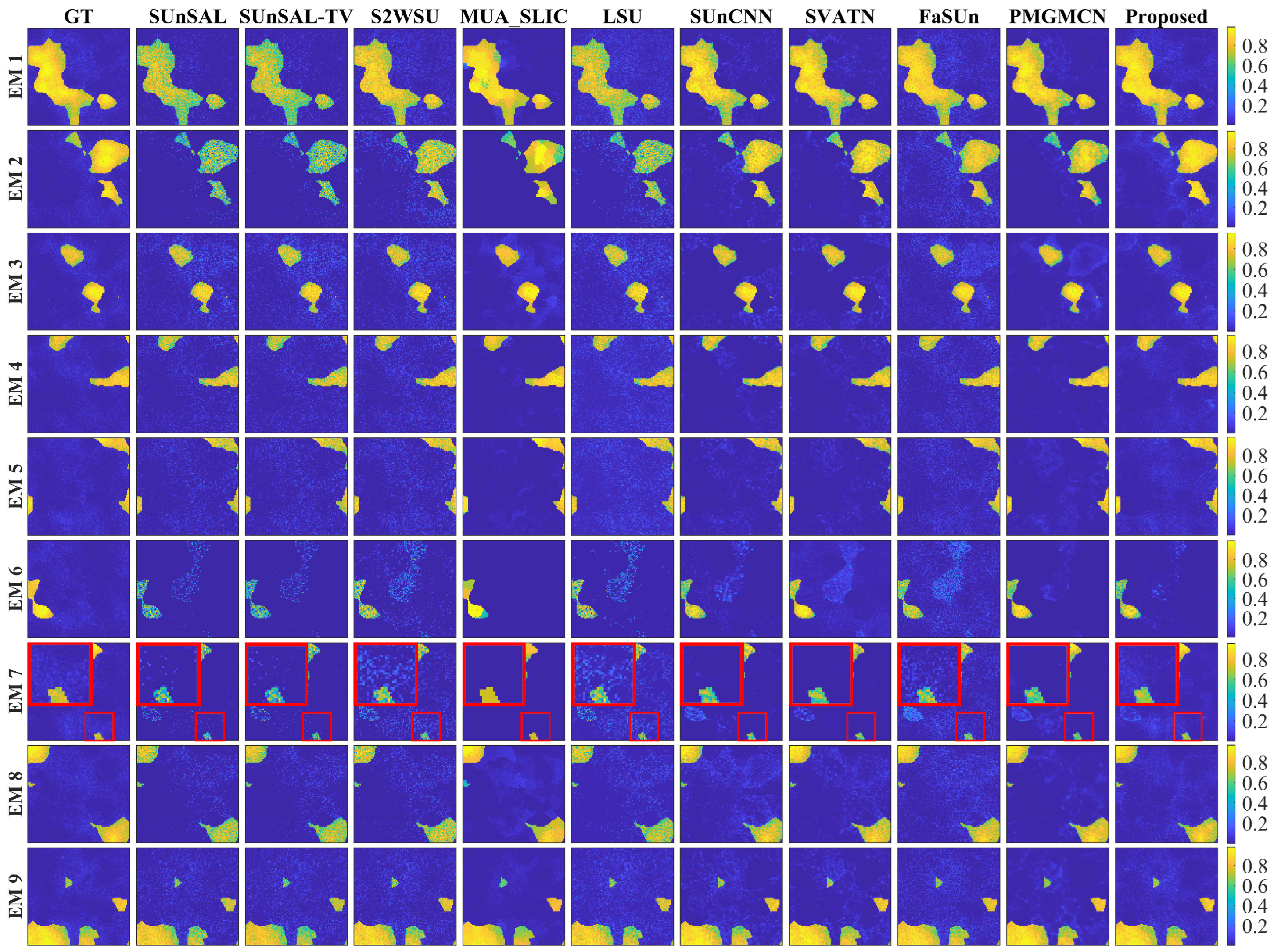
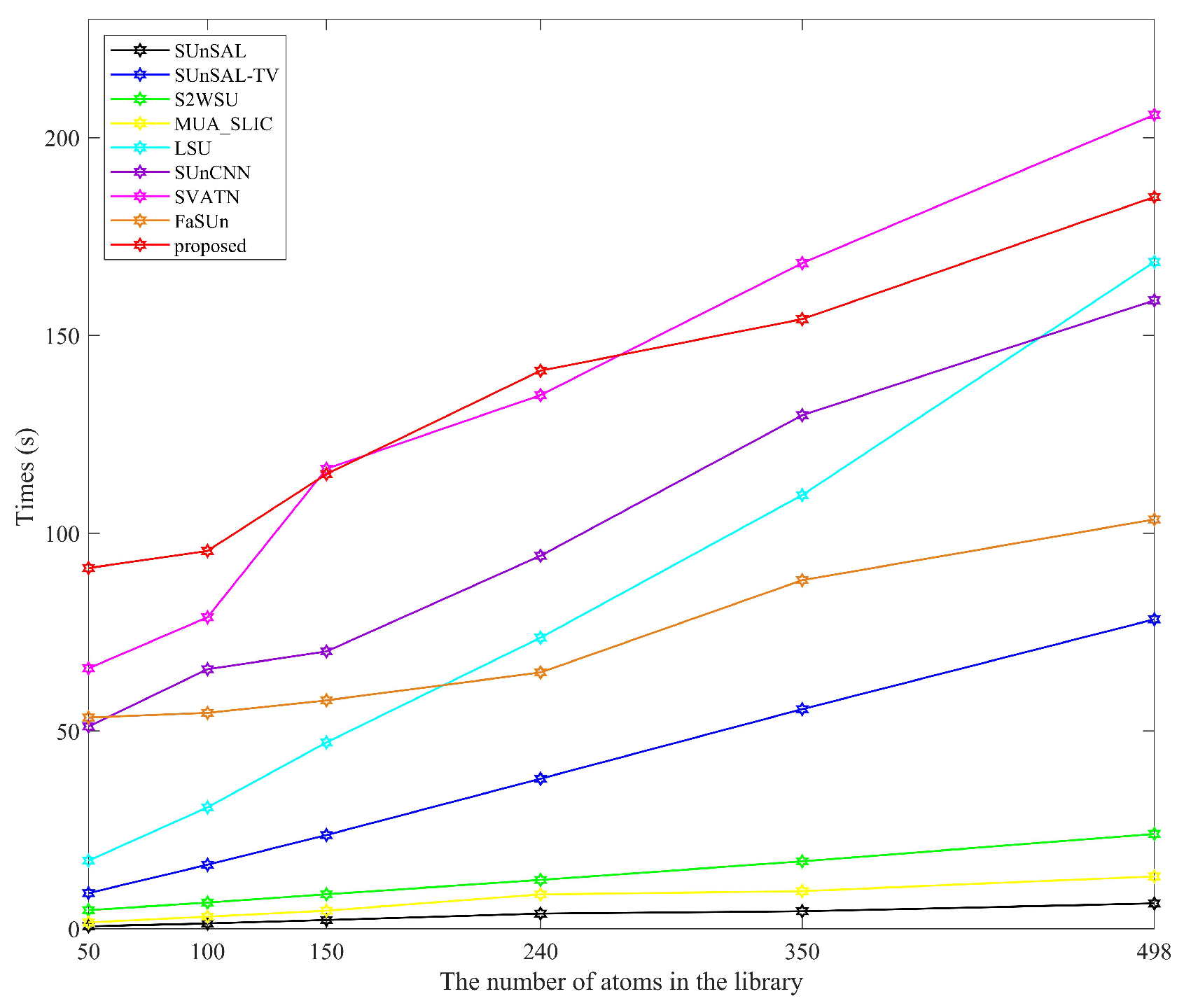
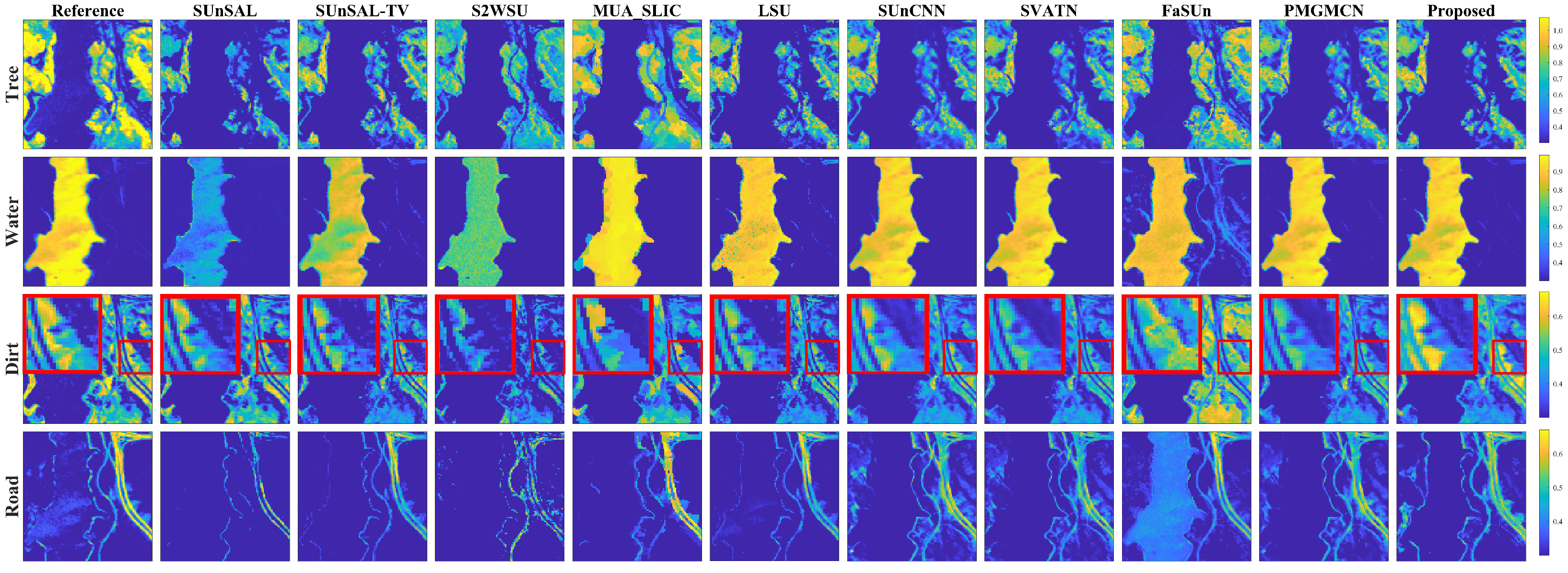
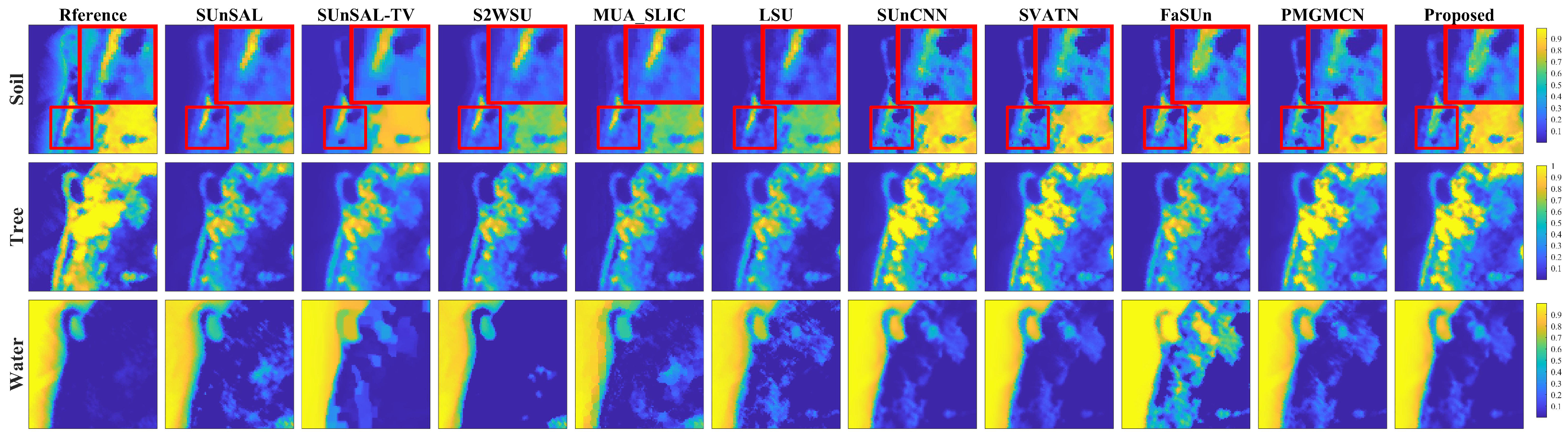
1. Introduction
With the rapid development of hyperspectral imaging technology, modern hyperspectral sensors are capable of capturing increasingly detailed spectral information [
1]. However, the mixed-pixel problem severely hinders the performance of HSIs in tasks like precise classification [
2], anomaly detection [
3], and other related applications. Consequently, hyperspectral unmixing (HU) has emerged as a crucial technique for analyzing hyperspectral data. The main goal of HU is to decompose each mixed pixel into a set of pure spectral signatures (i.e., endmembers) and their corresponding proportions (i.e., abundances) [
4].
