Combining Open-Source Machine Learning and Publicly Available Aerial Data (NAIP and NEON) to Achieve High-Resolution High-Accuracy Remote Sensing of Grass–Shrub–Tree Mosaics
Abstract
1. Introduction
2. Methods
2.1. Site Description
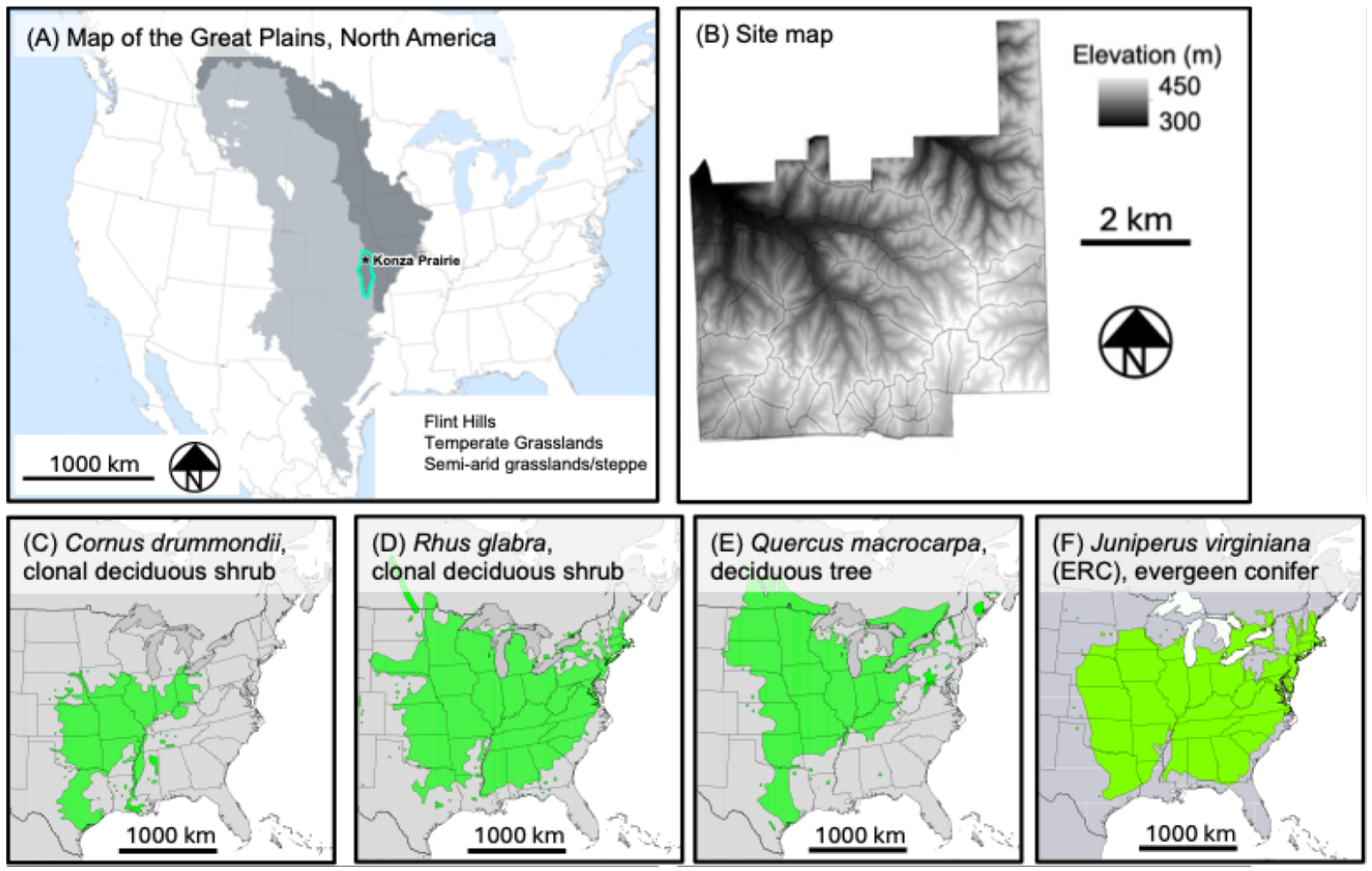
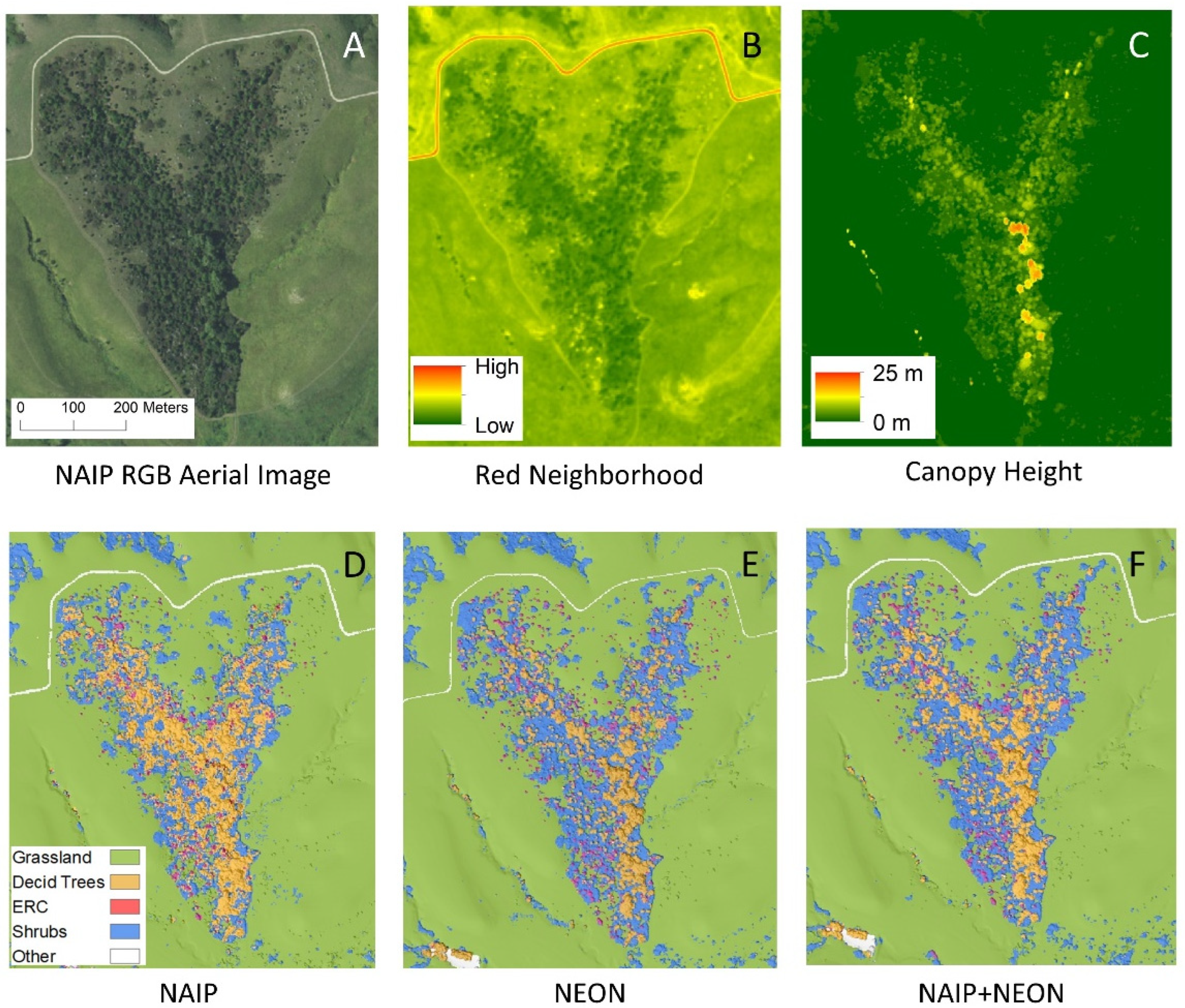
2.2. Imagery
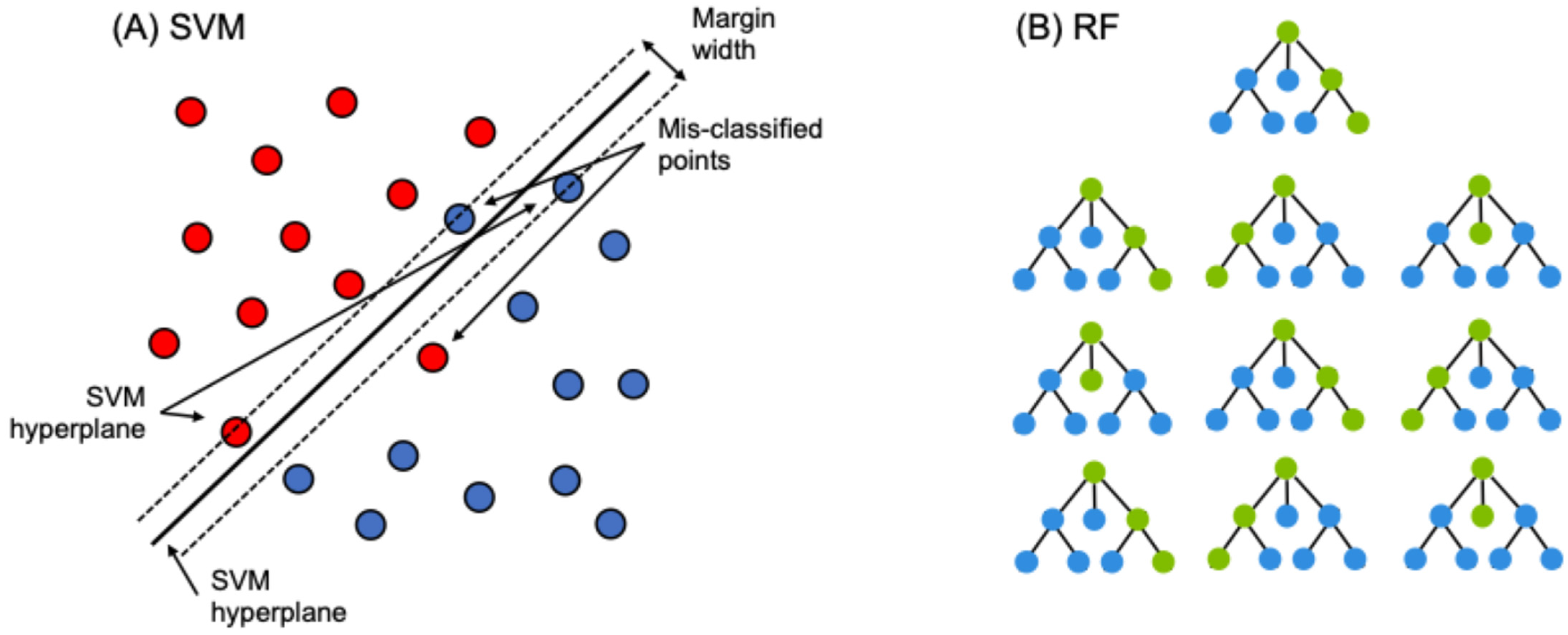
2.3. Machine Learning Methods
2.4. Training Data
2.5. Assessing Accuracy
2.6. Run Time and Other Logistics
3. Results
3.1. Model Accuracy Overview
3.2. Importance of Different Imagery Data Inputs
3.3. Model Run Time and Other Logistics
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briggs, J.M.; Knapp, A.K.; Blair, J.M.; Heisler, J.L.; Hoch, G.A.; Lett, M.S.; McCarron, J.K. An ecosystem in transition: Causes and consequences of the conversion of mesic grassland to shrubland. BioScience 2005, 55, 243–254. [Google Scholar] [CrossRef]
- Brandt, J.S.; Haynes, M.A.; Kuemmerle, T.; Waller, D.M.; Radeloff, V.C. Regime shift on the roof of the world: Alpine meadows converting to shrublands in the southern Himalayas. Biol. Conserv. 2013, 158, 116–127. [Google Scholar] [CrossRef]
- Moser, W.K.; Hansen, M.H.; Atchison, R.L.; Butler, B.J.; Crocker, S.J.; Domke, G.; Kurtz, C.M.; Lister, A.; Miles, P.D. Kansas’ forests 2010. In Resource Bulletin; NRS-85; USDA Forest Service, Northern Research Station: Newtown Square, PA, USA, 2013; Volume 63. [Google Scholar]
- Twidwell, D.; Rogers, W.E.; Fuhlendorf, S.D.; Wonkka, C.L.; Engle, D.M.; Weir, J.R.; Kreuter, U.P.; Taylor, C.A. The rising Great Plains fire campaign: Citizens’ response to woody plant encroachment. Front. Ecol. Environ. 2013, 11 (Suppl. S1), e64–e71. [Google Scholar] [CrossRef]
- Galgamuwa, G.P.; Wang, J.; Barden, C.J. Expansion of Eastern Redcedar (Juniperus virginiana L.) into the Deciduous Woodlands within the Forest–Prairie Ecotone of Kansas. Forests 2020, 11, 154. [Google Scholar] [CrossRef]
- Archer, S.R.; Andersen, E.M.; Predick, K.I.; Schwinning, S.; Steidl, R.J.; Woods, S.R. Woody plant encroachment: Causes and consequences. In Rangeland Systems: Processes, Management and Challenges; Springer: Berlin/Heidelberg, Germany, 2017; pp. 25–84. [Google Scholar]
