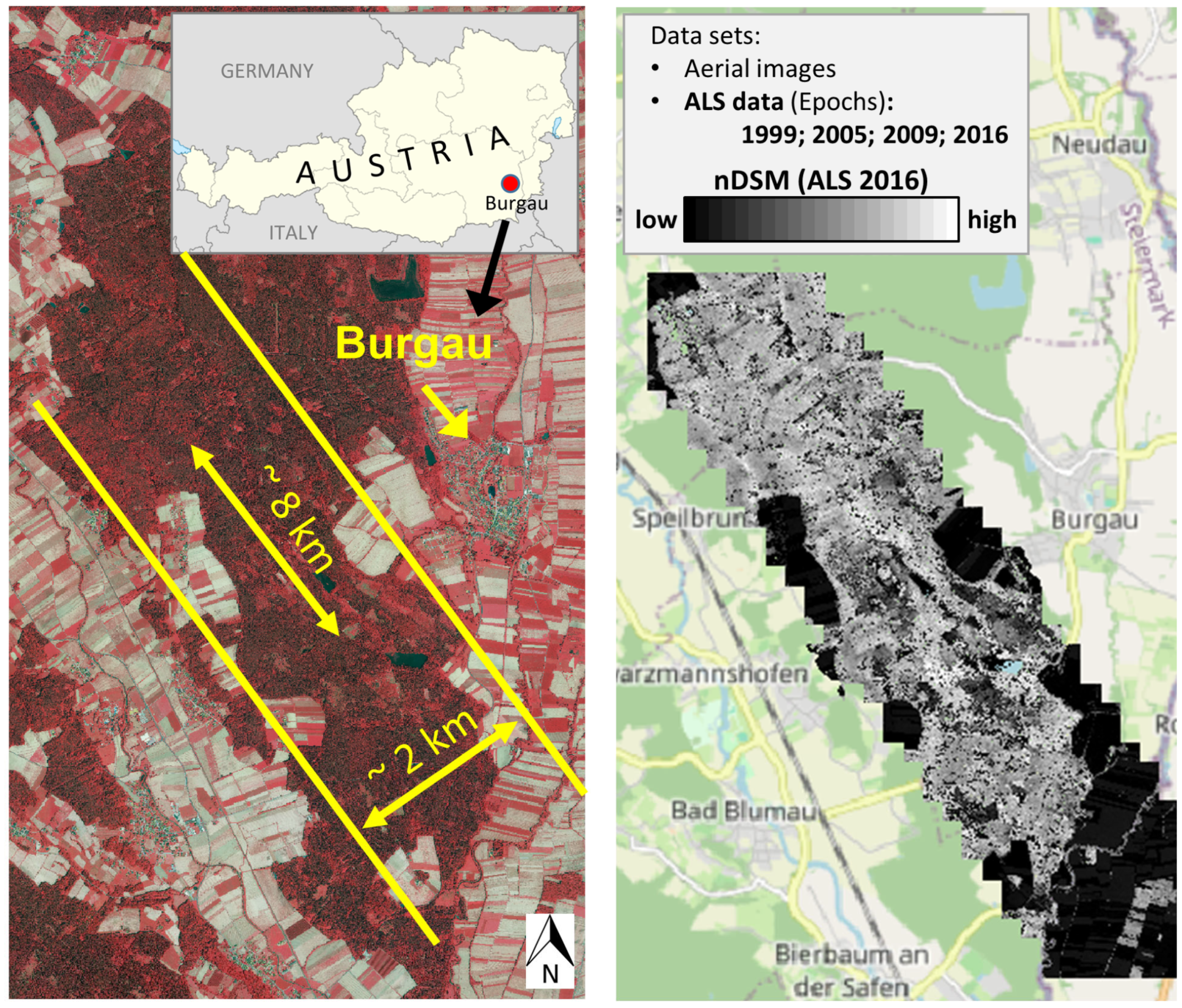
2.1. Study Site
2.2. Data
2.2.1. ALS Data
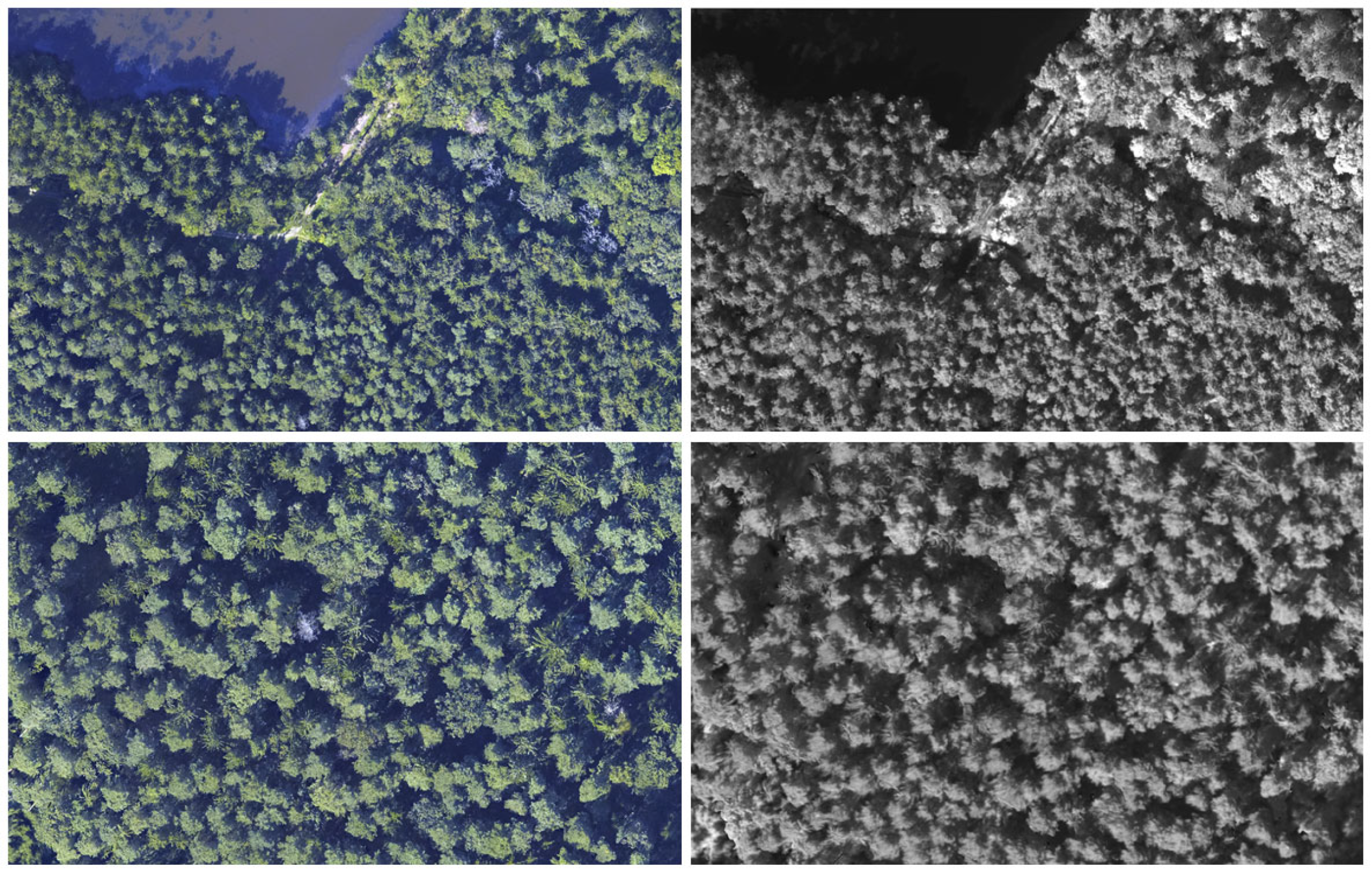
2.2.2. Aerial Images
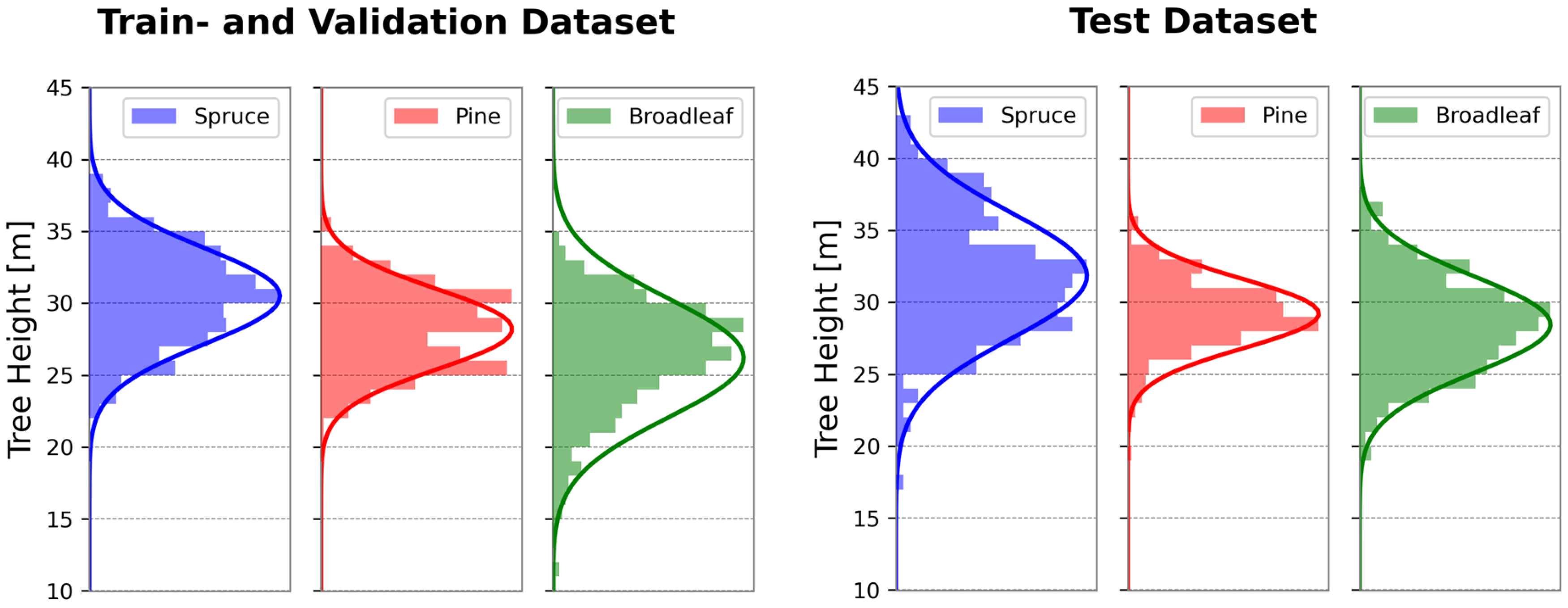
2.3. Training Data Collection
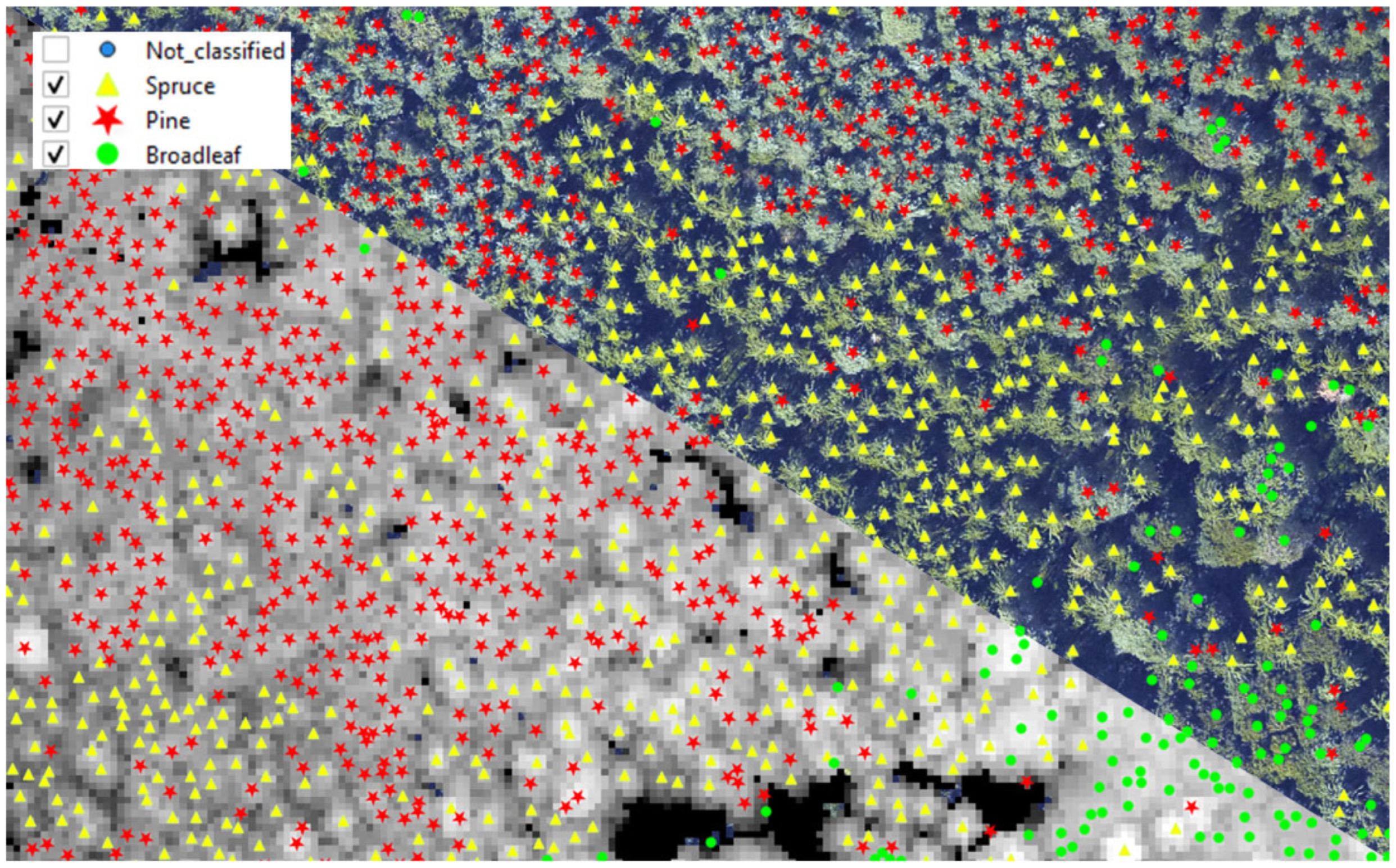
2.3.1. Ground Truth Data Collection in the Field
2.3.2. Collection of Data from Aerial Imagery and ALS Data
- About a quarter of the trees originated from the same area as the training data;
- Another quarter came from nearby forests with similar structural and species characteristics;
- The remaining half came from adjacent forests that differed in density, species mixture, and crown structure of broadleaf trees.
3. Methods
3.1. Pre-Processing and Preparation of Data for ML- and DL-Based Tree Type Classification
3.1.1. Individual Tree Detection (ITD)
3.1.2. Extraction of Tree Crown
3.1.3. Computation of Normals and Curvature