Overall, spectral unmixing models are generally categorized into two types based on pixel mixing mechanisms: the linear spectral mixture model (LMM) [
5] and the nonlinear spectral mixture model (NLMM) [
6]. An LMM models each pixel as a linear combination of pure endmember spectra, while an NLMM considers nonlinear effects, like photon scattering, offering better performance in complex scenes. Despite this, the LMM remains widely used owing to its simplicity and efficiency.
Currently, various unmixing models have been developed based on the NLMM. The Hapke [
6] model, derived from radiative transfer theory, effectively addresses intimate mixtures but is limited by the need for accurate physical parameters. To reduce this dependency, bilinear mixture models (BMMs) [
7] introduce bilinear terms to capture second-order scattering effects. Building on this, GCBAE-FCLS [
8] mitigates collinearity between real and virtual endmembers, improving the linear unmixing performance. With deep learning’s success in modeling nonlinear patterns, combined NLMM–deep learning approaches have gained attention. For instance, a deep multitask BMM [
9] puts forward an unsupervised framework based on a hierarchical BMM architecture, effectively addressing nonlinear mixing in complex scenarios, while HapkeCNN [
10] embeds nonlinear components into an LMM, enhancing the adaptability and expressiveness. Overall, the above NLMMs significantly enhance the unmixing performance and interpretability by taking nonlinear factors into account.
Up to now, LMMs can be broadly categorized into geometric- [
11,
12], statistical- [
13,
14], sparse regression (SR)- [
15,
16,
17], tensor-decomposition- [
18], and neural network (NN)-based [
19,
20,
21] methods. Specifically, geometric methods exploit the geometric properties of HSIs in the feature space and can be further classified into maximum simplex and minimum simplex approaches. Representative algorithms encompass N-FINDR [
11], vertex component analysis [
12], minimum volume simplex analysis [
22], and simplex identification via a split-augmented Lagrangian [
23]. Notably, maximum simplex methods rely on the pure-pixel assumption, whereas minimum simplex methods estimate the vertices of the simplex directly, rendering them more suitable for analyzing highly mixed hyperspectral data.
The statistical methods are grounded in mathematical statistical theory and describe the unmixing problem as a statistical model. Among these, non-negative matrix factorization (NMF) [
13] is one of the most widely adopted models, as it enables the simultaneous estimation of endmembers and abundances, making it particularly effective at handling highly mixed pixels. However, the objective function of NMF exhibits non-convexity, resulting in an extremely large solution space where the algorithm is highly susceptible to being trapped in local minima. Consequently, standard NMF cannot be directly employed for the unmixing problem. To address this, various regularizations have been introduced, such as the
norm for sparsity [
13], total variation (TV) for spatial smoothness [
14], and manifold regularization for preserving data geometry [
24]. Yet, the design of such regularizations often relies on substantial prior knowledge and extensive manual feature engineering. Moreover, traditional NMF methods flatten HSI data into matrices, resulting in the loss of spatial structure information. To mitigate this, tensor-based methods [
18] have been proposed to preserve spatial–spectral structures, while selecting an appropriate tensor rank remains a key challenge affecting both the accuracy and convergence.
Over the past few years, inspired by compressed sensing theory [
25] and SR theory, SR-based methods have attracted substantial attention in HU. These methods leverage pre-existing spectral libraries and regard the unmixing task as the selection of an optimal subset of endmembers from a large-scale dictionary. Compared with traditional methods, SR approaches do not require endmember extraction or pre-estimation of the number of endmembers, demonstrating strong practicality. The SR-based unmixing framework was initially introduced in [
15]. It utilized the
regularization together with variable splitting and the augmented Lagrangian method to address the sparse unmixing (SU) problem. Building on this foundation, a weighted
regularization was proposed in [
26] to further enhance the sparsity of abundances estimation. Considering that materials in natural scenes typically exhibit smoothly varying spatial distributions with abrupt changes only at boundaries, abundance maps often possess piecewise smooth characteristics. To exploit this prior, TV regularization [
27] was introduced to enforce spatial smoothness. However, since TV regularization operates solely in the spatial domain, it leads to a decoupling between spatial and spectral domains, increasing the model complexity and computational cost. To address this, the multiscale unmixing algorithm (MUA) [
28] segments the HSI into superpixels via simple linear iterative clustering (SLIC) [
29], performs coarse unmixing, and uses the initial abundances to guide fine-scale unmixing, effectively ensuring piecewise smoothness while improving the efficiency. Additionally, manifold-based methods leveraging graph Laplacian regularization have been proposed to better model local spatial correlations and preserve smoothness. For instance, SBGLSU [
30] applies a graph Laplacian on superpixels to capture spatial similarity, while [
31] incorporates spatial prior weights to exploit spectral similarity within local subspaces, enhancing unmixing accuracy.
The aforementioned SU methods have improved the unmixing accuracy by integrating various regularizations. However, they largely depend on large spectral libraries, which increase the computational costs and degrade performance due to the high coherence between spectral library endmembers. To mitigate these issues, various approaches have been put forward to reduce the spectral library coherence and redundancy, thereby improving the stability and robustness of the unmixing. For example, [
16] proposed a hierarchical framework that adaptively prunes the library using the row-wise sparsity of the abundances matrix. FaSUn [
32] dynamically adjusts the atom activity based on the pixel features and jointly optimizes the spectral library and contribution matrix; however, the latter lacks clear physical meaning. Building on this, [
17] introduced a novel two-stage unmixing strategy. The method first generates a representative reduced endmember set during a coarse unmixing stage. Subsequently, this set is employed in the fine-scale stage to estimate abundances. This staged processing cuts the computational complexity while improving the accuracy. Although the aforementioned methods improve the unmixing efficiency and accuracy, they often neglect modeling small target regions during spectral library pruning, potentially removing them and limiting further performance gains.
NNs have shown great promise in HSI processing due to their powerful nonlinear modeling and feature representation capabilities. Because the hidden layers of the autoencoder (AE) [
19] can effectively extract high-level features from observed data, and abundance estimation in HU essentially aims to find a low-dimensional representation of the HSI, the AE is especially suitable for HU. The first AE-based HU algorithm was introduced in [
33], which combined the AE framework with an
constraint to achieve sparse and discriminative abundance estimation. Building on this, EndNet [
34] enhanced the AE with additional layers and tailored loss functions to improve the sparsity and accuracy. TANet [
35] further improved the representational capacity and physical interpretability by designing symmetric AE structures. Despite their strong performance in abundance estimation, these methods predominantly employ a pixel-wise training paradigm, focusing solely on spectral information while neglecting the critical spatial structural information inherent in HSIs. To address this limitation, [
36] integrated convolutional neural networks (CNNs) into the AE framework to model the local spatial features of HSIs. Based on this, a plethora of unmixing methods combining a CNN with an AE have been proposed. For instance, MiSiCNet [
37] introduced residual connections to stabilize training and deepen feature learning, and DIFCNN [
38] incorporated endmember geometry, spectral variability, and noise modeling to enhance robustness and accuracy.
Recently, Transformer [
20,
39] architectures have received increasing attention in HU due to their self-attention mechanisms, which enable flexible interaction across feature positions and achieve global feature learning. Deep-Trans [
20] first demonstrated the feasibility of applying Transformers to model complex mixing relationships in HSIs. ULA-Net [
40] further partitioned HSIs into patches and applied local attention modules to capture neighborhood dependencies, enabling a more discriminative local feature extraction. UST-Net [
41] leverages the hierarchical sliding window mechanism of the Swin Transformer to effectively capture deeper spatial–spectral contextual features, enhancing the unmixing precision. Moreover, CNN–Transformer [
39] combined frameworks have been proposed to jointly exploit the local feature encoding of CNNs and the global modeling capacity of Transformers. Although Transformers excel at capturing long-range spatial–spectral dependencies, their self-attention mechanism has quadratic computational complexity, resulting in substantial computational and memory overhead when processing high-dimensional spectral data. To address this issue, Mamba [
42] was proposed. Based on state space models (SSMs), Mamba employs efficient linear recurrences along temporal or spatial dimensions to model sequential dependencies, reducing the computational complexity to a linear level. This makes Mamba particularly suitable for long sequence or large image tasks, and it has demonstrated strong performance in HSI classification [
43], HU, and related fields. UNMamba [
44] first introduced Mamba into the HU domain, leveraging its strength in modeling long-range dependencies to capture complex spatial–spectral features in HSIs. The introduction of UNMamba not only validates the feasibility of applying Mamba to HU but also provides new research directions for integrating Mamba with spectral mixture models and spatial structure modeling methods.