- Brunsell, N.A.; Van Vleck, E.S.; Nosshi, M.; Ratajczak, Z.; Nippert, J.B. Assessing the roles of fire frequency and precipitation in determining woody plant expansion in central US grasslands. J. Geophys. Res. Biogeosci. 2017, 122, 2683–2698. [Google Scholar] [CrossRef]
- Nippert, J.B.; Ocheltree, T.W.; Orozco, G.L.; Ratajczak, Z.; Ling, B.; Skibbe, A.M. Evidence of physiological decoupling from grassland ecosystem drivers by an encroaching woody shrub. PLoS ONE 2013, 8, e81630. [Google Scholar] [CrossRef]
- Miller, J.E.; Damschen, E.I.; Ratajczak, Z.; Özdoğan, M. Holding the line: Three decades of prescribed fires halt but do not reverse woody encroachment in grasslands. Landsc. Ecol. 2017, 32, 2297–2310. [Google Scholar] [CrossRef]
- Collins, S.L.; Nippert, J.B.; Blair, J.M.; Briggs, J.M.; Blackmore, P.; Ratajczak, Z. Fire frequency, state change and hysteresis in tallgrass prairie. Ecol. Lett. 2021, 24, 636–647. [Google Scholar] [CrossRef]
- Ratajczak, Z.; D’Odorico, P.; Nippert, J.B.; Collins, S.L.; Brunsell, N.A.; Ravi, S. Changes in spatial variance during a grassland to shrubland state transition. J. Ecol. 2017, 105, 750–760. [Google Scholar] [CrossRef]
- Ratajczak, Z.; D’Odorico, P.; Collins, S.L.; Bestelmeyer, B.T.; Isbell, F.I.; Nippert, J.B. The interactive effects of press/pulse intensity and duration on regime shifts at multiple scales. Ecol. Monogr. 2017, 87, 198–218. [Google Scholar] [CrossRef]
- Swengel, A.B. Effects of fire and hay management on abundance of prairie butterflies. Biol. Conserv. 1996, 76, 73–85. [Google Scholar] [CrossRef]
- Lettow, M.C.; Brudvig, L.A.; Bahlai, C.A.; Gibbs, J.; Jean, R.P.; Landis, D.A. Bee community responses to a gradient of oak savanna restoration practices. Restor. Ecol. 2018, 26, 882–890. [Google Scholar] [CrossRef]
- Engle, D.M.; Coppedge, B.R.; Fuhlendorf, S.D. From the dust bowl to the green glacier: Human activity and environmental change in Great Plains grasslands. In Western North American Juniperus Communities; Springer: New York, NY, USA, 2008; pp. 253–271. [Google Scholar]
- Silber, K.M.; Hefley, T.J.; Castro-Miller, H.N.; Ratajczak, Z.; Boyle, W.A. The long shadow of woody encroachment: An integrated approach to modeling grassland songbird habitat. Ecol. Appl. 2024, 34, e2954. [Google Scholar] [CrossRef] [PubMed]