3.1.4. Normalization of Data
3.2. 1D Vector-Based Methods
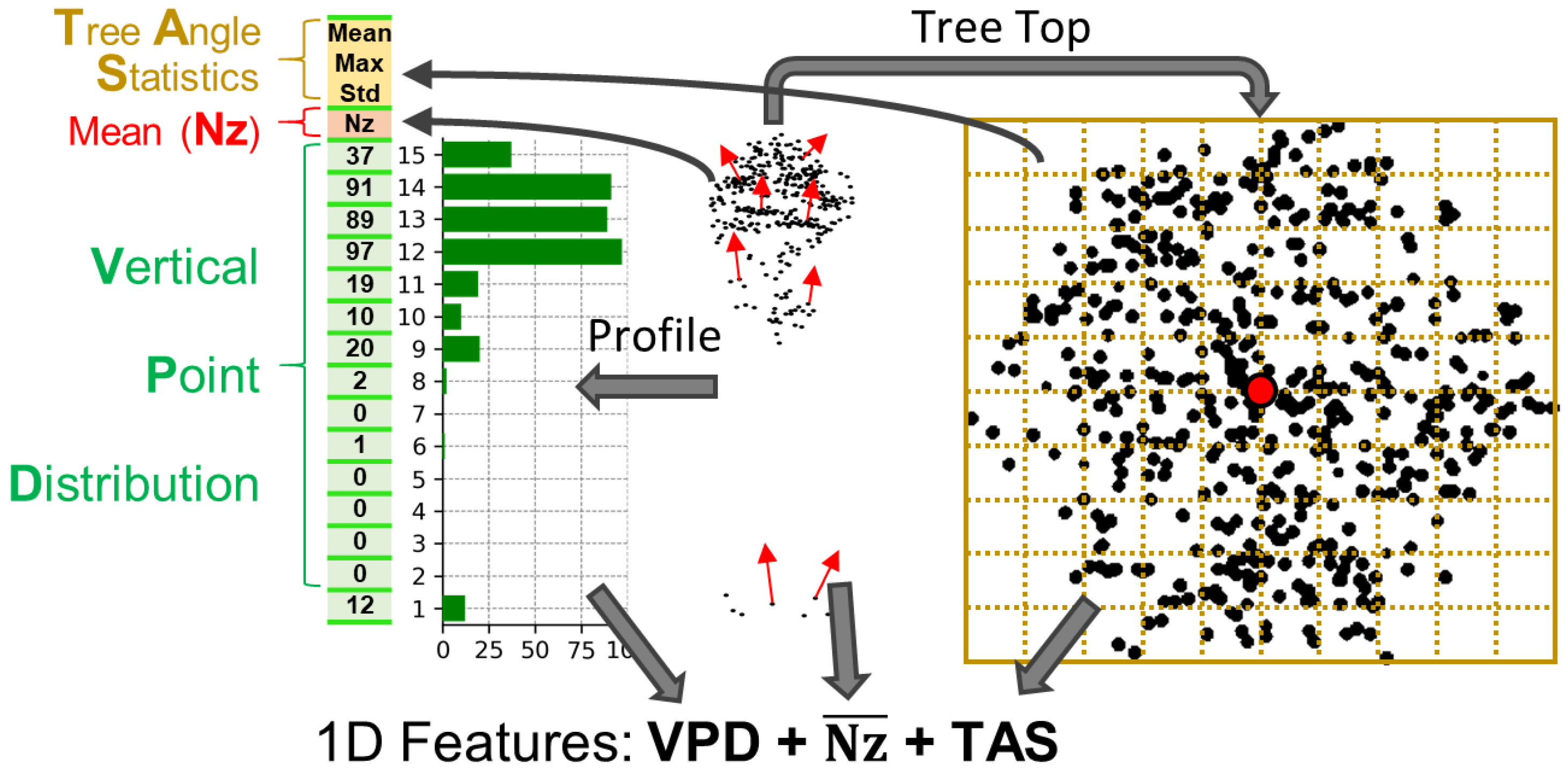
3.2.1. 1D Specific Data Preparation
3.2.2. ML Algorithms
3.2.3. DL Algorithms
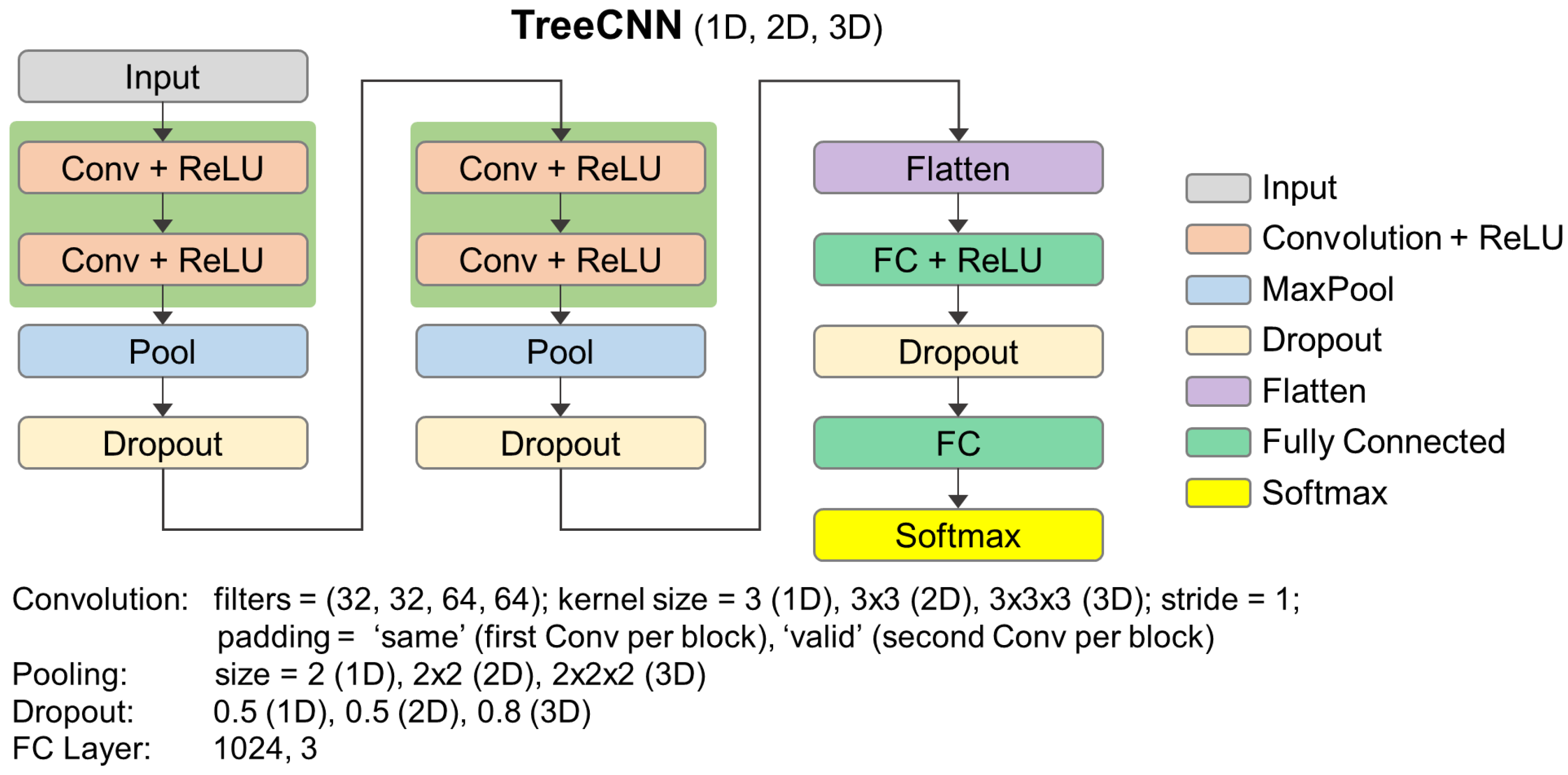
3.3. 2D Raster-Based Methods
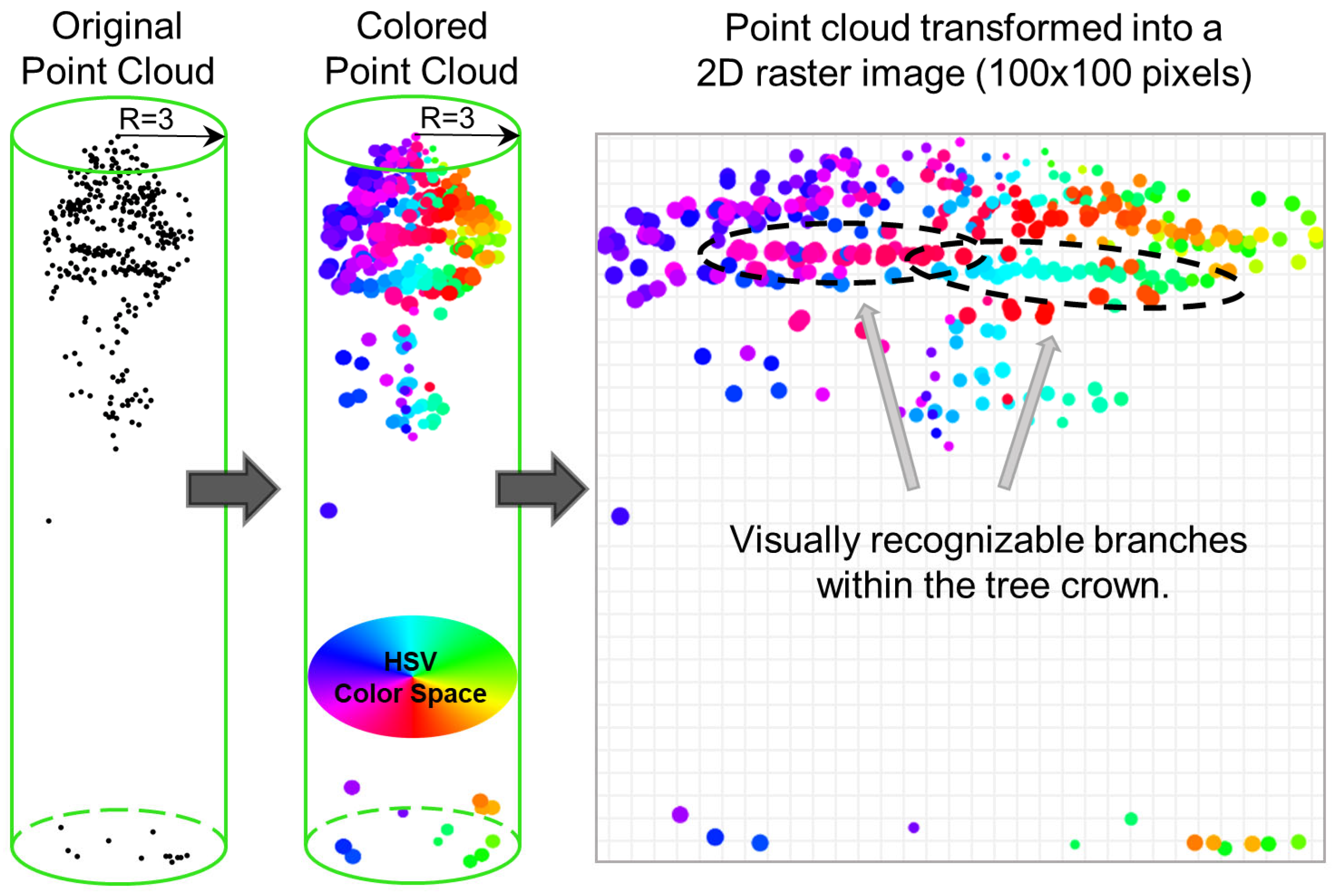
3.3.1. 2D Specific Data Preparation
3.3.2. ML Algorithms
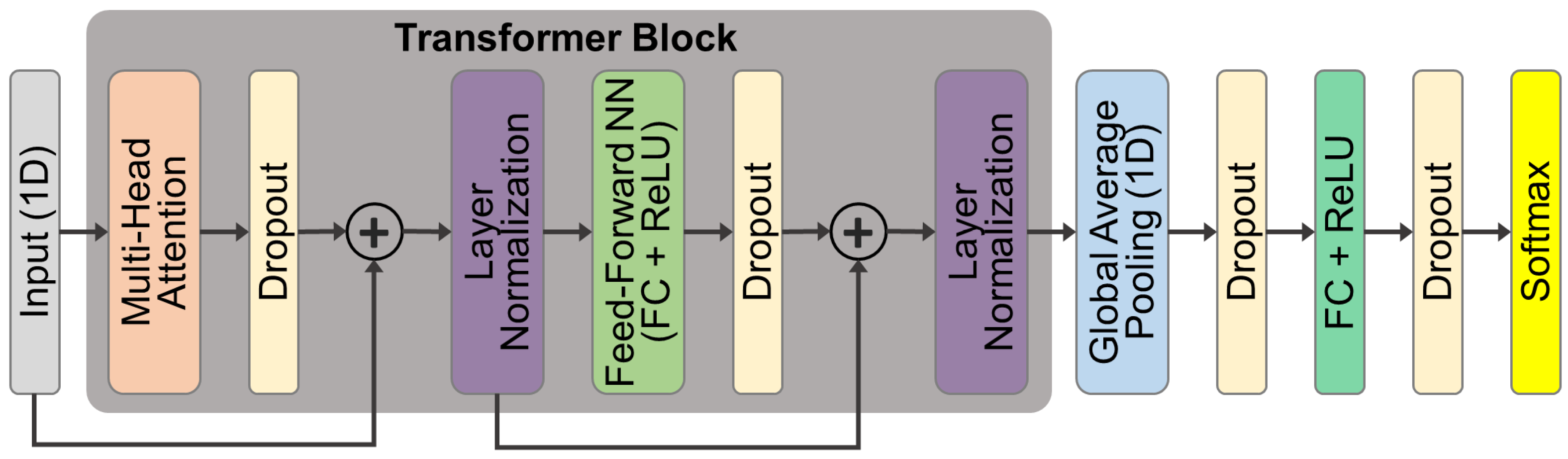
3.3.3. DL Algorithms
- Number of training epochs: 50;
- Batch size: 16;
- Checkpointing: Best model saved after each epoch.
- Random zoom adjustments of the input images up to ±10%;
- Randomly masking 25% of each image by setting the corresponding pixels to white.
3.4. 3D Voxel-Based Methods
3.4.1. 3D Specific Data Preparation
3.4.2. ML Algorithms
3.4.3. DL Algorithms
3.5. 3D Point Cloud-Based Methods
DL Algorithms
4. Results
4.1. Validation Methods
4.2. Illustrative Results
4.3. Quantitative Results