Despite the notable progress of NN-based HU methods, they typically require manual determination of the number of endmembers, and the extracted spectral signatures are often mathematical approximations rather than true reflectance values. To address this, SUnCNN [
21] integrates a CNN with SR to effectively capture local spatial structures. It reconstructs an HSI by combining a prior spectral library with estimated abundance maps, proposing the first sparse unmixing network model. Building on this, various combined SU-CNN methods have been proposed [
45]. However, CNNs’ limited receptive fields constrain their capacity to model long-range spatial dependencies. To alleviate this limitation, GACAE [
46] incorporates superpixel segmentation and graph attention networks (GATs) to enhance global spatial modeling, thereby boosting the unmixing accuracy. Yet, it has shortcomings in capturing fine-grained local details compared with a CNN. Furthermore, PMGMCN [
47] adopts a dual-branch architecture to separately extract global and local spatial features. This collaborative strategy significantly improves the accuracy and performs well across multiple benchmark datasets. However, the number of hops in the multi-hop graph used in the model must be determined experimentally, and multi-level, multi-scale convolutions, while enhancing local features, also lead to a significant computational overhead.
1.1. Motivation
The aforementioned SR–NN collaborative unmixing methods integrate prior endmember knowledge from SR with the nonlinear feature learning capabilities of an NN, forming a complementary framework that excels in spatial–spectral feature modeling. However, these approaches still face numerous challenges and limitations in practical applications. From the SR perspective, collaborative unmixing relies on large-scale spectral libraries that offer rich endmember candidates but also incur substantial computational and memory burdens. Furthermore, the high coherence of spectral libraries also compromises the unmixing accuracy. From the standpoint of NN feature modeling, these methods typically employ multi-layer convolutional structures with various-scale kernels [
21], graph attention mechanisms [
46], or Transformer architectures [
48] to extract deep features across spatial and spectral dimensions. Although these techniques improve feature representation, each has notable limitations. Multi-layer CNNs are constrained by fixed receptive fields, limiting the long-range dependency modeling and introducing redundant computation. A GAT models complex spatial structures but incurs heavy memory and computation costs due to adjacency matrix construction and attention calculations. Transformers offer strong global modeling but suffer from quadratic complexity in processing long sequences, limiting their efficiency in large-scale HSIs. Therefore, within the context of collaborative unmixing frameworks, effectively balancing the comprehensive modeling of key information—including global spatial structures, local spatial details, and spectral features—while controlling memory usage and computational complexity has become a critical challenge.
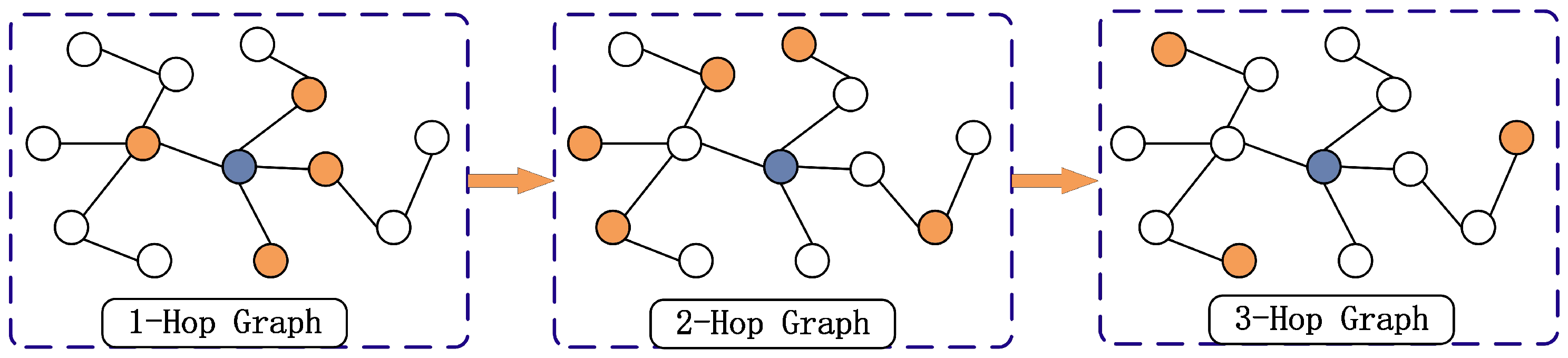
To address the aforementioned challenges, this paper proposes a dual-branch sparse unmixing network named DGMNet. Built upon the core principles of a low parameter count and minimal memory footprint, the model systematically balances the unmixing performance and computational resources by efficiently modeling and fusing spatial–spectral features and reducing the spectral library redundancy. DGMNet consists of two synergistic branches. The first branch employs an adaptive hop-neighborhood-aware GCN (AHNAGC) to capture global spatial structures. Superpixel segmentation is first applied to reduce the redundancy, followed by a local spatial correlation graph optimization (LSCGO) mechanism that dynamically refines the adjacency matrix using local spectral variations. An adaptive hop graph structure (AHGS) then constructs a lightweight multi-hop graph [
49] to model global spatial dependencies. The second branch introduces the neighborhood spatial offset Mamba (NSOM) module, leveraging Mamba’s linear complexity advantage. It partitions the HSI into patches, then integrates a multi-scale dynamic offset mechanism (MSDOM) with bidirectional offset learning Mamba (BOL-Mamba) to capture fine-grained local spatial structures. To integrate spatial–spectral features extracted from both branches, a Mamba-enhanced dual-stream feature fusion (MEDFF) module is designed to simultaneously fuse global and local spatial features while introducing spectral attention mechanisms to strengthen the spectral feature representation. Residual connections are employed to alleviate the feature degradation and information loss, thereby enhancing the overall stability and robustness of the feature representation. Finally, DGMNet pioneers the integration of sparse unmixing networks with spectral library pruning. To prevent the mispruning of small targets, a new strategy is proposed to mitigate spectral coherence and reduce the redundancy. Furthermore, to evaluate the applicability of HU methods in complex natural scenes, experiments were conducted on a real dataset acquired by the GF-5 satellite over the Shahu region. The results based on this dataset not only validate the superior performance of the proposed algorithm under challenging environments but also provide fresh perspectives for research in the HU domain.
1.2. Novelty and Contribution
The main contributions of this paper are summarized as follows:
This paper proposes a hyperspectral unmixing dual-branch network integrating an adaptive hop-aware GCN and neighborhood offset Mamba that is termed DGMNet. It integrates AHNAGC, NSOM, MEDFF, and a new spectral-library-pruning strategy, greatly reducing the parameter count and computational redundancy while fully considering the spatial–spectral feature extraction.
An AHNAGC module was designed not only to optimize the adjacency graph by capturing spectral variations within spatial neighborhoods, thereby enhancing the model’s focus on spectrally similar regions, but also to integrate an adaptive hop graph structure that dynamically selects and weights multi-hop features, reducing the manual feature design and improving the generalization.
A NSOM module was developed to employ bidirectional Mamba to capture sequence dependencies within each HSI block and incorporate a multi-scale offset mechanism to alleviate Mamba’s limitations in neighborhood feature extraction for non-sequential tasks, thereby enhancing the perception of local structures at different scales. These two components work synergistically to jointly model local spatial information from both the sequence and spatial neighborhood perspectives.
The MEDFF module is devised not only fuses the global and local spatial features from the dual branches but also incorporates a bidirectional Mamba (D-Mamba) to implement a spectral attention mechanism.
The spectral-library-pruning strategy with a new pruning rule was innovatively integrated into the SU network models, which simultaneously considers atom activity and small target protection. In addition, the ESS-Loss was carefully designed by combining ETV and sparsity constraint, which effectively utilizes prior knowledge to improve the unmixing performance.
The remainder of this paper is organized as follows. The related work is described in
Section 2. The proposed DGMNet model and its key module designs are detailed in
Section 3. Experimental results and performance evaluations on simulated and real datasets are presented in
Section 4. A discussion on the model’s key mechanisms and potential improvements is given in
Section 5. Finally, conclusions and future research directions are summarized in
Section 6.
3. Proposed Approach
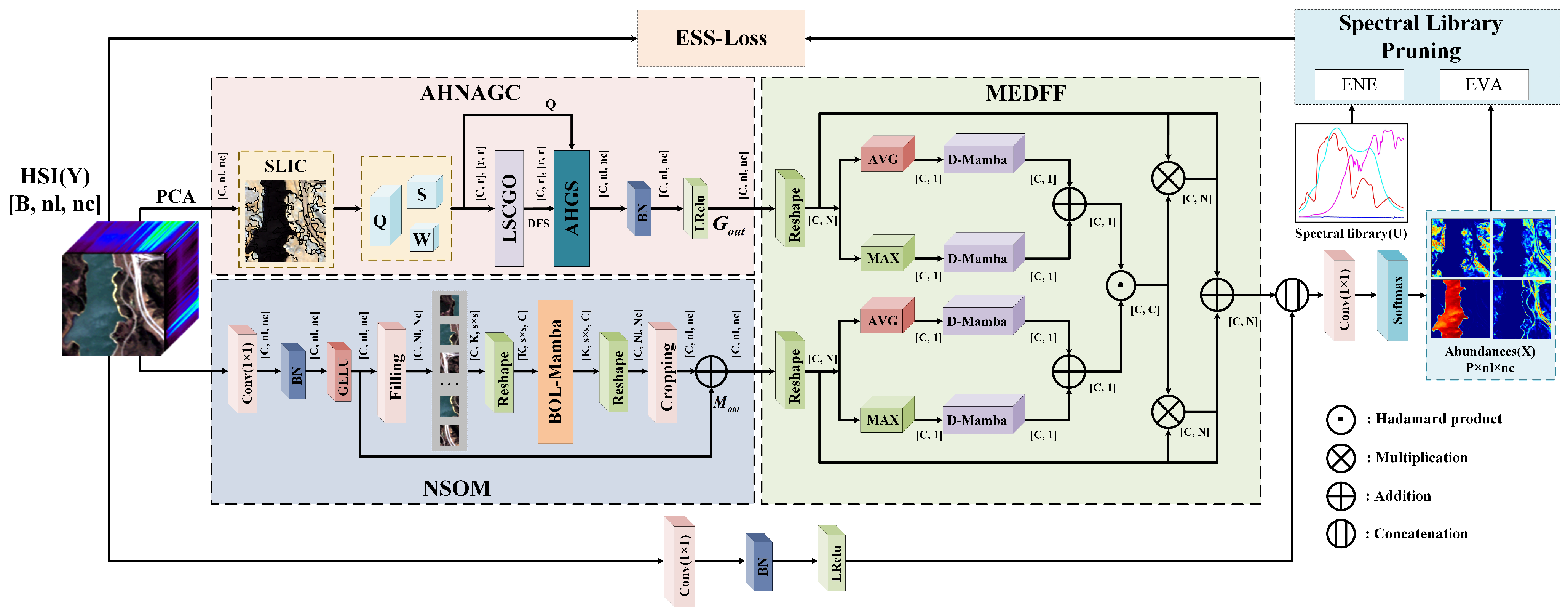
Next, we provide a detailed description of the DGMNet method in this chapter, and the overall architecture of this network is illustrated in
Figure 2.