- Lautenbach, J.M.; Plumb, R.T.; R.n, S.G.; Hagen, C.A.; Haukos, D.A.; Pitman, J.C. Lesser prairie-chicken avoidance of trees in a grassland landscape. Rangel. Ecol. Manag. 2017, 70, 78–86. [Google Scholar] [CrossRef]
- Albrecht, M.A.; Becknell, R.E.; Long, Q. Habitat change in insular grasslands: Woody encroachment alters the population dynamics of a rare ecotonal plant. Biol. Conserv. 2016, 196, 93–102. [Google Scholar] [CrossRef]
- Keen, R.M.; Nippert, J.B.; Sullivan, P.L.; Ratajczak, Z.; Ritchey, B.; O’Keefe, K.; Dodds, W.K. Impacts of Riparian and Non-riparian Woody Encroachment on Tallgrass Prairie Ecohydrology. Ecosystems 2022, 26, 290–301. [Google Scholar] [CrossRef]
- Dodds, W.K.; Ratajczak, Z.; Keen, R.M.; Nippert, J.B.; Grudzinski, B.; Veach, A.; Taylor, J.H.; Kuhl, A. Trajectories and state changes of a grassland stream and riparian zone after a decade of woody vegetation removal. Ecol. Appl. 2023, 33, e2830. [Google Scholar] [CrossRef]
- Morford, S.L.; Allred, B.W.; Twidwell, D.; Jones, M.O.; Maestas, J.D.; Roberts, C.P.; Naugle, D.E. Herbaceous production lost to tree encroachment in United States rangelands. J. Appl. Ecol. 2022, 59, 2971–2982. [Google Scholar] [CrossRef]
- Ratajczak, Z.; Nippert, J.B.; Briggs, J.M.; Blair, J.M. Fire dynamics distinguish grasslands, shrublands and woodlands as alternative attractors in the Central Great Plains of North America. J. Ecol. 2014, 102, 1374–1385. [Google Scholar] [CrossRef]
- Meneguzzo, D.M.; Liknes, G.C. Status and Trends of Eastern Redcedar (Juniperus virginiana) in the Central United States: Analyses and Observations Based on Forest Inventory and Analysis Data. J. For. 2015, 113, 325–334. [Google Scholar] [CrossRef]
- Briggs, J.; Hoch, G.; Johnson, L. Assessing the Rate, Mechanisms, and Consequences of the Conversion of Tallgrass Prairie to Juniperus virginiana Forest. Ecosystems 2002, 5, 578–586. [Google Scholar] [CrossRef]
- Ratajczak, Z.; Briggs, J.M.; Goodin, D.G.; Luo, L.; Mohler, R.L.; Nippert, J.B.; Obermeyer, B. Assessing the potential for transitions from tallgrass prairie to woodlands: Are we operating beyond critical fire thresholds? Rangel. Ecol. Manag. 2016, 69, 280–287. [Google Scholar] [CrossRef]