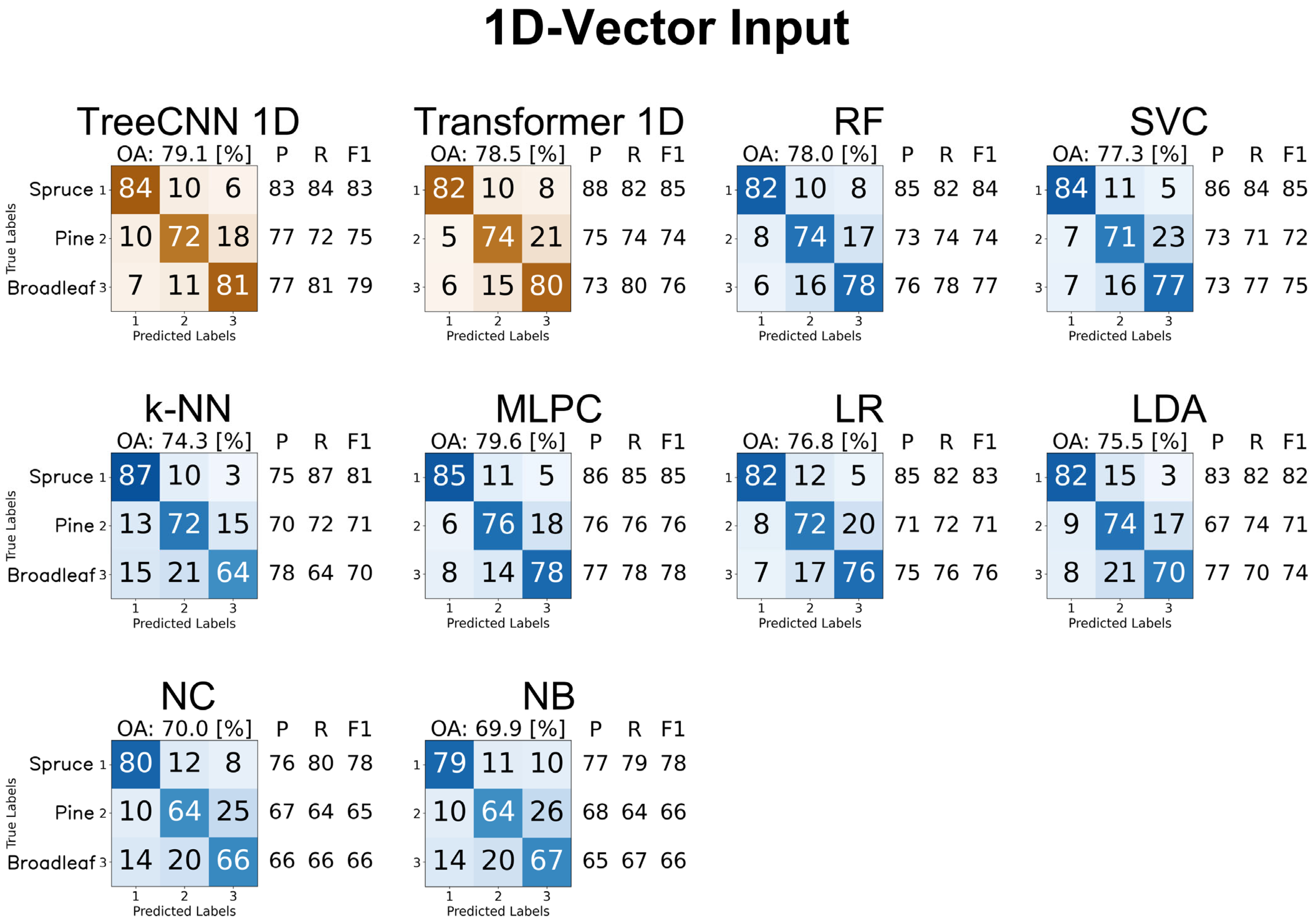
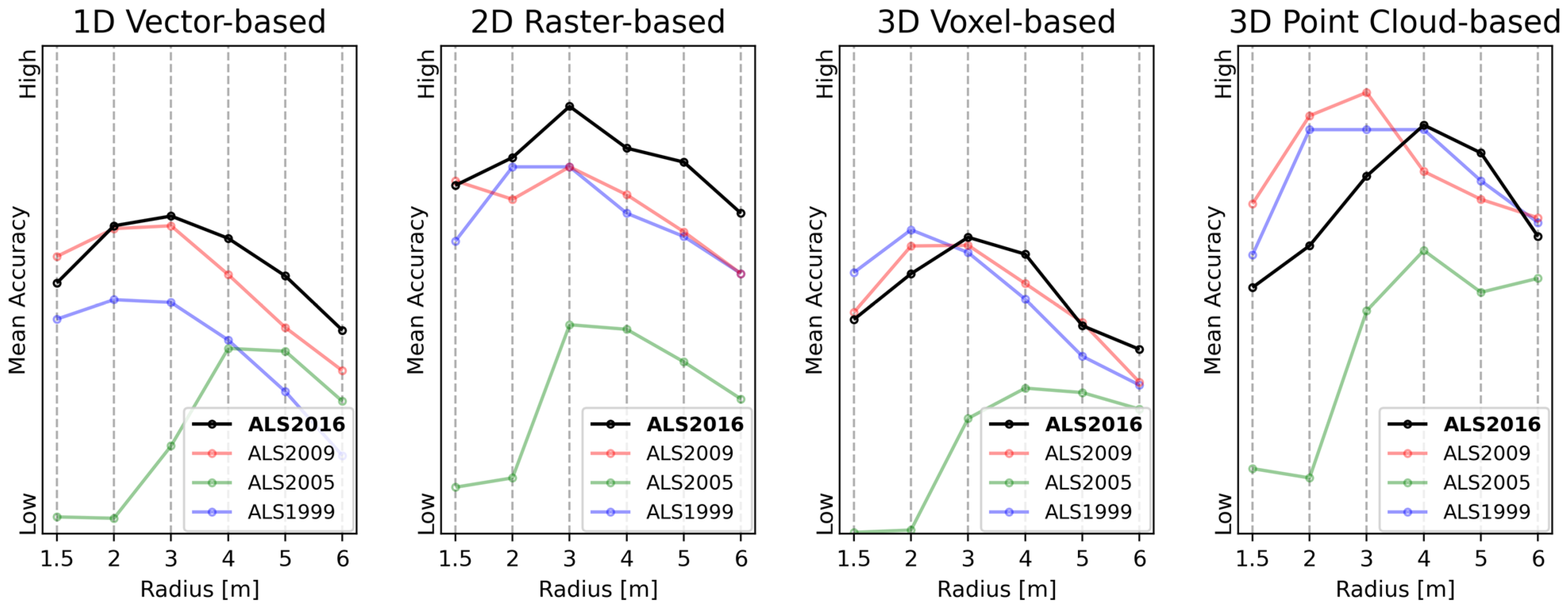
4.3.1. 1D Feature Vectors
4.3.2. 2D Raster Profiles
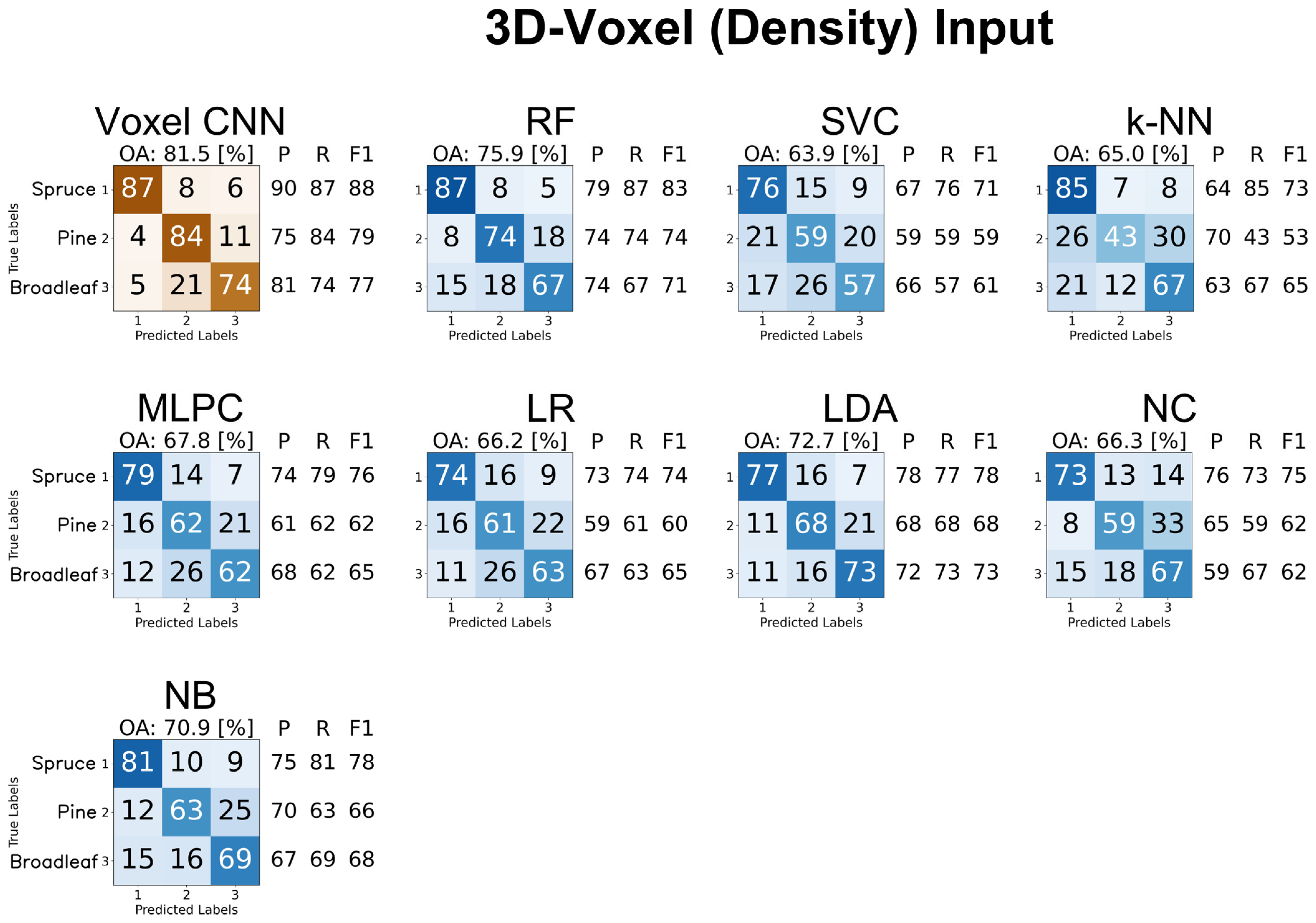
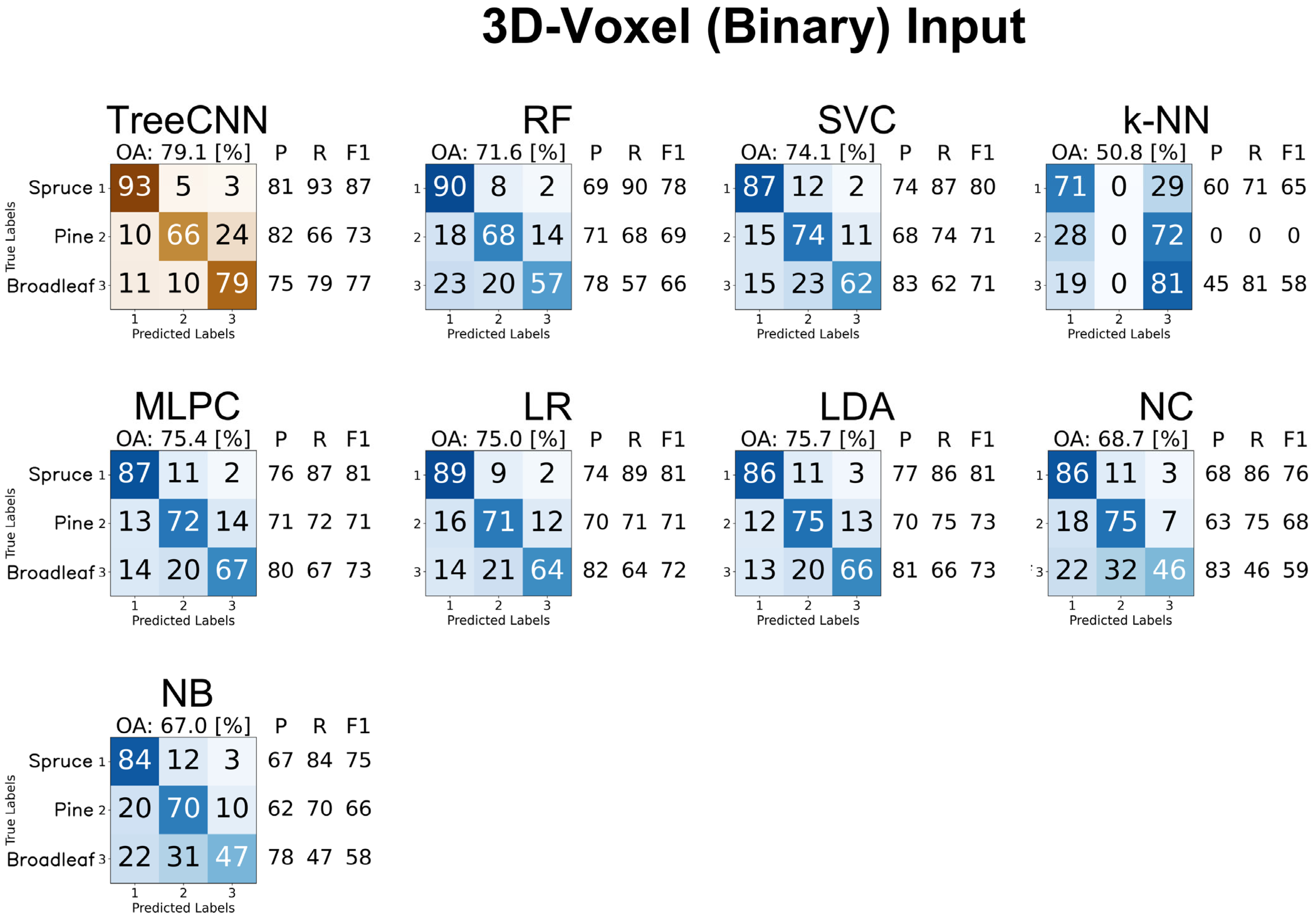
4.3.3. 3D Voxel Representations
4.3.4. 3D Point Clouds
4.3.5. Summary of Results
5. Discussion
5.1. Model Behavior and Interpretation
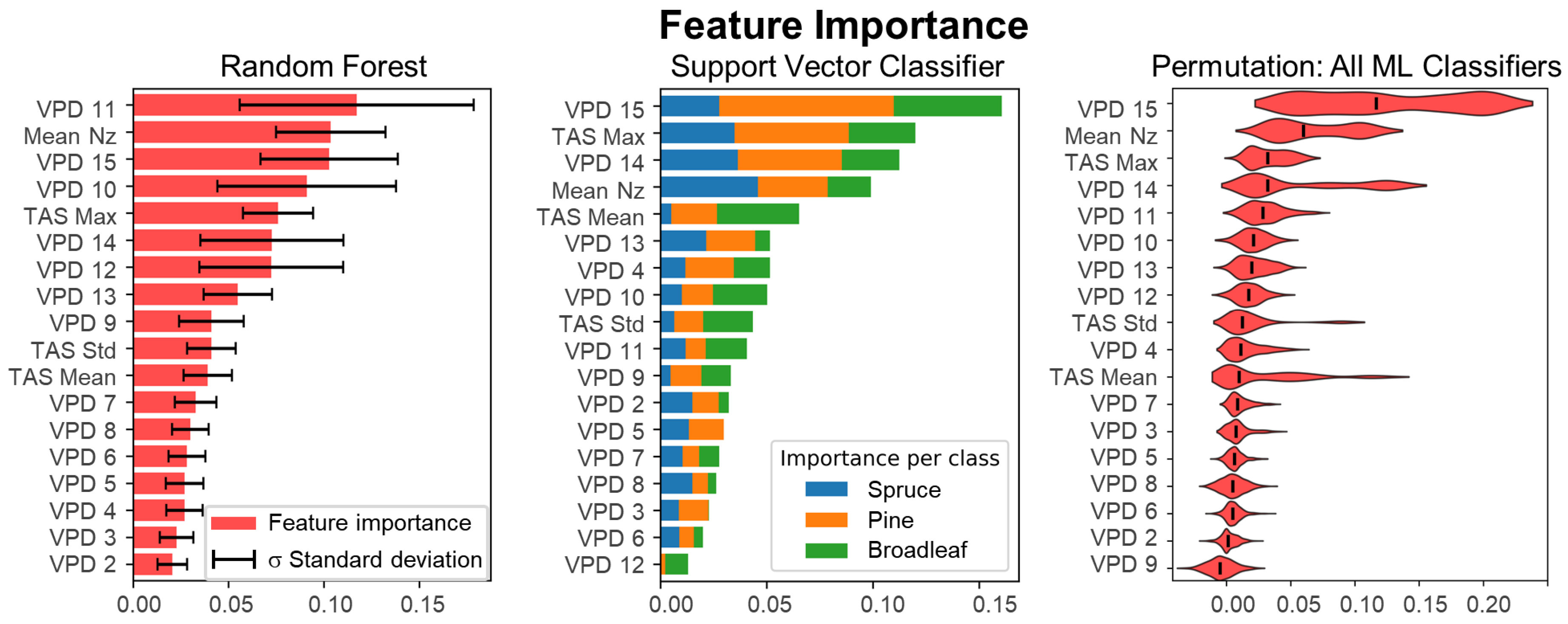
5.1.1. Feature Importance for 1D Data Structure
- VPD upper layers (especially layers 10–15) had the highest discriminative power across all classifiers. These layers capture structural characteristics near the top of the crown, where ALS point density is highest and species-specific shapes are most distinct.
- Nz (normal z-component) emerged as a highly informative horizontal descriptor, likely reflecting branching orientation differences, e.g., the uniform horizontal layering in spruce and pine vs. the irregular crowns of broadleaf species.