Specifically, the input HSI is first divided into global and local parallel branches to comprehensively capture its spatial information. The AHNAGC module extracts global spatial features through an adaptive hopping-neighborhood-aware graph convolution mechanism, while the NSOM focuses on accurately modeling the fine-grained local spatial structure. Subsequently, the MEDFF module leverages a spectral attention mechanism to enhance the spectral feature learning and effectively fuse the global and local spatial features, thereby strengthening the interaction and representation of spatial–spectral information. The fused features are subsequently concatenated with four dimension-reduced channels derived from the original HSI, and then projected into the abundance matrix through a 1D convolution followed by a softmax layer. This process is optimized under the guidance of a carefully designed loss function. Notably, during the model iteration, we introduce a spectral-library-pruning mechanism based on a sparse NN, which dynamically selects spectral features. This strategy significantly reduces redundant computation while maintaining the accuracy and robustness of the unmixing performance.
3.1. Adaptive Hop-Neighborhood-Aware GCN (AHNAGC)
To effectively extract global spatial features, we designed the AHNAGC module, which consists of two components: LSCGO and an AHGS.
After constructing the superpixel graph, the original GCN propagates information across all connected superpixels; however, some connections are based solely on spatial adjacency and may not reflect the true land-cover relationships. Such indiscriminate feature propagation can cause confusion between different land-cover classes, negatively affecting the HU accuracy. To mitigate this, the proposed LSCGO module enhances the regional spectral representation by averaging the local neighborhood. It then refines adjacency by removing connections with large spectral differences, allowing the model to focus on spectrally similar regions and improving the unmixing stability and accuracy.
Multi-hop graph structures have been widely used in HSI classification and unmixing [
47,
50], but they typically rely on preset hop numbers. This not only increases the feature engineering workload but also limits the generalization, as suboptimal hop settings may lead to information loss or redundancy, degrading the performance. To overcome this, we propose the AHGS module, which adaptively selects hop counts during each iteration without fixed manual settings. This dynamic mechanism captures global information more effectively, enhances the model expressiveness, and reduces the tuning effort.
Traditional convolution operations treat each pixel in the HSI as a node, resulting in an excessive number of nodes and greatly increasing the computational complexity of the adjacency matrix. Therefore, DGMNet employs the SLIC algorithm for superpixel segmentation, effectively reducing the number of nodes and generating three matrices: the transformation matrix between the pixels and superpixels
, the superpixel adjacency matrix
, and the superpixel feature matrix
, where
N denotes the number of pixels with
,
r is the number of superpixels, and
C represents the number of channels after a dimensionality reduction.
where
represents the dimensionality-reduced HSI,
(·) refers to the process of flattening the
along its spatial dimensions, and
represents the
i-th pixel of the flattened matrix.
represents the set of elements for the
j-th superpixel, and
represents the value at the coordinates (
,
) in the adjacency relationship matrix.
(1) LSCGO: In this module, we optimize the correlation graph between superpixels through the constructed matrices W and S to better enhance the local spatial correlations.
First, the adjacency matrix
and the identity matrix
are added. Then, a dot-product operation is performed with
. That is, each row of
is multiplied element-wise with the first dimension of
to obtain
, where
and ⊙ denotes element-wise multiplication. Next,
is again added to the identity matrix
, and then multiplied with
to compute the average feature values over the neighborhood of each superpixel, i.e.,
, where
. Subsequently, the number of neighbors for the
i-th superpixel, including itself, is computed as
, where
denotes the
-norm, which is used to count the number of non-zero elements in a row. Finally,
is divided by
to obtain the normalized matrix
, as shown in Equation (
11):
where each element in
reflects the mean feature value within the neighborhood region of a superpixel and is used to represent the overall characteristics of the neighborhood.
Subsequently, each row of
is subtracted from the corresponding non-zero elements in
, with the self-relations of each superpixel removed. The resulting tensor, denoted as
, reflects the strength of correlation between each pixel and its neighborhood in each dimension. The greater the correlation, the smaller the value in
. The detailed formulation is illustrated in Equation (
12):
where
. Next, a channel-wise mean operation is performed on
to obtain
, as shown in Equation (
13).
Then, we introduce a soft thresholding strategy. Based on the distribution of values in
, a random threshold parameter
was designed within the range
. By iterating through
element-wise, values greater than
are set to 0, while those smaller than
are set to 1. As a result, a locally enhanced adjacency matrix
is obtained, as shown in Equation (
14). This matrix disconnects neighboring superpixels that deviate significantly from the overall feature level of each local region, thereby enhancing the local spatial correlation.
Finally, the average feature representation of each superpixel, denoted as
, is computed, as shown in Equation (
15), and the weight relationships between the superpixel nodes for non-zero elements in
is calculated using a Gaussian kernel function. The resulting weighted adjacency matrix is denoted as
, as illustrated in Equation (
16), where
is a tunable hyperparameter.
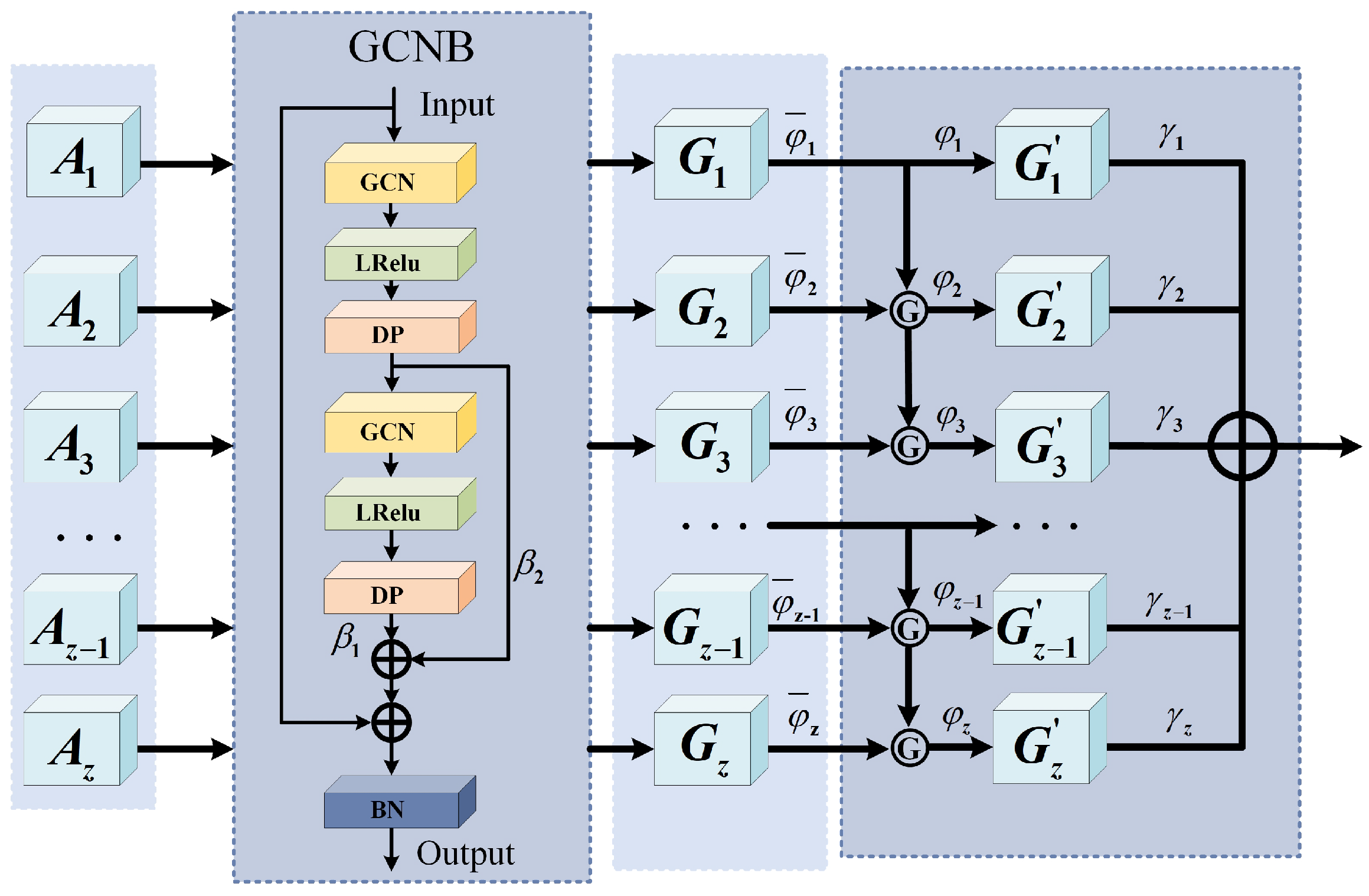
(2) The AHGS: In this module, a multi-hop graph structure is created based on
, enabling adaptive selection and weighting of multi-hop features. The AHGS module is shown in
Figure 3.