- Loss, S.R.; Noden, B.H.; Fuhlendorf, S.D. Woody plant encroachment and the ecology of vector-borne diseases. J. Appl. Ecol. 2022, 59, 420–430. [Google Scholar] [CrossRef]
- Veach, A.M.; Dodds, W.K.; Skibbe, A. Fire and grazing influences on rates of riparian woody plant expansion along grassland streams. PLoS ONE 2014, 9, e106922. [Google Scholar] [CrossRef]
- Laliberte, A.S.; Rango, A.; Havstad, K.M.; Paris, J.F.; Beck, R.F.; McNeely, R.; Gonzalez, A.L. Object-oriented image analysis for mapping shrub encroachment from 1937 to 2003 in southern New Mexico. Remote Sens. Environ. 2004, 93, 198–210. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Allred, B.W.; Bestelmeyer, B.T.; Boyd, C.S.; Brown, C.; Davies, K.W.; Duniway, M.C.; Ellsworth, L.M.; Erickson, T.A.; Fuhlendorf, S.D.; Griffiths, T.V.; et al. Improving Landsat predictions of rangeland fractional cover with multitask learning and uncertainty. Methods Ecol. Evol. 2021, 12, 841–849. [Google Scholar] [CrossRef]
- Kranjčić, N.; Medak, D.; Župan, R.; Rezo, M. Support vector machine accuracy assessment for extracting green urban areas in towns. Remote Sens. 2019, 11, 655. [Google Scholar] [CrossRef]
- Nguyen, H.T.T.; Doan, T.M.; Tomppo, E.; McRoberts, R.E. Land Use/land cover mapping using multitemporal Sentinel-2 imagery and four classification methods—A case study from Dak Nong, Vietnam. Remote Sens. 2020, 12, 1367. [Google Scholar] [CrossRef]
- Thanh Noi, P.; Kappas, M. Comparison of Random Forest, k-Nearest Neighbor, and Support Vector Machine Classifiers for Land Cover Classification Using Sentinel-2 Imagery. Sensors 2018, 18, 18. [Google Scholar] [CrossRef]
- Freitag, M.; Kamp, J.; Dara, A.; Kuemmerle, T.; Sidorova, T.V.; Stirnemann, I.A.; Velbert, F.; Hölzel, N. Post-soviet shifts in grazing and fire regimes changed the functional plant community composition on the Eurasian steppe. Glob. Change Biol. 2021, 27, 388–401. [Google Scholar] [CrossRef] [PubMed]
- Toth, C.; Jóźków, G. Remote sensing platforms and sensors: A survey. ISPRS J. Photogramm. Remote Sens. 2016, 115, 22–36. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Vanderbilt, B.C.; Ramezan, C.A. Land Cover Classification and Feature Extraction from National Agriculture Imagery Program (NAIP) Orthoimagery: A Review. Photogramm. Eng. Remote Sens. 2017, 83, 737–747. [Google Scholar] [CrossRef]