- TAS metrics (mean, median, standard deviation of crown surface angles) added complementary geometric information, improving classification, particularly when combined with VPD.
5.1.2. Limitations of ML on 2D-Raster and 3D-Voxel Data Structures
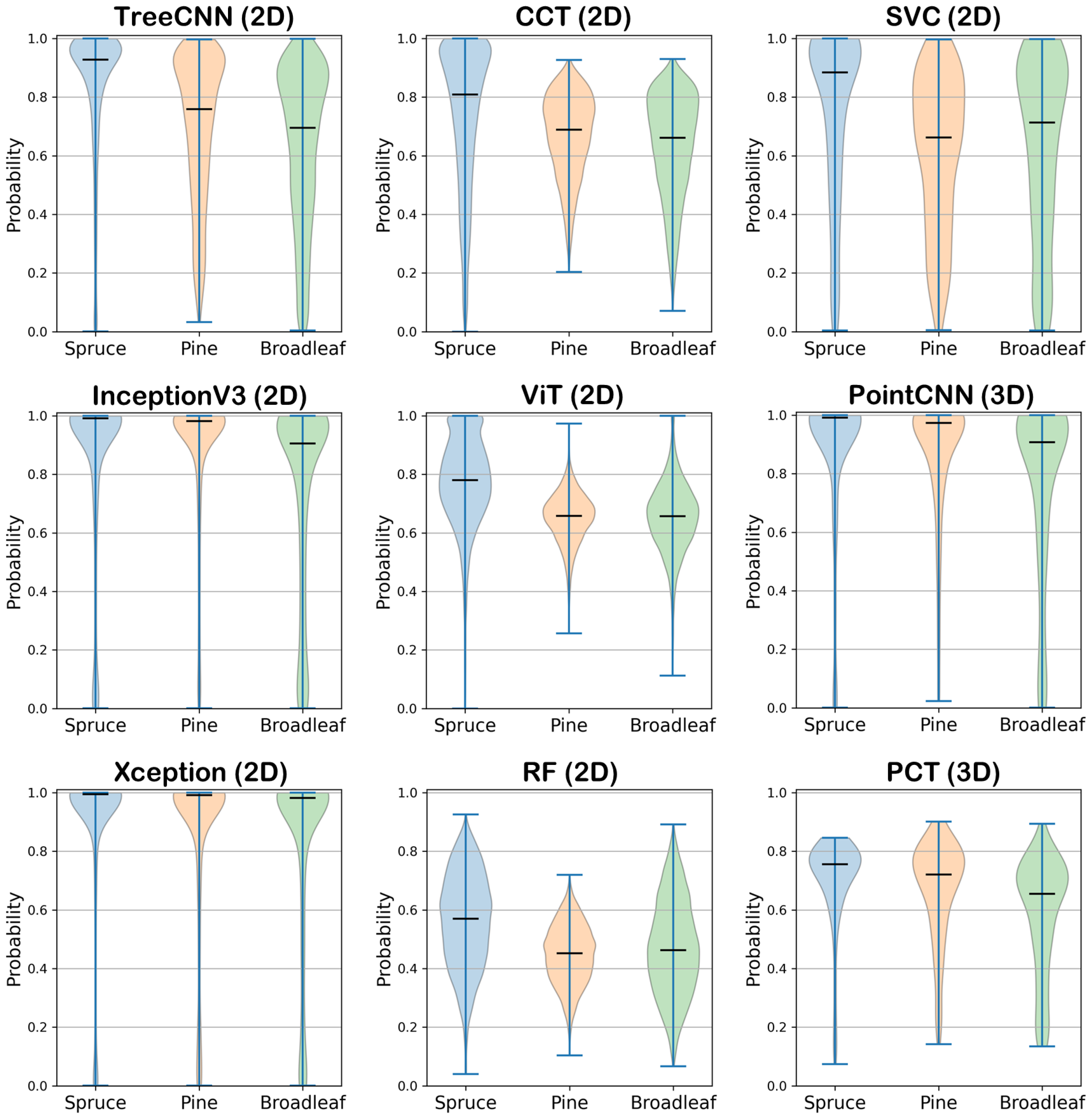
5.1.3. Reliability and Probabilistic Confidence
- TreeCNN (2D) shows high reliability for spruce, with probabilities clustered near 1.0. However, predictions for pine and broadleaf are more dispersed, indicating lower confidence and greater uncertainty in these classes.
- The InceptionV3, Xception, and PointCNN (3D) models exhibit very narrow distributions near 1.0 for all three classes. These models provide consistently high prediction confidence.
- CCT (2D) and SVC (2D) demonstrate moderate confidence, with predicted probabilities typically ranging from 0.6 to 0.9. Notably, SVC exhibits slightly broader distributions for pine and broadleaf, suggesting more variability and lower certainty for these more ambiguous classes.
- ViT (2D) and PCT (3D) fall into an intermediate range. Both show peaked but wider distributions, particularly for pine, reflecting moderate confidence in predictions. Between the two, PCT tends to produce slightly higher median probabilities, particularly for spruce.
- RF (2D) displays the least reliable outputs, with all three classes showing broad, flat distributions and the lowest median probabilities. This suggests a general tendency toward low certainty and less discriminative output compared to DL models.
5.2. Ensemble Modeling
- Intra-structure ensembles, combining models of the same data type (e.g., multiple 2D-based models);
- Cross-structure ensembles, combining models across different data representations (e.g., 1D, 2D, and 3D).