First, the adjacency weight matrix of the first-order multi-hop graph is defined as
. DFS is applied to each superpixel node to identify and record all the paths that are
z hops away from the selected central node. The endpoints of these paths are then marked as
. Accordingly, the adjacency matrix of the
z-th hop can be defined as
where
represents the intermediate nodes along the path. Therefore, the
z-hop adjacency feature matrix is obtained by summing and averaging the
adjacency feature matrices, which encompasses all the features of paths that range from 1 hop to
z hops. Based on this method, a series of
z-hop graph adjacency matrices can be generated, denoted as
. These matrices form the foundation of the multi-hop graph structure. In this structure, the direct neighbors of a central superpixel node are considered the 1-hop graph, the neighbors of neighbors form the 2-hop graph, and so on. This effectively avoids redundancy, expands the receptive field of a single node, and allows it to connect to more distant nodes via multi-hop propagation, thereby enhancing the network’s global perception capability. The results after applying graph convolution to the 1-hop, 2-hop, and
z-hop adjacency feature matrices are as follows:
where
denotes the feature matrix obtained after the
i-hop graph convolution, and
represents batch normalization.
is a carefully designed dual-layer graph convolution feature extraction module, which innovatively employs two cascaded graph convolution layers to achieve multi-level feature learning. A key advantage of this module lies in the introduction of a learnable dynamic weight matrix, which enables the model to adaptively balance the contribution of different feature levels through
. Meanwhile, a skip connection mechanism is incorporated to deeply fuse the original input features. This design not only preserves hierarchical information but also significantly enhances the model’s representational power and training stability, thereby delivering superior feature extraction performance when dealing with complex graph-structured data.
Next, two sets of learnable parameters are initialized, denoted as
, representing the hop selection factors and feature contribution factors, respectively. Meanwhile, a parameter matrix
is initialized with the same dimensions as
to control the hop activation. During training, for each element in
, we introduced a fixed threshold (set to 0.3) to determine whether the corresponding hop feature is activated. Specifically, the hop control parameter matrix
is computed as follows:
where
denotes the sigmoid activation function,
represents the logical operator that returns 1 if the condition inside the brackets is true and 0 otherwise, and ∏ denotes the product operation. The above design enforces that the first-hop feature is always activated, while the activation of subsequent hops depends on whether all previous hops have been selected. Specifically, the
z-th hop can only be activated if all preceding
hops are also activated, ensuring the continuity of the feature selection. This operation is indicated by Ⓖ in
Figure 3. Accordingly, the global spatial feature extracted by the AHGS module can be formulated as Equation (
20):
To obtain the final pixel-level global spatial feature matrix
for subsequent feature fusion, the fused global spatial features are first projected back to the pixel domain via the pixel transformation matrix
, and then refined through
and LeakyReLU (
), as defined in Equation (
21):
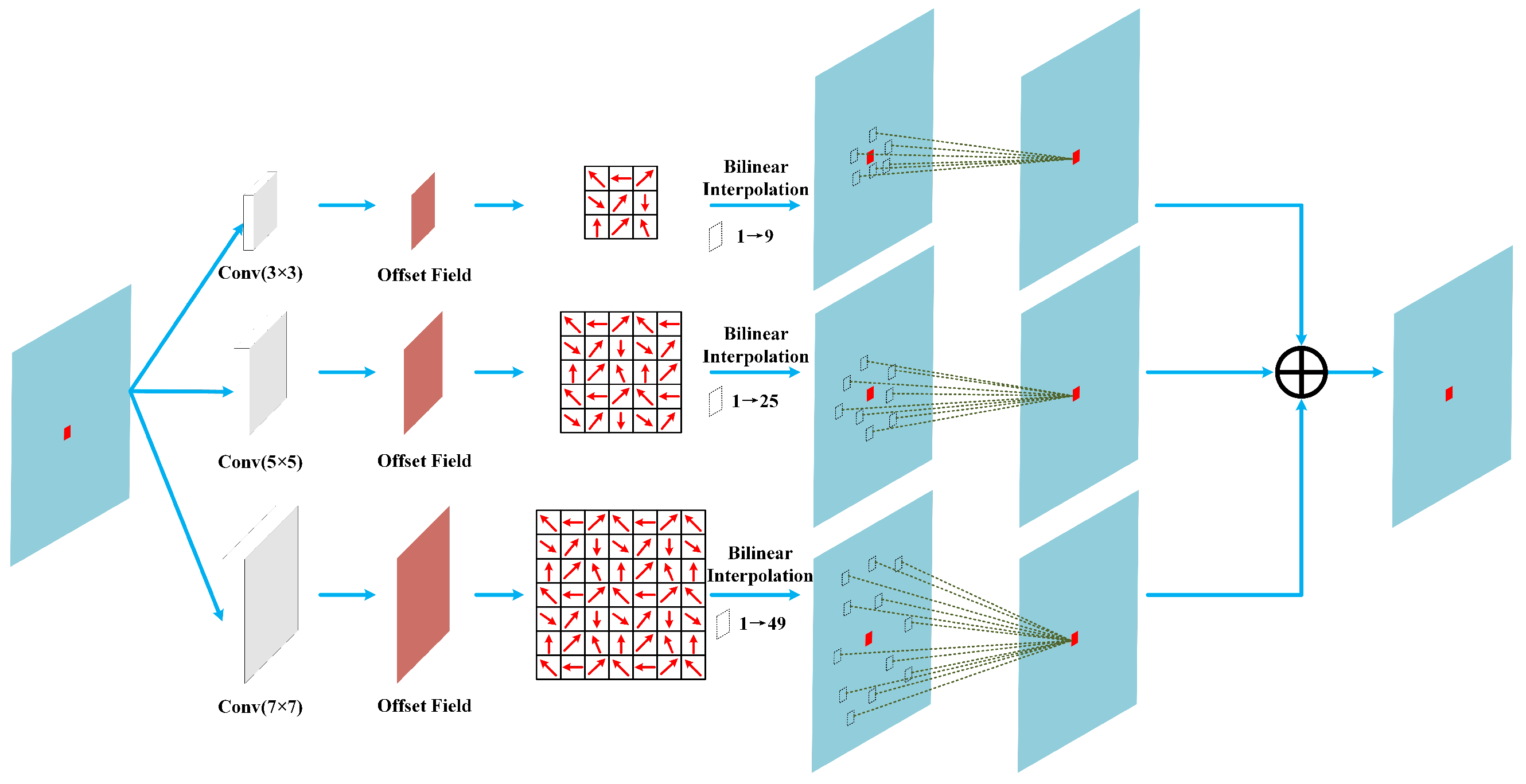
3.2. Neighborhood Spatial Offset Mamba (NSOM)
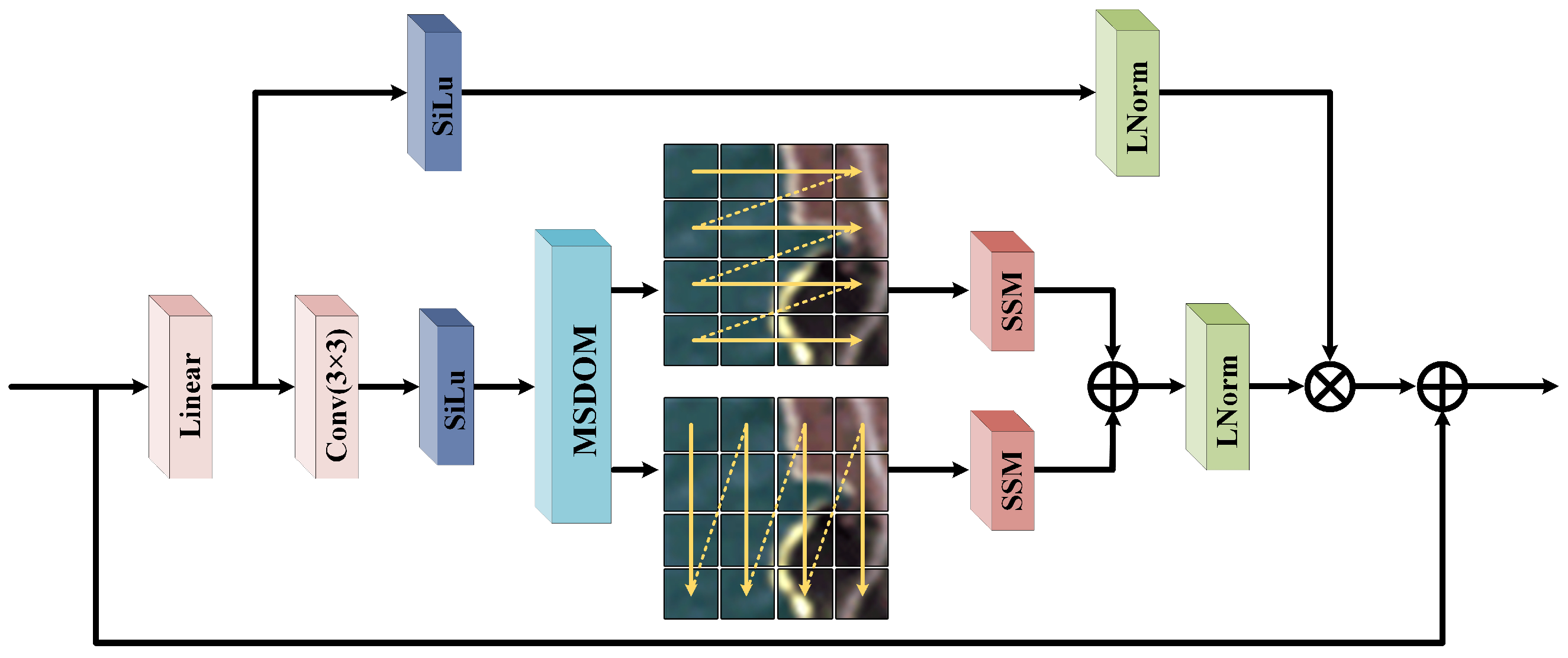
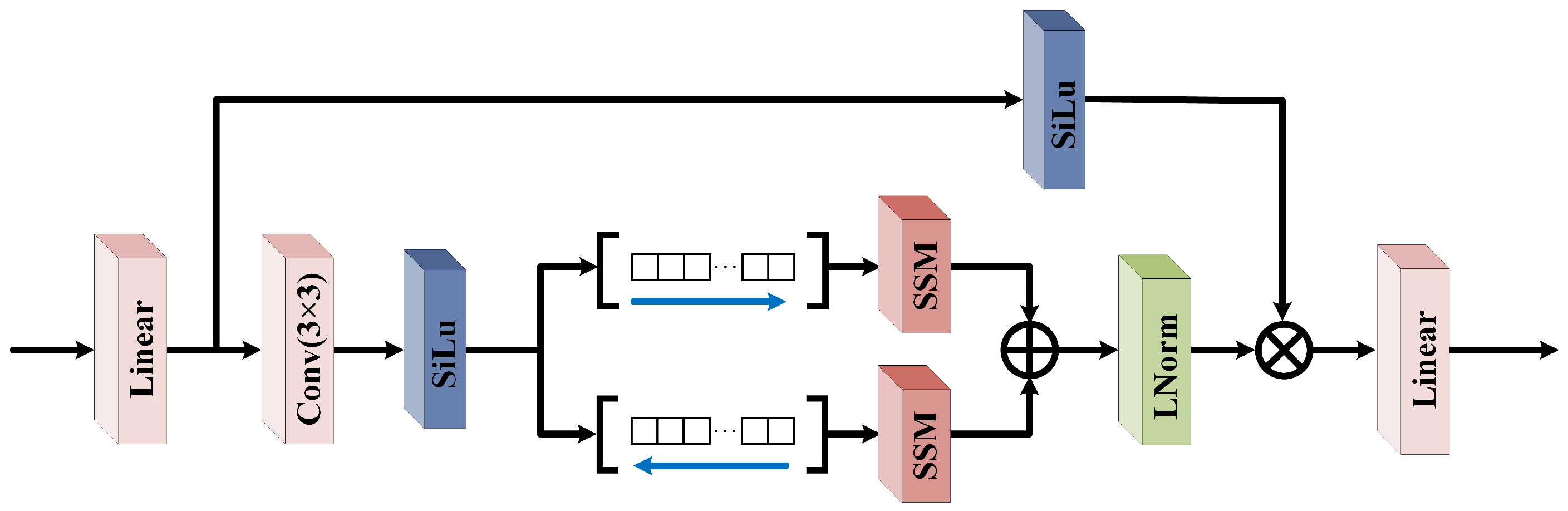
In HU tasks, the fine spectral information of local spatial features plays a crucial role in accurately analyzing the composition of mixed pixels. To enhance the model’s capability for modeling local spatial structures and to collaboratively extract multi-level spatial information with the AHNAGC module, we designed the NSOM module. Within this module, we propose BOL-Mamba, as illustrated in
Figure 4. BOL-Mamba integrates MSDOM (shown in
Figure 5) with a bidirectional Mamba architecture, enabling the fine-grained characterization and modeling of spatial information.