- Nagy, R.C.; Balch, J.K.; Bissell, E.K.; Cattau, M.E.; Glenn, N.F.; Halpern, B.S.; Ilangakoon, N. Harnessing the NEON data revolution to advance open environmental science with a diverse and data-capable community. Ecosphere 2021, 12, e03833. [Google Scholar] [CrossRef]
- Soubry, I.; Guo, X. Quantifying woody plant encroachment in grasslands: A review on remote sensing approaches. Can. J. Remote Sens. 2022, 48, 337–378. [Google Scholar] [CrossRef]
- Brandt, M.; Tucker, C.J.; Kariryaa, A.; Rasmussen, K.; Abel, C.; Small, J.; Chave, J.; Rasmussen, L.V.; Hiernaux, P.; Diouf, A.A.; et al. An unexpectedly large count of trees in the West African Sahara and Sahel. Nature 2020, 587, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E.; Kellner, J.R.; Wright, S.J. Operational tree species mapping in a diverse tropical forest with airborne imaging spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.M.; Byrd, K.B.; Anderegg, L.D.; Allan, E.; Gatziolis, D.; Roberts, D.; Yacoub, R.; Nemani, R.R. Capturing patterns of evolutionary relatedness with reflectance spectra to model and monitor biodiversity. Proc. Natl. Acad. Sci. USA 2023, 120, e2215533120. [Google Scholar] [CrossRef]
- Pau, S.; Nippert, J.B.; Slapikas, R.; Griffith, D.; Bachle, S.; Helliker, B.R.; O’Connor, R.C.; Riley, W.J.; Still, C.J.; Zaricor, M. Poor relationships between NEON Airborne Observation Platform data and field-based vegetation traits at a mesic grassland. Ecology 2022, 103, e03590. [Google Scholar] [CrossRef]
- Tooley, E.G.; Nippert, J.B.; Ratajczak, Z. Evaluating methods for measuring the leaf area index of encroaching shrubs in grasslands: From leaves to optical methods, 3-D scanning, and airborne observation. Agric. For. Meteorol. 2024, 349, 109964. [Google Scholar] [CrossRef]
- Klodd, A.E.; Nippert, J.B.; Ratajczak, Z.; Waring, H.; Phoenix, G.K. Tight coupling of leaf area index to canopy nitrogen and phosphorus across heterogeneous tallgrass prairie communities. Oecologia 2016, 182, 889–898. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Zare, A.; Bohlman, S.A.; Singh, A.; Graves, S.J.; Magee, L.; Johnson, D.J.; Record, S.; Rubio, V.E.; et al. Individual canopy tree species maps for the National Ecological Observatory Network. PLoS Biol. 2024, 22, e3002700. [Google Scholar] [CrossRef]