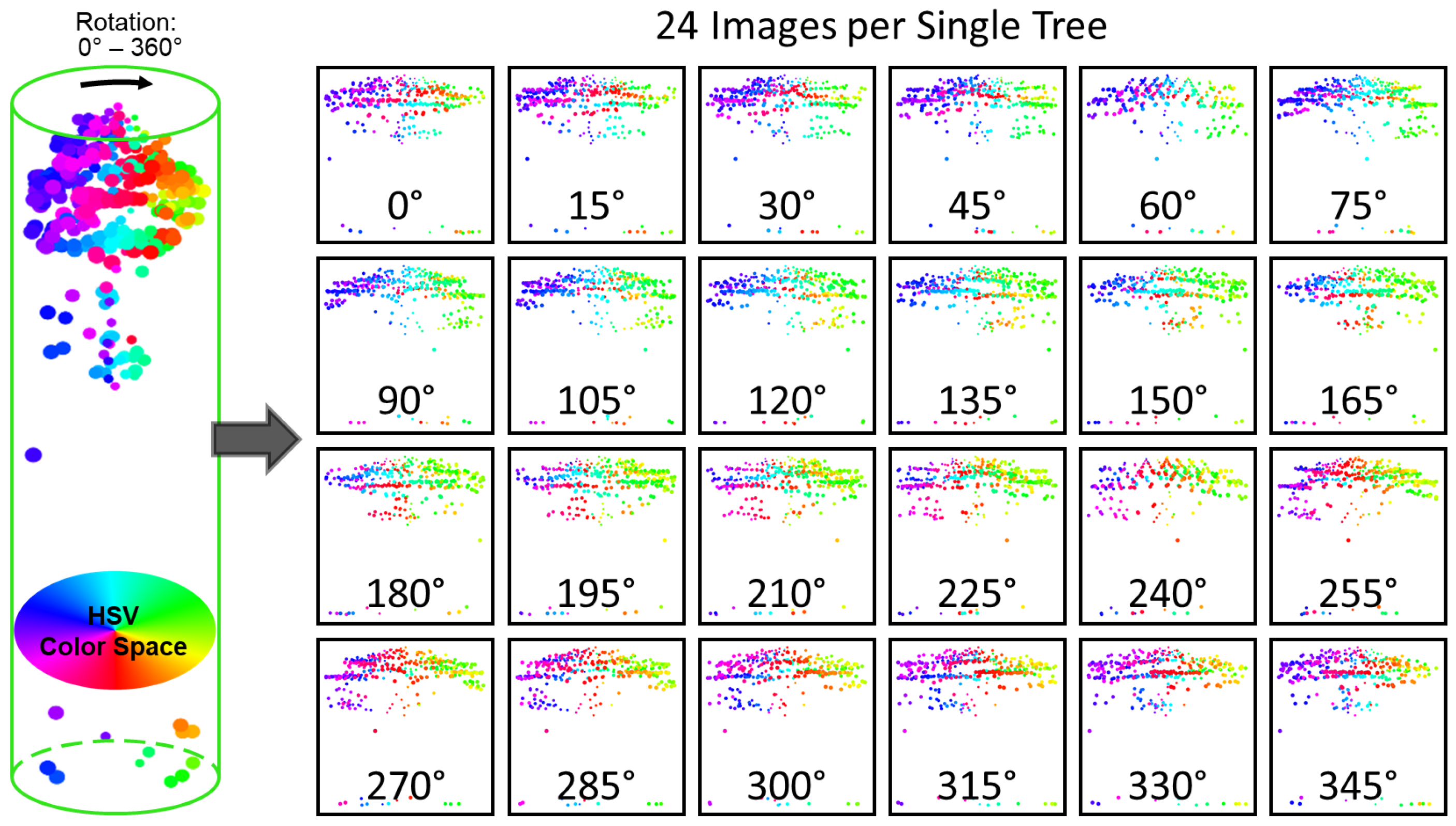
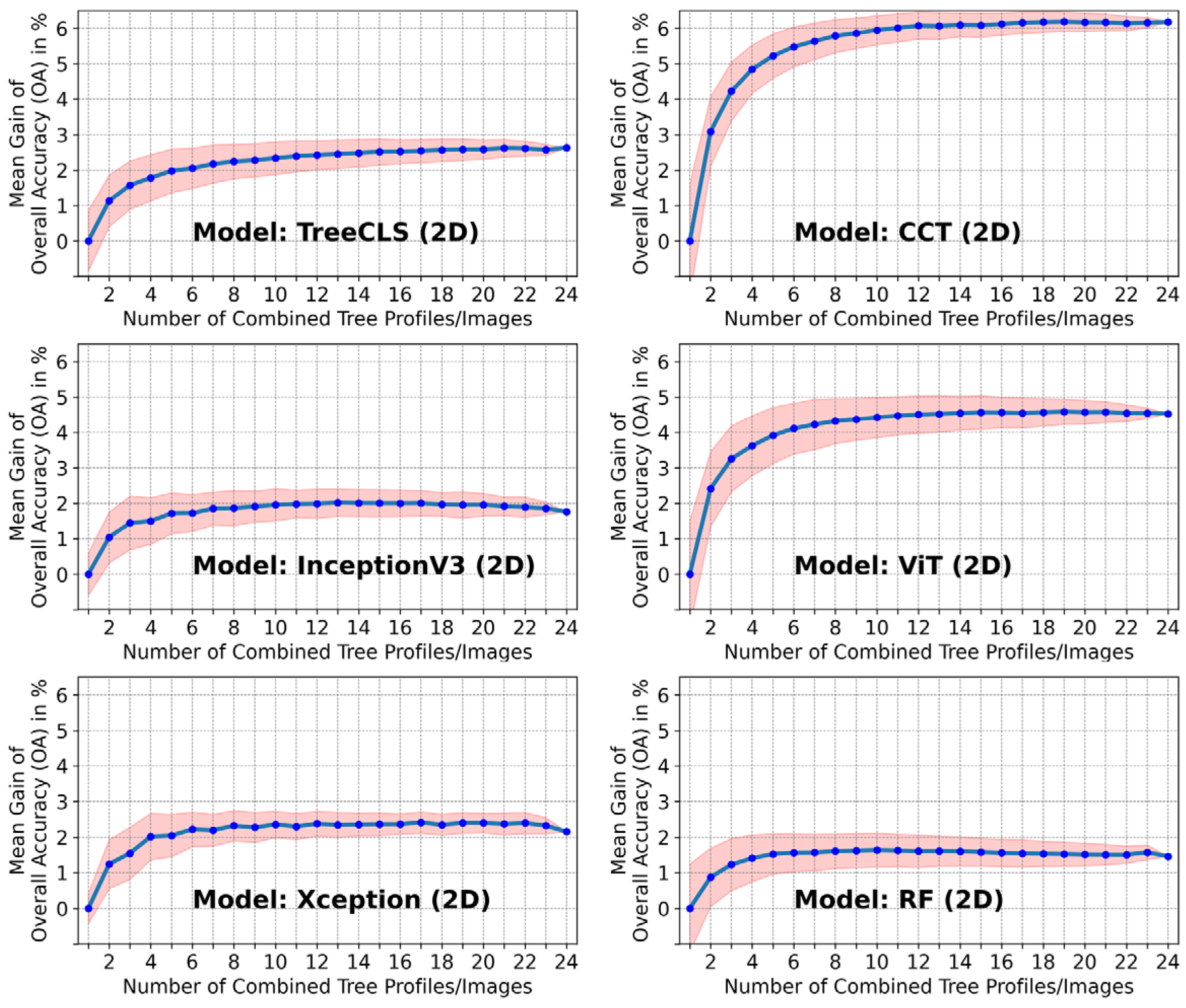
5.3. Accuracy Improvement Using Multi-View Profiles (MVPs) of a Single Tree
- The largest gains occurred when combining the first few additional views. On average, adding just one more profile led to a 1–3% accuracy increase.
- After approximately eight views, the improvement curve began to flatten, suggesting that the most relevant structural information is captured within the first 8 orientations.
- Transformer-based DL models (e.g., ViT, CCT) exhibited the highest relative gains (4–6.5%), supporting the hypothesis that they benefit more from larger and more diverse input distributions, possibly compensating for limited training data.
- CNN-based models (e.g., Xception, TreeCNN) and traditional ML classifiers (e.g., RF, SVC) also benefited, though with smaller relative gains (1.5–3.5%).
5.4. Implications for Application and Method Selection
5.5. Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Airborne Laser Scanning |
| CCT | Compact Convolutional Transformer |
| CHM | Canopy Height Model |
| CIR | Color-Infrared |
| CNN | Convolutional Neural Network |
| CP | Colored Profile |
| CPU | Central Processing Unit |
| DBH | Diameter at Breast Height |
| DL | Deep Learning |
| DNN | Deep Neural Network |
| FC | Fully Connected |
| FFN | Feed-Forward Network |
| GPS | Global Positioning System |
| GPU | Graphics Processing Unit |
| Hmean | mean Height |
| Hp | Height percentiles |
| HSV | Hue–Saturation–Value |
| ID | Identifier |
| ITD | Individual Tree Detection |
| k-NN | k-Nearest Neighbors |
| LDA | Linear Discriminant Analysis |
| LiDAR | Light Detection And Ranging |
| LIME | Local Interpretable Model-agnostic Explanations |
| LR | Logistic Regression |
| ML | Machine Learning |
| MLPC | Multilayer Perceptron Classifier |
| MVP | Multi-View Profiles |
| NB | Naive Bayes Classifier |
| NC | Nearest Centroid |
| nDSM | normalized Digital Surface Model |
| NIR | Near-Infrared |
| NLP | Natural Language Processing |
| Nz | Normal vector component in the z-direction |
| OA | Overall Accuracy |
| PCA | Principal Component Analysis |
| PCT | Point Cloud Transformer |
| ReLU | Rectified Linear Unit |
| RF | Random Forest |
| RGB | Red–Green–Blue |
| RMSProp | Root Mean Square Propagation |
| SGD | Stochastic Gradient Descent |
| SVC | Support Vector Classifier |
| SVM | Support Vector Machines |
| SwinT | Swin Transformer |
| TAS | Tree Angle Statistic |
| ViT | Vision Transformer |
| VPD | Vertical Point Distribution |
Appendix A


References
- Leuschner, C.; Homeier, J. Global Forest Biodiversity: Current State, Trends, and Threats. In Progress in Botany; Lüttge, U., Cánovas, F.M., Risueño, M.-C., Leuschner, C., Pretzsch, H., Eds.; Springer International Publishing: Cham, Switzerland, 2023; Volume 83, pp. 125–159. ISBN 978-3-031-12782-3. [Google Scholar]
- Merritt, M.; Maldaner, M.E.; de Almeida, A.M.R. What Are Biodiversity Hotspots? Front. Young Minds 2019, 7, 29. [Google Scholar] [CrossRef]
- Hyde, P.; Dubayah, R.; Peterson, B.; Blair, J.B.; Hofton, M.; Hunsaker, C.; Knox, R.; Walker, W. Mapping Forest Structure for Wildlife Habitat Analysis Using Waveform Lidar: Validation of Montane Ecosystems. Remote Sens. Environ. 2005, 96, 427–437. [Google Scholar] [CrossRef]
- Li, J.; Hu, B.; Noland, T.L. Classification of Tree Species Based on Structural Features Derived from High Density LiDAR Data. Agric. For. Meteorol. 2013, 171–172, 104–114. [Google Scholar] [CrossRef]
- Luo, S.; Wang, C.; Xi, X.; Nie, S.; Fan, X.; Chen, H.; Ma, D.; Liu, J.; Zou, J.; Lin, Y.; et al. Estimating Forest Aboveground Biomass Using Small-Footprint Full-Waveform Airborne LiDAR Data. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101922. [Google Scholar] [CrossRef]
- Yao, W.; Krzystek, P.; Heurich, M. Tree Species Classification and Estimation of Stem Volume and DBH Based on Single Tree Extraction by Exploiting Airborne Full-Waveform LiDAR Data. Remote Sens. Environ. 2012, 123, 368–380. [Google Scholar] [CrossRef]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A Segmentation-Based Method to Retrieve Stem Volume Estimates from 3-D Tree Height Models Produced by Laser Scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Leutner, B.F.; Reineking, B.; Müller, J.; Bachmann, M.; Beierkuhnlein, C.; Dech, S.; Wegmann, M. Modelling Forest α-Diversity and Floristic Composition—On the Added Value of LiDAR plus Hyperspectral Remote Sensing. Remote Sens. 2012, 4, 2818–2845. [Google Scholar] [CrossRef]
- Amiri, N.; Yao, W.; Heurich, M.; Krzystek, P.; Skidmore, A.K. Estimation of Regeneration Coverage in a Temperate Forest by 3D Segmentation Using Airborne Laser Scanning Data. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 252–262. [Google Scholar] [CrossRef]
- Hasenauer, H. Ein Einzelbaumwachstumssimulator für Ungleichaltrige Fichten-, Kiefern- und Buchen-Fichtenmischbestände; Forstliche Schriftenreihe Universität für Bodenkultur: Vienna, Austria; Österr. Ges. für Waldökosystemforschung und Experimentelle Baumforschung: Vienna, Austria, 1994; ISBN 978-3-900865-07-8. [Google Scholar]
- Kraus, K. Photogrammetrie: Geometrische Informationen Aus Photographien Und Laserscanneraufnahmen; De Gruyter Lehrbuch, De Gruyter: Berlin, Germany, 2012; ISBN 978-3-11-090803-9. [Google Scholar]
- Næsset, E. Effects of Different Sensors, Flying Altitudes, and Pulse Repetition Frequencies on Forest Canopy Metrics and Biophysical Stand Properties Derived from Small-Footprint Airborne Laser Data. Remote Sens. Environ. 2009, 113, 148–159. [Google Scholar] [CrossRef]
- Budei, B.C.; St-Onge, B.; Hopkinson, C.; Audet, F.-A. Identifying the Genus or Species of Individual Trees Using a Three-Wavelength Airborne Lidar System. Remote Sens. Environ. 2018, 204, 632–647. [Google Scholar] [CrossRef]
- Lindberg, E.; Briese, C.; Doneus, M.; Hollaus, M.; Schroiff, A.; Pfeifer, N. Multi-Wavelength Airborne Laser Scanning for Characterization of Tree Species. In Proceedings of the SilviLaser 2015: 14th Conference on Lidar Applications for Assessing and Managing Forest Ecosystems, La Grande Motte, France, 28–30 September 2015. [Google Scholar]
- Yu, X.; Hyyppä, J.; Litkey, P.; Kaartinen, H.; Vastaranta, M.; Holopainen, M. Single-Sensor Solution to Tree Species Classification Using Multispectral Airborne Laser Scanning. Remote Sens. 2017, 9, 108. [Google Scholar] [CrossRef]
- Briechle, S.; Krzystek, P.; Vosselman, G. Classification of Tree Species and Standing Dead Trees by Fusing UAV-Based Lidar Data and Multispectral Imagery in the 3D Deep Neural Network Pointnet++. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, V-2–2020, 203–210. [Google Scholar] [CrossRef]
- Zhang, C.; Qiu, F. Mapping Individual Tree Species in an Urban Forest Using Airborne Lidar Data and Hyperspectral Imagery. Photogramm. Eng. Remote Sens. 2012, 78, 1079–1087. [Google Scholar] [CrossRef]
- Holmgren, J.; Persson, Å. Identifying Species of Individual Trees Using Airborne Laser Scanner. Remote Sens. Environ. 2004, 90, 415–423. [Google Scholar] [CrossRef]
- Vaughn, N.R.; Moskal, L.M.; Turnblom, E.C. Tree Species Detection Accuracies Using Discrete Point Lidar and Airborne Waveform Lidar. Remote Sens. 2012, 4, 377–403. [Google Scholar] [CrossRef]
- Yu, X.; Litkey, P.; Hyyppä, J.; Holopainen, M.; Vastaranta, M. Assessment of Low Density Full-Waveform Airborne Laser Scanning for Individual Tree Detection and Tree Species Classification. Forests 2014, 5, 1011–1031. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, J.; Wang, H.; Tan, T.; Cui, M.; Huang, Z.; Wang, P.; Zhang, L. Multi-Species Individual Tree Segmentation and Identification Based on Improved Mask R-CNN and UAV Imagery in Mixed Forests. Remote Sens. 2022, 14, 874. [Google Scholar] [CrossRef]
- Vermeer, M.; Hay, J.A.; Völgyes, D.; Koma, Z.; Breidenbach, J.; Fantin, D.S.M. Lidar-Based Norwegian Tree Species Detection Using Deep Learning. arXiv 2023, arXiv:2311.06066. [Google Scholar] [CrossRef]