The MSDOM module employs multi-scale deformable convolutions for dynamic sampling, overcoming the limitations of fixed receptive fields and enabling the adaptive capture of fine textures and multi-scale spatial patterns. To overcome the global perception limitations of MSDOM, a bidirectional Mamba is introduced to model long-range dependencies along horizontal and vertical directions within HSI blocks. Leveraging Mamba’s efficient sequence modeling, this design enhances the intra-block spatial perception and global context modeling. The synergy between MSDOM and bidirectional Mamba enables comprehensive and structured spatial feature extraction. Meanwhile, its lightweight design ensures a low computational and parameter overhead, providing a robust foundation for downstream tasks.
The input HSI
is first passed through a
convolution to reduce its dimensionality to
C channels, followed by normalization and activation. To ensure that the spatial dimensions of the feature map can be evenly divided into
K fixed-size blocks, reflection padding is applied along the height and width dimensions. This padding strategy not only ensures tidy block partitioning but also mitigates modeling bias caused by edge effects. Finally, the padded feature map is reshaped into non-overlapping blocks. The process can be formally described as
where
and
represent the reflection padding operation and the activation function, respectively.
denotes the resulting 3D feature tensor.
and
represent the height and width of the image after padding, respectively.
s denotes the width of each HSI block, and
.
is subsequently fed into the BOL-Mamba module for further feature learning. Within BOL-Mamba, the channel dimension of
is first expanded to twice its original size through a linear transformation, and then evenly split into two feature sub-tensors, denoted as
and
. To enhance the representation capability of the local spatial context, we introduce a
convolution operation after the linear mapping, and after passing through the activation function, the result is fed into our designed MSDOM module. The formula is shown as follows:
where
represents the resulting feature map. MSDOM first extracts local information under different receptive fields through parallel single-layer convolutions with
,
, and
kernels, and combines a learnable offset strategy to achieve dynamic adaptation to the local spatial structure.
To enhance the capability of sequential modeling along different spatial dimensions,
is input into the SSM module through two parallel paths: the original form and a variant with height and width swapped. This allows the model to capture long-range dependencies along different spatial directions, improving the completeness of the spatial context modeling. After processing, the output of the variant path is restored to the original dimensions and summed with the output of the original path, achieving effective bidirectional spatial feature fusion. The specific formulation is as follows:
Furthermore,
is first normalized and then element-wise multiplied with
, which has been processed by activation and normalization. This facilitates feature refinement guided by attention signals. To preserve the original feature information and avoid potential information loss, a residual connection is employed. The above process can be formulated as follows:
where ⊙ denotes element-wise multiplication. This mechanism not only enhances the model’s ability to extract key features but also improves the training stability and feature robustness of the deep network structure. Finally, to ensure that the output features maintain the same spatial dimensions as the original input, spatial dimension restoration is performed by removing the previously added padding. The resulting features are then fused with shallow features to produce the final output features
, which can be formulated as
In summary, the NSOM module enhances the spatial discriminative capability and adaptability of the model in HU tasks by integrating Mamba with low computational complexity, thereby enabling dynamic perception, multi-scale fusion, and sequential modeling of local spatial features.
3.3. Mamba-Enhanced Dual-Stream Feature Fusion (MEDFF)
To further enhance the model’s ability to capture complex image features, we propose the MEDFF module. This module significantly improves the model performance by fusing global and local spatial features and integrating a spectral attention mechanism, along with a residual structure. The spectral attention mechanism adaptively adjusts the weights of the individual spectral channels, enabling the model to focus on critical information and enhancing its sensitivity to important features. The introduction of the residual structure ensures effective information flow across different layers of the network, which mitigates the vanishing gradient problem and improves the model stability. Through this combination of designs, MEDFF not only enhances the feature representation capability but also improves the model’s robustness and generalization ability.
First, the global spatial features
and local spatial features
are processed using average pooling and max pooling, respectively, to extract the overall information along the channel dimension and capture the most salient features. The specific computation is as follows:
Through pooling operations, the features of each channel are compressed into a single value, forming a serialized feature representation. To efficiently learn these sequential features, we leveraged the advantages of the Mamba model and designed a D-Mamba module, as shown in
Figure 6. This module performs bidirectional sequence learning in both forward and backward directions, enabling it to simultaneously capture complex dependencies between channels and promote feature interaction and fusion. The specific computation process is as follows:
Subsequently, the features processed by D-Mamba are fused effectively by performing element-wise addition and multiplication operations:
Finally, the spectral attention mechanism adds
to the original HSI, and then concatenates it with partial spectral features
from the original HSI to obtain the final feature map
. The specific formula is as follows:
3.4. Spectral Library Pruning
We innovatively integrated a spectral-library-pruning strategy into NN unmixing models. This innovative design effectively addresses the high coherence and large-scale drawbacks commonly encountered in traditional unmixing methods [
16], while significantly reducing the model’s parameter count and computational complexity. The implementation of this strategy can be divided into two key steps: endmember number estimation (ENE) and endmember validity assessment (EVA).
(1) ENE: Singular value decomposition (SVD) can decompose a matrix into orthogonal bases with clear statistical meaning, where the magnitude of singular values reflects the importance of each component. Therefore, this paper uses SVD to determine the number of endmembers to retain. Specifically,
P singular values are computed via SVD, then a threshold
is set, and the number of singular values exceeding this threshold is taken as the final number of retained endmembers. The calculation formula is as follows:
where
denotes the singular values. The effectiveness of this method is further discussed in the
Section 5.
(2) EVA: During the unmixing process, the abundances matrix is typically sparse, with most row vectors approaching zero and only a few containing significant non-zero elements. These non-zero rows correspond to endmember components that are actually present in the HSI, while the near-zero rows represent dictionary atoms that contribute little to the unmixing process. To quantify the activity level of each endmember, the
norm is applied to each row of the abundances matrix for the activity assessment, and then min–max normalization is performed to obtain standardized activity indicators, as shown in Equation (
35). The min–max normalization (MNorm) is defined in Equation (
34):
In addition, inspired by [
51], we propose a new effectiveness evaluation strategy from the perspective of energy, aiming to prevent small-target endmembers from being pruned during the iterative process. Specifically, the contribution of each endmember to the reconstructed image is calculated, and can be reflected by the difference between the original image and the reconstructed image obtained after removing the corresponding endmember. The larger the difference, the greater the contribution of that endmember. Subsequently, the contribution values are normalized, as formulated below:
where
denotes the matrix
with its
i-th column removed. Next, the maximum value of each row in the abundances matrix (i.e., the energy) is computed and multiplied with the corresponding contribution value, which serves as a new evaluation criterion, as defined in Equation (
39):
The final evaluation strategy is as follows:
This evaluation strategy effectively addresses the issues of high spectral library coherence and large-scale complexity by jointly considering the atom activity, correlation, and a small-target preservation mechanism. At the same time, it significantly reduces the model’s parameter count.
(3) Pruning process: Specifically, during the first stage of iteration (), the need for feature dimension reduction is dynamically determined based on the SVD singular value analysis results. If , the first principal columns and rows of and are retained, namely, and . Otherwise, the process enters the second stage () to retain P key features.