- Wedel, E.R.; Ratajczak, Z.; Tooley, E.G.; Nippert, J.B. Divergent resource-use strategies of encroaching shrubs: Can traits predict encroachment success in tallgrass prairie? J. Ecol. 2025, 113, 339–352. [Google Scholar] [CrossRef]
- Scholtz, R.; Prentice, J.; Tang, Y.; Twidwell, D. Improving on MODIS MCD64A1 burned area estimates in grassland systems: A case study in Kansas Flint Hills tall grass prairie. Remote Sens. 2020, 12, 2168. [Google Scholar] [CrossRef]
- Ratajczak, Z.; Collins, S.L.; Blair, J.M.; Nippert, J.B. Reintroducing bison results in long-running and resilient increases in grassland diversity. Proc. Natl. Acad. Sci. USA 2022, 119, e2210433119. [Google Scholar] [CrossRef]
- Ratajczak, Z.; Nippert, J.B.; Hartman, J.C.; Ocheltree, T.W. Positive feedbacks amplify rates of woody encroachment in mesic tallgrass prairie. Ecosphere 2011, 2, 121. [Google Scholar] [CrossRef]
- VanderWeide, B.L.; Hartnett, D.C. Fire resistance of tree species explains historical gallery forest community composition. For. Ecol. Manag. 2011, 261, 1530–1538. [Google Scholar] [CrossRef]
- Thompson, R.S.; Anderson, K.H.; Bartlein, P.J. Digital representations of tree species range maps from “Atlas of United States trees” by Elbert L. Little, Jr. (and other publications). In Atlas of Relations Between Climatic Parameters and Distributions of Important Trees and Shrubs in North America; U.S. Geological Survey, Information Services (Producer): Denver, CO, USA, 1999; On file at: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station, Fire Sciences Laboratory: Missoula, MT, USA; FEIS files. [82831]. [Google Scholar]
- Hulslander, D. NEON Normalized Difference Vegetation Index (NDVI), Enhanced Vegetation Index (EVI), Atmospherically Resistant Vegetation Index (ARVI), Canopy Xanthophyll Cycle (PRI), and Canopy Lignin (NDLI) Algorithm Theoretical Basis Document (NEON Doc. #: NEON.DOC.002391, Rev. A). NEON Inc. 2016. Available online: https://data.neonscience.org/data-products/DP3.30026.001 (accessed on 26 May 2025).