- Zhao, K.; Popescu, S. Hierarchical Watershed Segmentation of Canopy Height Model for Multi-Scale Forest Inventory. In Proceedings of the ISPRS Workshop on Laser Scanning, Espoo, Finland, 12–14 September 2007; pp. 12–14. [Google Scholar]
- Wang, Y.; Weinacker, H.; Koch, B.; Sterenczak, K. Lidar Point Cloud Based Fully Automatic 3D Single Tree Modelling in Forest and Evaluations of the Procedure. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 37, 45–51. [Google Scholar]
- Harikumar, A.; Bovolo, F.; Bruzzone, L. A Local Projection-Based Approach to Individual Tree Detection and 3-D Crown Delineation in Multistoried Coniferous Forests Using High-Density Airborne LiDAR Data. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1168–1182. [Google Scholar] [CrossRef]
- Williams, J.; Schönlieb, C.-B.; Swinfield, T.; Lee, J.; Cai, X.; Qie, L.; Coomes, D.A. 3D Segmentation of Trees Through a Flexible Multiclass Graph Cut Algorithm. IEEE Trans. Geosci. Remote Sens. 2020, 58, 754–776. [Google Scholar] [CrossRef]
- Wack, R.; Schardt, M.; Lohr, U.; Barrucho, L.; Oliveira, T. Forest Inventory for Eucalyptus Plantations Based on Airborne Laserscanner Data. In Proceedings of the ISPRS Workshop 3-D Reconstruction from Airborne Laserscanner and InSAR Data, Dresden, Germany, 8–10 October 2003; pp. 40–46. [Google Scholar]
- Wack, R.; Stelzl, H. Assessment of Forest Stand Parameters from Laserscanner Data in Mixed Forests. Proc. For. 2005, 56–60. [Google Scholar]
- Persson, Å.; Holmgren, J.; Söderman, U. Detecting and Measuring Individual Trees Using an Airborne Laser Scanner. Photogramm. Eng. Remote Sens. 2002, 68, 925–932. [Google Scholar]
- Hamraz, H.; Jacobs, N.B.; Contreras, M.A.; Clark, C.H. Deep Learning for Conifer/Deciduous Classification of Airborne LiDAR 3D Point Clouds Representing Individual Trees. ISPRS J. Photogramm. Remote Sens. 2019, 158, 219–230. [Google Scholar] [CrossRef]
- Braga, J.R.G.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragão, L.E.O.C.; Velho, H.F.d.C.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef]
- Wang, Z.; Li, P.; Cui, Y.; Lei, S.; Kang, Z. Automatic Detection of Individual Trees in Forests Based on Airborne LiDAR Data with a Tree Region-Based Convolutional Neural Network (RCNN). Remote Sens. 2023, 15, 1024. [Google Scholar] [CrossRef]
- Zhao, H.; Morgenroth, J.; Pearse, G.; Schindler, J. A Systematic Review of Individual Tree Crown Detection and Delineation with Convolutional Neural Networks (CNN). Curr. For. Rep. 2023, 9, 149–170. [Google Scholar] [CrossRef]
- Reitberger, J.; Krzystek, P.; Stilla, U. Analysis of Full Waveform LIDAR Data for the Classification of Deciduous and Coniferous Trees. Int. J. Remote Sens. 2008, 29, 1407–1431. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, T.; Skidmore, A.K.; Heurich, M. Important LiDAR Metrics for Discriminating Forest Tree Species in Central Europe. ISPRS J. Photogramm. Remote Sens. 2018, 137, 163–174. [Google Scholar] [CrossRef]
- Höfle, B.; Hollaus, M.; Hagenauer, J. Urban Vegetation Detection Using Radiometrically Calibrated Small-Footprint Full-Waveform Airborne LiDAR Data. ISPRS J. Photogramm. Remote Sens. 2012, 67, 134–147. [Google Scholar] [CrossRef]
- Koenig, K.; Höfle, B. Full-Waveform Airborne Laser Scanning in Vegetation Studies—A Review of Point Cloud and Waveform Features for Tree Species Classification. Forests 2016, 7, 198. [Google Scholar] [CrossRef]
- Amiri, N.; Heurich, M.; Krzystek, P.; Skidmore, A.K. Feature Relevance Assessment of Multispectral Airborne Lidar Data for Tree Species Classification. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, 42, 31–34. [Google Scholar] [CrossRef]
- Axelsson, A.; Lindberg, E.; Olsson, H. Exploring Multispectral ALS Data for Tree Species Classification. Remote Sens. 2018, 10, 183. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. A Comprehensive but Efficient Framework of Proposing and Validating Feature Parameters from Airborne LiDAR Data for Tree Species Classification. Int. J. Appl. Earth Obs. Geoinf. 2016, 46, 45–55. [Google Scholar] [CrossRef]
- You, H.T.; Lei, P.; Li, M.S.; Ruan, F.Q. Forest Species Classification Based on Three-Dimensional Coordinate and Intensity Information of Airborne Lidar Data with Random Forest Method. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLII-3/W10, 117–123. [Google Scholar] [CrossRef]
- Cao, L.; Coops, N.C.; Innes, J.L.; Dai, J.; Ruan, H.; She, G. Tree Species Classification in Subtropical Forests Using Small-Footprint Full-Waveform LiDAR Data. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 39–51. [Google Scholar] [CrossRef]
- Duong, V. Processing and Application of ICESat Large Footprint Full Waveform Laser Range Data. Ph.D. Thesis, Delft University of Technology, Delft, The Netherlands, 2010. [Google Scholar]
- Heinzel, J.; Koch, B. Exploring Full-Waveform LiDAR Parameters for Tree Species Classification. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 152–160. [Google Scholar] [CrossRef]
- Mustafic, S.; Schardt, M. Deep Learning-Basierte Baumartenklassifizierung auf Basis von ALS-Daten; Dreiländertagung 2019, Photogrammetrie, Fernerkundung und Geoinformation, OVG–DGPF–SGPF; Publikationen der DGPF: Vienna, Austria, 2019; Volume 28, pp. 527–536. [Google Scholar]
- Mustafic, S.; Schardt, M. Deep-Learning-basierte Baumartenklassifizierung auf Basis von multitemporalen ALS-Daten. Agit. J. Angew. Geoinformatik 2019, 5, 329–337. [Google Scholar]
- Hell, M.; Brandmeier, M.; Briechle, S.; Krzystek, P. Classification of Tree Species and Standing Dead Trees with Lidar Point Clouds Using Two Deep Neural Networks: PointCNN and 3DmFV-Net. PFG–J. Photogramm. Remote Sens. Geoinf. Sci. 2022, 90, 103–121. [Google Scholar] [CrossRef]
- Ørka, H.O.; Næsset, E.; Bollandsås, O.M. Utilizing Airborne Laser Intensity for Tree Species Classification. In Proceedings of the ISPRS Workshop on Laser Scanning 2007 and SilviLaser 2007, Espoo, Finland, 12–14 September 2007. [Google Scholar]
- Briechle, S.; Krzystek, P.; Vosselman, G. Silvi-Net–A Dual-CNN Approach for Combined Classification of Tree Species and Standing Dead Trees from Remote Sensing Data. Int. J. Appl. Earth Obs. Geoinf. 2021, 98, 102292. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. PointNet++: Deep Hierarchical Feature Learning on Point Sets in a Metric Space. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Auckland, New Zealand, 2–6 December 2024; Curran Associates Inc.: New York, NY, USA, 2017; pp. 5105–5114. [Google Scholar]
- Li, Y.; Bu, R.; Sun, M.; Wu, W.; Di, X.; Chen, B. PointCNN: Convolution on X-Transformed Points. In Proceedings of the Neural Information Processing Systems, Montréal, QC, Canada, 2–8 December 2018. [Google Scholar]
- Briechle, S.; Krzystek, P.; Vosselman, G. Semantic Labeling of ALS Point Clouds for Tree Species Mapping Using the Deep Neural Network Pointnet++. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, XLII-2/W13, 951–955. [Google Scholar] [CrossRef]
- Korpela, I.; Ørka, H.O.; Maltamo, M.; Tokola, T.; Hyyppä, J. Tree Species Classification Using Airborne LiDAR–Effects of Stand and Tree Parameters, Downsizing of Training Set, Intensity Normalization, and Sensor Type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef]
- Yang, G.; Zhao, Y.; Li, B.; Ma, Y.; Li, R.; Jing, J.; Dian, Y. Tree Species Classification by Employing Multiple Features Acquired from Integrated Sensors. J. Sens. 2019, 2019, 3247946. [Google Scholar] [CrossRef]
- Ba, A.; Dufour, S.; Laslier, M.; Hubert-Moy, L. Riparian Trees Genera Identification Based on Leaf-on/Leaf-off Airborne Laser Scanner Data and Machine Learning Classifiers in Northern France. Int. J. Remote Sens. 2020, 41, 1645–1667. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the Utility of Airborne Hyperspectral and LiDAR Data for Species Distribution Mapping in the Coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- Nguyen, H.M.; Demir, B.; Dalponte, M. Weighted Support Vector Machines for Tree Species Classification Using Lidar Data. In Proceedings of the IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 6740–6743. [Google Scholar]