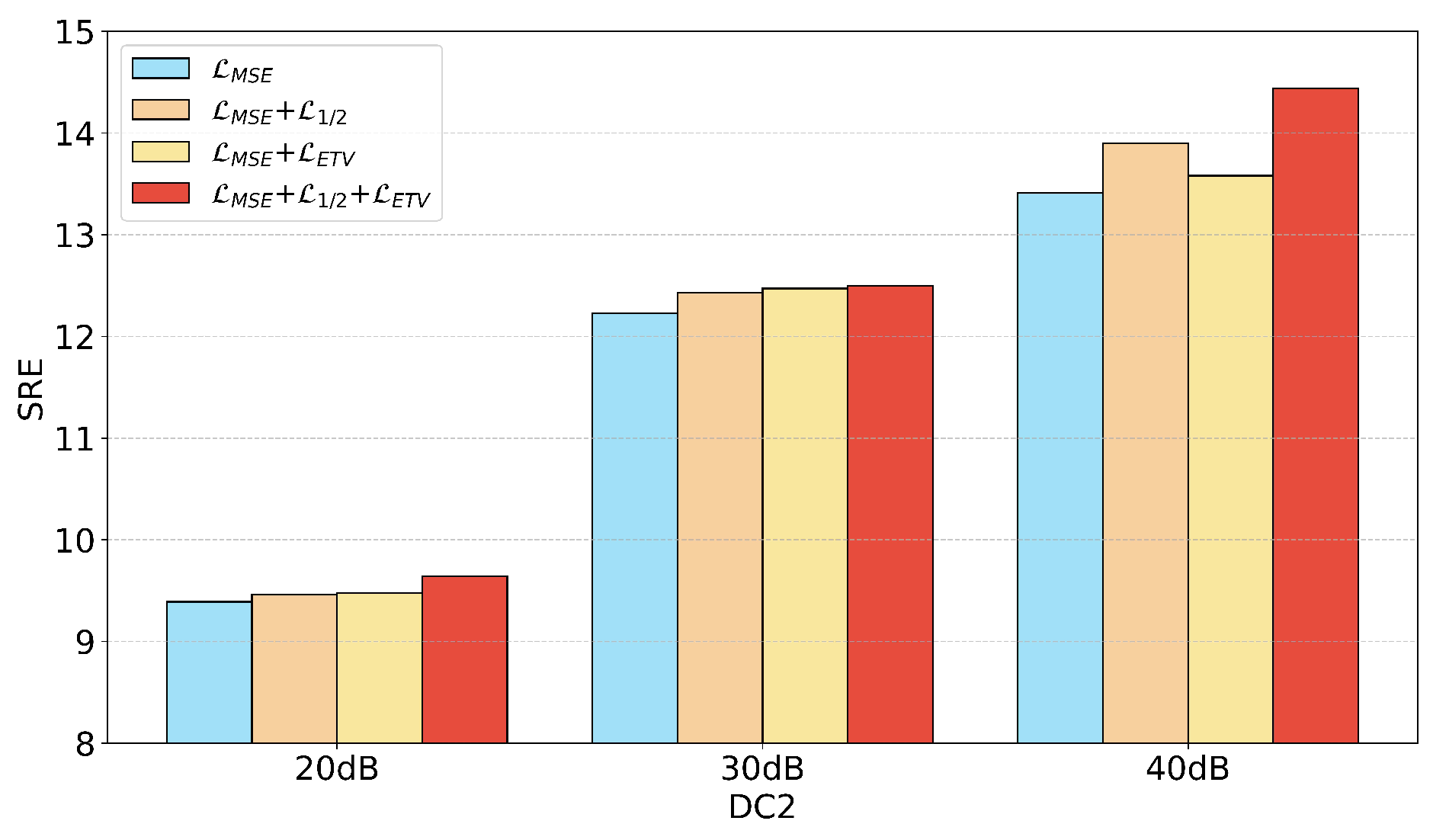
3.5. Enhanced Sparsity Smoothing Loss (ESS-Loss)
This paper proposes the ESS-Loss, which significantly improves the model performance by combining the reconstruction error with multiple prior conditions of the HSI. To measure the reconstruction quality, the mean squared error (MSE) is adopted as the loss function. The MSE is a widely used metric in regression tasks that aims to minimize the difference between the reconstructed image and the original image. Its formula is as follows, where
and
represent the spectral vectors of the original and estimated images, respectively:
Based on the sparsity prior of the abundances matrix, [
13] proposed an
sparse-constrained NMF, which enhances the abundances sparsity through an
regularization term. They demonstrated that the
regularizer is easier to optimize than the
regularizer and achieves stronger sparsity than the
regularizer. Therefore, this paper adopts the
norm as the sparsity loss, formulated as follows:
To impose smoothness constraints on the abundances, this paper extends the traditional TV loss by introducing diagonal directions and incorporating a pixel-wise local gradient-based adaptive weighting mechanism. Traditional TV loss typically considers only pixel differences in the horizontal and vertical directions, overlooking diagonal structures within the image. To enhance the model’s ability to perceive structural information, we propose enhanced total variation (ETV). This loss calculates pixel gradient differences in four directions: horizontal, vertical, diagonal, and anti-diagonal. The pixel gradient differences in these four directions are defined as follows:
Based on the gradient information in the four directions mentioned above, the average gradient magnitude of each pixel is defined as follows:
This gradient value is used to construct the pixel-level adaptive weight matrix
, which is defined as follows:
where the parameter
controls the width of the Gaussian function. This adaptive weighting mechanism assigns weights to each pixel based on the local gradient: regions with large gradients are assigned smaller weights to avoid the excessive smoothing of image edges, while flat regions are given larger weights to enhance the smoothness. Through this dynamic adjustment, the loss function can be flexibly optimized according to pixel values, preserving important edge structures while suppressing unnecessary noise.
Considering the contribution of different directions to the image structure, we divided the loss into two parts: the horizontal and vertical direction loss
, and the diagonal direction loss
:
The final ETV loss function is defined as follows:
where
and
are hyperparameters that control the balance between the two directions. The final loss can be expressed as
5. Discussion
In this section, we delve into the details of the proposed model while also identifying potential areas for future improvements.
5.1. Number of Retained Endmembers Determined by SVD
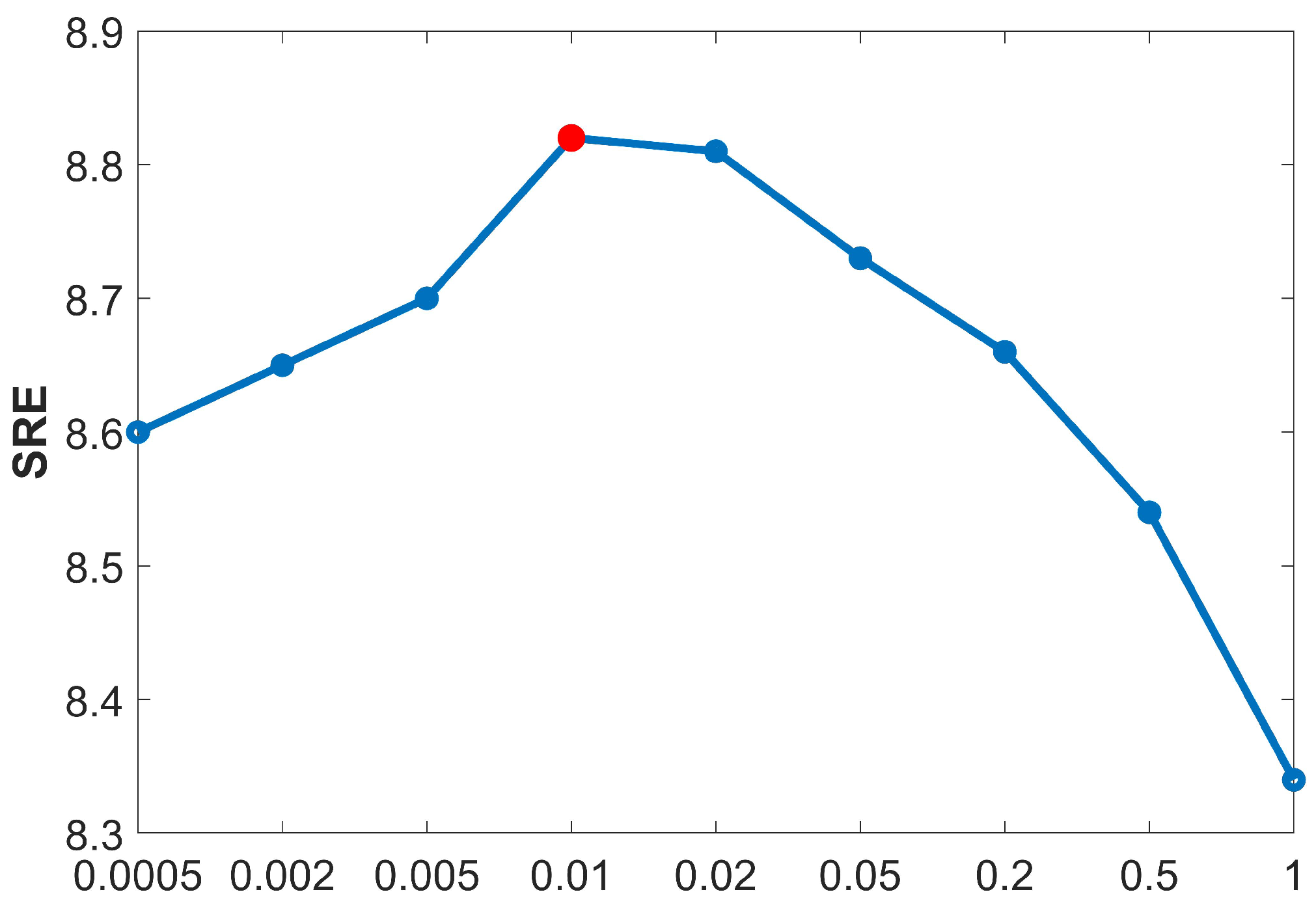
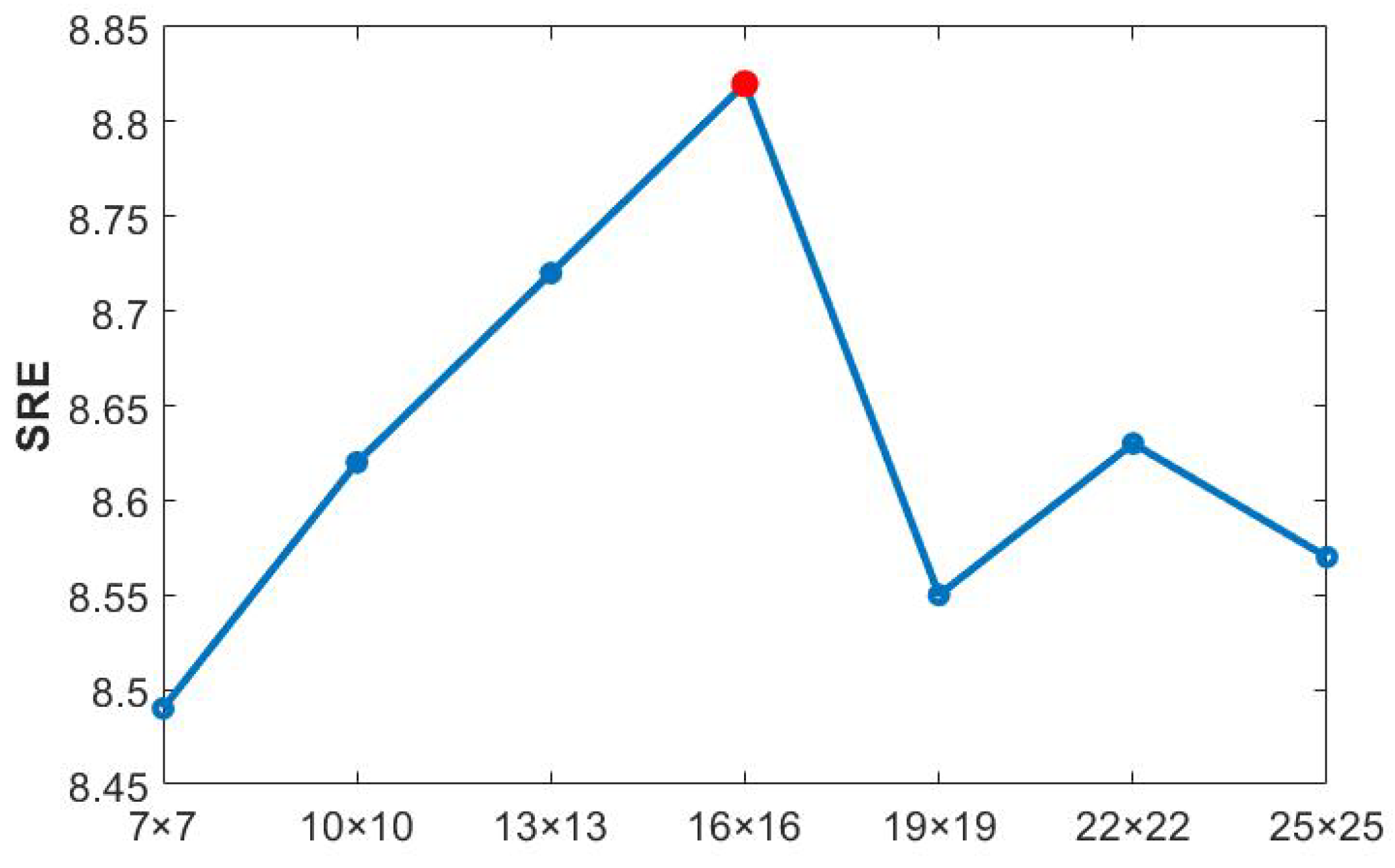
Currently, HU methods based on spectral library pruning have achieved remarkable progress. Approaches like LSU, FaSUn, and NeSU-LP typically retain 30 atoms per dataset, reducing complexity while maintaining endmember coverage. In this paper, we propose an improved and more interpretable method within the SR framework: extracting principal endmember components from HSIs using SVD.