- USDA Geospatial Data Gateway. NAIP Orthorectified Kansas Color Infrared 161 2019. Available online: https://nrcs.app.box.com/v/naip/file/579854034024 (accessed on 1 November 2020).
- USDA Geospatial Data Gateway. NAIP Orthorectified Kansas Natural Color 161 2019. Available online: https://nrcs.app.box.com/v/naip/file/578936034378 (accessed on 1 November 2020).
- NEON (National Ecological Observatory Network). Ecosystem structure (DP3.30015.001) 2020. RELEASE-2022. Available online: https://data.neonscience.org (accessed on 8 December 2022). [CrossRef]
- NEON (National Ecological Observatory Network). Vegetation indices—Spectrometer—Mosaic (DP3.30026.001) 2020. RELEASE-2022. Available online: https://data.neonscience.org (accessed on 8 December 2022). [CrossRef]
- Noble, B.; Ratajczak, Z. WPE01 Assessing the value added of NEON for using machine learning to quantify vegetation mosaics and woody plant encroachment at Konza Prairie ver 1. Environ. Data Initiat. 2022. [Google Scholar] [CrossRef]
- Noble, B.; Ratajczak, Z. Combining machine learning and publicly available aerial data (NAIP and NEON) to achieve high-resolution remote sensing of grass-shrub-tree mosaics in the Central Great Plains (USA). bioRxiv 2025. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support vector machine versus random forest for remote sensing image classification: A meta-analysis and systematic review. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Burges, C.J. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Mantero, P.; Moser, G.; Serpico, S.B. Partially supervised classification of remote sensing images through SVM-based probability density estimation. IEEE Trans. Geosci. Remote Sens. 2005, 43, 559–570. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; V4.0.5; R Foundation for Statistical Computing: Vienna, Austria, 2021. Available online: https://www.R-project.org/ (accessed on 1 November 2020).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R package version 1.7-6. 2021. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 1 November 2020).
- Evans, J.S.; Murphy, M.A.; Holden, Z.A.; Cushman, S.A. Modeling species distribution and change using random forest. In Predictive Species and Habitat Modeling in Landscape Ecology; Springer: New York, NY, USA, 2011; pp. 139–159. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22, R package version 4.6.14. Available online: https://CRAN.R-project.org/doc/Rnews/ (accessed on 1 November 2020).