- Guo, M.-H.; Cai, J.; Liu, Z.-N.; Mu, T.-J.; Martin, R.R.; Hu, S. PCT: Point Cloud Transformer. Comput. Vis. Media 2020, 7, 187–199. [Google Scholar] [CrossRef]
- Hirschmugl, M. Derivation of Forest Parameters from UltracamD Data. Ph.D. Thesis, Graz University of Technology, Graz, Austria, 2008. [Google Scholar]
- Heurich, M.; Schneider, T.; Kennel, E. Laser Scanning for Identification of Forest Structures in the Bavarian Forest National Park; European Association of Remote Sensing Laboratories: Paris, France, 2003. [Google Scholar]
- Hans-Jörg, F. Methodische Ansätze Zur Erfassung von Waldbäumen Mittels Digitaler Luftbildauswertung; Georg-August Universität: Gottingen, Germany, 2004. [Google Scholar]
- Buslaev, A.; Iglovikov, V.I.; Khvedchenya, E.; Parinov, A.; Druzhinin, M.; Kalinin, A.A. Albumentations: Fast and Flexible Image Augmentations. Information 2020, 11, 125. [Google Scholar] [CrossRef]
- Mustafic, S.; Schardt, M. Methode Für Die Automatische Verifizierung Der Ergebnisse Der Einzelbaumdetektion, Baumartenklassifizierung Und Baumkronengrenzen Aus LiDAR-Daten. AGIT J. Angew. Geoinform. 2016, 2, 600–605. [Google Scholar]
- Eysn, L.; Hollaus, M.; Lindberg, E.; Berger, F.; Monnet, J.-M.; Dalponte, M.; Kobal, M.; Pellegrini, M.; Lingua, E.; Mongus, D.; et al. A Benchmark of Lidar-Based Single Tree Detection Methods Using Heterogeneous Forest Data from the Alpine Space. Forests 2015, 6, 1721–1747. [Google Scholar] [CrossRef]
- Dalponte, M.; Reyes, F.; Kandare, K.; Gianelle, D. Delineation of Individual Tree Crowns from ALS and Hyperspectral Data: A Comparison among Four Methods. Eur. J. Remote Sens. 2015, 48, 365–382. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A Survey of Remote Sensing-Based Aboveground Biomass Estimation Methods in Forest Ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- Ke, Y.; Quackenbush, L.J. A Review of Methods for Automatic Individual Tree-Crown Detection and Delineation from Passive Remote Sensing. Int. J. Remote Sens. 2011, 32, 4725–4747. [Google Scholar] [CrossRef]
- Dalponte, M.; Coomes, D.A. Tree-Centric Mapping of Forest Carbon Density from Airborne Laser Scanning and Hyperspectral Data. Methods Ecol. Evol. 2016, 7, 1236–1245. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.; Heyder, U.; Weinacker, H. Detection of Individual Tree Crowns in Airborne Lidar Data. Photogramm. Eng. Remote Sens. 2006, 72, 357–363. [Google Scholar] [CrossRef]
- Reitberger, J. 3D-Segmentierung von Einzelbäumen und Baumartenklassifikation Aus Daten Flugzeuggetragener Full Waveform Laserscanner. Ph.D. Thesis, Technische Universität München, Munich, Germany, 2010. [Google Scholar]
- Dersch, S.; Heurich, M.; Krueger, N.; Krzystek, P. Combining Graph-Cut Clustering with Object-Based Stem Detection for Tree Segmentation in Highly Dense Airborne Lidar Point Clouds. ISPRS J. Photogramm. Remote Sens. 2021, 172, 207–222. [Google Scholar] [CrossRef]
- Kamińska, A.; Lisiewicz, M.; Stereńczak, K. Single Tree Classification Using Multi-Temporal ALS Data and CIR Imagery in Mixed Old-Growth Forest in Poland. Remote Sens. 2021, 13, 5101. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 1984. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Representations by Back-Propagating Errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Cover, T.; Hart, P. Nearest Neighbor Pattern Classification. IEEE Trans. Inf. Theory 1967, 13, 21–27. [Google Scholar] [CrossRef]
- Walker, S.H.; Duncan, D.B. Estimation of the Probability of an Event as a Function of Several Independent Variables. Biometrika 1967, 54, 167. [Google Scholar] [CrossRef]
- Fisher, R.A. The Use of Multiple Measurements in Taxonomic Problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Rocchio, J.J. Relevance Feedback in Information Retrieval. In The Smart Retrieval System-Experiments in Automatic Document Processing; Prentice Hall: Hoboken, NJ, USA, 1971. [Google Scholar]
- Friedman, N.; Geiger, D.; Goldszmidt, M. Bayesian Network Classifiers. Mach. Learn. 1997, 29, 131–163. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Tan, M.; Le, Q. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the 38th International Conference on Machine Learning, Vienna, Austria, 18–24 July 2021; Volume 139, pp. 10096–10106. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Hassani, A.; Walton, S.; Shah, N.; Abuduweili, A.; Li, J.; Shi, H. Escaping the Big Data Paradigm with Compact Transformers. arXiv 2021, arXiv:2104.05704. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 9992–10002. [Google Scholar]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation Importance: A Corrected Feature Importance Measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef]
- Azmat, M.; Alessio, A.M. Feature Importance Estimation Using Gradient Based Method for Multimodal Fused Neural Networks. In Proceedings of the 2022 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Milano, Italy, 5–12 November 2022; pp. 1–5. [Google Scholar]
- Iranzad, R.; Liu, X. A Review of Random Forest-Based Feature Selection Methods for Data Science Education and Applications. Int. J. Data Sci. Anal. 2024, 1–15. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why Should I Trust You?”: Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Martinez, A.M.; Kak, A.C. PCA versus LDA. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 228–233. [Google Scholar] [CrossRef]





| ML Algorithm | Modified Parameter | Value/Name |
|---|---|---|
| Random Forest (RF) [73,74] | Number of trees: | 500 |
| Split criterion: | Entropy | |
| Support Vector Classifier (SVC) [75] | Regularization strength (C): | 100 |
| Gamma: | Auto (1/n_features) | |
| Multilayer Perceptron Classifier (MLPC) [76] | Hidden layers: | 2 (200 neurons each) |
| Optimizer: | Lbfgs | |
| Learning rate: | Invscaling | |
| Max. training epochs: | 1000 | |
| k-Nearest Neighbors (k-NN) [77] | Number of neighbors (k): | 10 |
| Logistic Regression (LR) [78] | Extension: | Multinomial |
| Regularization (C): | 0.2 | |
| Max. training epochs: | 2000 | |
| Linear Discriminant Analysis (LDA) [79] | Optimizer: | Lsqt |
| Shrinkage: | Auto | |
| Nearest Centroid/Min. Distance (NC) [80] | (Default parameters) | --- |
| Naive Bayes Classifier (NB) [81] | (Default parameters) | --- |
| Deep Neural Network (DNN) | Optimizer | Learning Rate | Weight Decay |
|---|---|---|---|
| Tree CNN_2D (see Section 3.2.3) | RMSprop | 1 × 10−4 | 1 × 10−5 |
| InceptionV3 [82] | RMSprop | 1 × 10−4 | 1 × 10−5 |
| Xception [83] | RMSprop | 1 × 10−4 | 1 × 10−5 |
| EfficientNet [84] | RMSprop | 1 × 10−4 | 1 × 10−5 |
| Vision Transformer (ViT) [85] | AdamW | 1 × 10−3 | 1 × 10−4 |
| Compact Convolutional Transformer (CCT) [86] | AdamW | 1 × 10−3 | 1 × 10−4 |
| Swin Transformer (SwinT) [87] | AdamW with label smoothing of 0.1 | 1 × 10−3 | 1 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustafić, S.; Schardt, M.; Perko, R. Tree Type Classification from ALS Data: A Comparative Analysis of 1D, 2D, and 3D Representations Using ML and DL Models. Remote Sens. 2025, 17, 2847. https://doi.org/10.3390/rs17162847
Mustafić S, Schardt M, Perko R. Tree Type Classification from ALS Data: A Comparative Analysis of 1D, 2D, and 3D Representations Using ML and DL Models. Remote Sensing. 2025; 17(16):2847. https://doi.org/10.3390/rs17162847
Chicago/Turabian StyleMustafić, Sead, Mathias Schardt, and Roland Perko. 2025. "Tree Type Classification from ALS Data: A Comparative Analysis of 1D, 2D, and 3D Representations Using ML and DL Models" Remote Sensing 17, no. 16: 2847. https://doi.org/10.3390/rs17162847
APA StyleMustafić, S., Schardt, M., & Perko, R. (2025). Tree Type Classification from ALS Data: A Comparative Analysis of 1D, 2D, and 3D Representations Using ML and DL Models. Remote Sensing, 17(16), 2847. https://doi.org/10.3390/rs17162847