The sizes of singular values directly indicate the importance of each component within the data matrix.
Table 9 presents the descending order of the singular values for each dataset. It can be observed that at the positions marked by black lines, the singular values exhibited a sharp drop of several orders of magnitude (e.g., for the Samson dataset, the values dropped abruptly from the
to the
magnitudes). Based on this observation, the threshold for principal component selection in this study was set to
, and the number of singular values greater than this threshold was taken as the number of endmembers to be retained.
As illustrated in
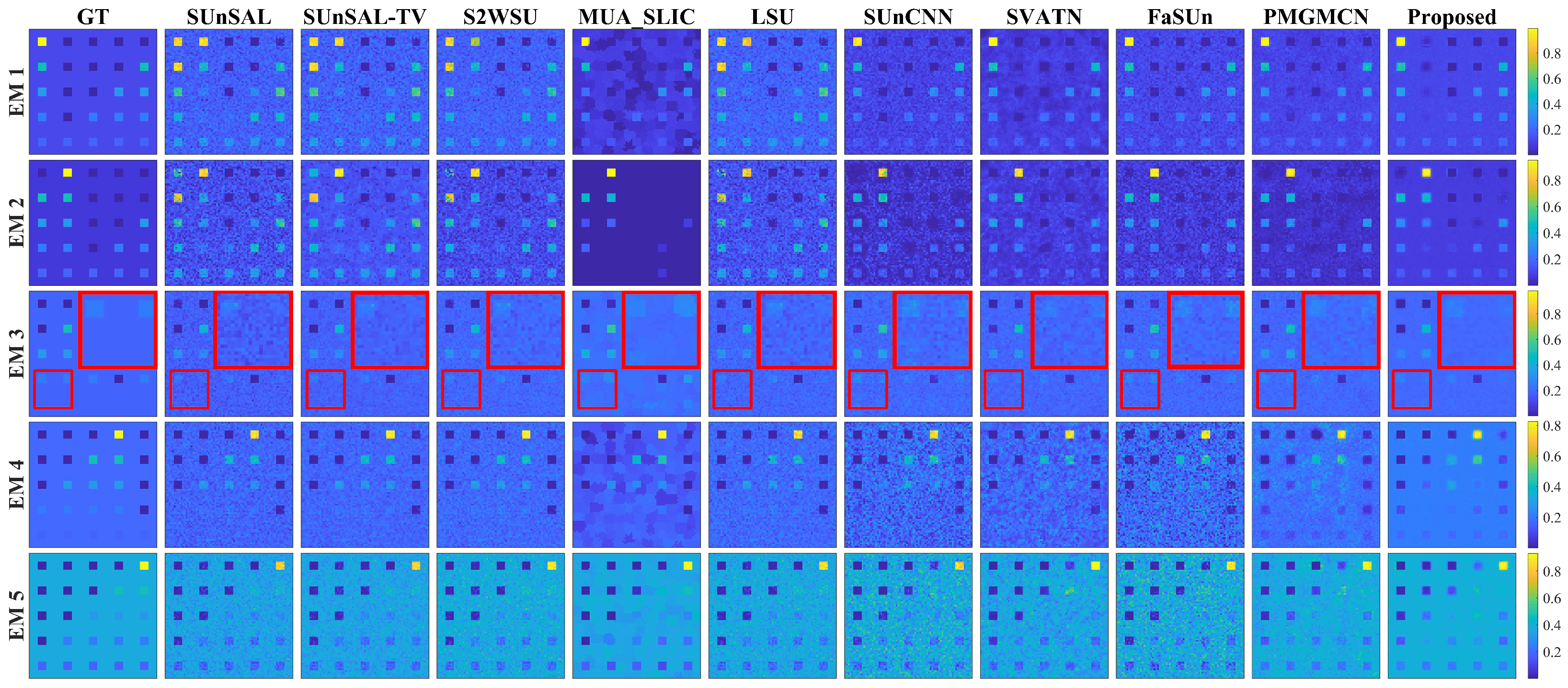
Table 10, when compared with the traditional methods, the number of endmembers retained by our proposed method was always greater than or equal to the referenced number of endmembers. Extensive experimental validations indicate that this method not only accurately identifies the commonly acknowledged endmember components in unmixing but also effectively avoids the mispruning of important endmembers by retaining more components with physical significance. This adaptive pruning strategy based on singular value statistics not only aligns with the physical characteristics of HSIs but also provides a reliable prior on the number of endmembers for SU.
5.2. Future Prospects of Mamba in HU
The Mamba model has brought new opportunities for HU due to its linear computational complexity and efficient sequence-modeling capabilities. Although Transformer has already achieved remarkable results in this field, Mamba still demonstrated a higher computational efficiency, making it particularly suitable for handling the long-sequence characteristics of HSIs. However, it still has limitations in two-dimensional spatial modeling and local feature capture. To address this, this paper introduces a multi-scale deformable convolution module to enhance the spatial structure perception while preserving Mamba’s advantages in global modeling.
Future research in HU can proceed in two directions: First, integrating GCN with Mamba to construct hybrid models that possess both non-Euclidean structural perception capabilities, thereby improving the unmixing ability for irregularly spatially distributed features. Second, in response to the potential information decay that may occur in Mamba’s long-sequence modeling, exploring the fusion with mechanisms such as local attention and adaptive gating to strengthen the response to critical spectral bands and regions, thereby alleviating the problem of representation degradation.
Although the current application of Mamba in HU is still in its early stages, its efficient modeling potential makes it promising as a key technology for breaking through computational bottlenecks.
5.3. Design Motivation of DGMNet
The structural motivation of DGMNet can be explained from the following three aspects.
First, from the perspective of global and local feature modeling capabilities, the GCN branch builds non-Euclidean relationships between nodes over the entire image or its constructed graph structure, which facilitates the capture of long-range dependencies and complex spatial structures in HSIs. This is particularly suitable for characterizing global associations between different land-cover types. In contrast, the Mamba branch demonstrates outstanding sequence modeling capabilities. Based on the SSM, it recursively fuses the features of all elements in the input sequence. This allows it to perceive long-range pixel relationships within local patches, effectively modeling the contextual dependencies and fine-grained features of local spatial sequences, thereby improving the accuracy of the abundance estimation and the consistency of intra-region feature representation. Together, these two branches form a complementary relationship in feature modeling: one enhances the understanding of global spatial structures, while the other strengthens the local detail perception, thus jointly improving the overall expressive power of the network.
Second, in the GCN branch, we employed a multi-hop mechanism based on the GCN. This mechanism adopts a hierarchical and recursive design, which reduces the memory consumption and computational redundancy. In the Mamba branch, we utilized its linear computational complexity to efficiently model local features. Both were essentially motivated by the goal of optimizing the model computation.
Finally, we further optimized the feature extraction strategies for both branches. In the GCN branch, in addition to refining the adjacency matrix, we introduced a multi-hop feature propagation mechanism and innovatively propose an adaptive hop selection strategy. This enables the model to dynamically adjust the number of hops based on different datasets, thereby improving the extraction of global spatial features and enhancing generalization ability. In the Mamba branch, considering the potential loss of spatial structure information when transforming input features into sequences, we incorporated a multi-scale deformable convolution strategy as a compensation mechanism. This was designed to comprehensively and precisely capture local spatial features, further enhancing the model’s capability to characterize internal structures within local regions.
5.4. Limitations of DGMNet and Future Work
Although DGMNet significantly improved the HU performance by introducing a dual-branch spatial feature learning and spectral feature fusion strategy, certain limitations remain in its design and implementation that warrant further improvement and exploration.
From the perspective of the GCN branch, we optimized the adjacency matrix during the graph construction phase by intentionally severing connections between spatially adjacent nodes that have low correlation with the local overall feature representation. Although this strategy enhances feature correlation within local regions to some extent, it may also lead to the neglect of some valuable cross-region information, thereby causing a potential weakening of feature learning capability and a risk of information loss.
From the perspective of the Mamba branch, although we introduced a multi-scale deformable convolution strategy to mitigate its limited capacity in modeling spatial structural information during sequence modeling, this approach remains an external compensatory mechanism rather than a deep optimization based on the internal mechanism of the Mamba model itself. Therefore, there is still potential room for improvement in its modeling of local spatial structures.
These limitations also provide clear directions and insights for future work.
Specifically, for the GCN branch, future research can explore more fine-grained weighted neighborhood aggregation mechanisms by introducing learnable adjacency weights or attention mechanisms to achieve discriminative modeling of the contributions of neighboring pixels. Additionally, the weighted neighborhood strategy can be combined with multi-hop feature propagation mechanisms to synergistically optimize the spatial modeling capability of the GCN at both the feature aggregation and global propagation levels.
Regarding the Mamba branch, since it transforms input features into sequences for feature learning, it inevitably loses some spatial structural information. Future work could consider embedding the local perception and structural modeling modules into the internal architecture of Mamba, realizing an organic fusion of positional awareness and sequence modeling. This would construct a unified network framework that collaboratively optimizes spatial information and sequential dependencies, thereby further enhancing the expressive power and consistency of the local spatial feature extraction.