- NEON (National Ecological Observatory Network). High-Resolution Orthorectified Camera Imagery (DP1.30010.001). 2025, RELEASE-2025. Available online: https://data.neonscience.org/data-products/DP1.30010.001/RELEASE-2025 (accessed on 28 May 2025). [CrossRef]
- Google. RGB Image Centered on Konza Prairie Biological Station, Credited to Maxar Technologies in 2020; Retrieved 1 September 2020; Google Earth; Google: Mountain View, CA, USA, 2020. [Google Scholar]
- Kaskie, K.D.; Wimberly, M.C.; Bauma, P.J.R. Rapid assessment of juniper distribution in prairie landscapes of the northern Great Plains. Int. J. Appl. Earth Obs. Geoinf. 2019, 83, 101946. [Google Scholar] [CrossRef]
- Kaskie, K.D.; Wimberly, M.C.; Bauman, P.J. Predictive Mapping of Low-Density Juniper Stands in Prairie Landscapes of the Northern Great Plains. Rangel. Ecol. Manag. 2022, 83, 81–90. [Google Scholar] [CrossRef]
- Bork, E.W.; Su, J.G. Integrating LIDAR data and multispectral imagery for enhanced classification of rangeland vegetation: A meta analysis. Remote Sens. Environ. 2007, 111, 11–24. [Google Scholar] [CrossRef]
- Scholl, V.M.; Cattau, M.E.; Joseph, J.B.; Balch, J.K. Integrating National Ecological Observatory Network (NEON) airborne remote sending in in-situ data for optimal tree species classification. Remote Sens. 2020, 12, 1414. [Google Scholar] [CrossRef]
- Pervin, R.; Robeson, S.M.; MacBean, N. Fusion of airborne hyperspectral and LiDAR canopy-height data for estimating fractional cover of tall woody plants, herbaceous vegetation, and other soil cover types in a semi-arid savanna ecosystem. Int. J. Remote Sens. 2022, 43, 3890–3926. [Google Scholar] [CrossRef]
- Jin, H.; Mountrakis, G. Fusion of optical, radar and waveform LiDAR observations for land cover classification. ISPRS J. Photogramm. Remote Sens. 2022, 187, 171–190. [Google Scholar] [CrossRef]
- Whiteman, G.; Brown, J.R. Assessment of a method for mapping woody plant density in a grassland matrix. J. Arid Environ. 1998, 38, 269–282. [Google Scholar] [CrossRef]
- Ayhan, B.; Kwan, C. Tree, shrub, and grass classification using only RGB images. Remote Sens. 2020, 12, 1333. [Google Scholar] [CrossRef]
- Zhong, B.; Yang, L.; Luo, X.; Wu, J.; Hu, L. Extracting Shrubland in Deserts from Medium-Resolution Remote-Sensing Data at Large Scale. Remote Sens. 2024, 16, 374. [Google Scholar] [CrossRef]
- Hayes, M.M.; Miller, S.N.; Murphy, M.A. High-resolution landcover classification using Random Forest. Remote Sens. Lett. 2014, 5, 112–121. [Google Scholar] [CrossRef]
- Tooley, E.G.; Nippert, J.B.; Bachle, S.; Keen, R.M. Intra-canopy leaf trait variation facilitates high leaf area index and compensatory growth in a clonal woody encroaching shrub. Tree Physiol. 2022, 42, 2186–2202. [Google Scholar] [CrossRef]
- Strand, E.K.; Robinson, A.P.; Bunting, S.C. Spatial patterns on the sagebrush steppe/Western juniper ecotone. Plant Ecol. 2007, 190, 159–173. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef]
- Tang, L.; Shao, G. Drone remote sensing for forestry research and practices. J. For. Res. 2015, 26, 791–797. [Google Scholar] [CrossRef]
- Bansod, B.; Singh, R.; Thakur, R.; Singhal, G. A comparison between satellite based and drone based remote sensing technology to achieve sustainable development: A review. J. Agric. Environ. 2017, 111, 383–407. [Google Scholar]
- Fassnacht, F.E.; White, J.C.; Wulder, M.A.; Næsset, E. Remote sensing in forestry: Current challenges, considerations and directions. For. Int. J. For. Res. 2024, 97, 11–37. [Google Scholar] [CrossRef]
- Rosen, A.; Jörg Fischer, F.; Coomes, D.A.; Jackson, T.D.; Asner, G.P.; Jucker, T. Tracking shifts in forest structural complexity through space and time in human-modified tropical landscapes. Ecography 2024, 2024, e07377. [Google Scholar] [CrossRef]
- Weisberg, P.J.; Lingua, E.; Pillai, R.B. Spatial patterns of pinyon–juniper woodland expansion in central Nevada. Rangel. Ecol. Manag. 2007, 60, 115–124. [Google Scholar] [CrossRef]
- Smith, A.M.; Strand, E.K.; Steele, C.M.; Hann, D.B.; Garrity, S.R.; Falkowski, M.J.; Evans, J.S. Production of vegetation spatial-structure maps by per-object analysis of juniper encroachment in multitemporal aerial photographs. Can. J. Remote Sens. 2008, 34, S268–S285. [Google Scholar] [CrossRef]
- Hanan, N.P.; Limaye, A.S.; Irwin, D.E. Use of Earth Observations for Actionable Decision Making in the Developing World. Front. Environ. Sci. 2020, 8, 601340. [Google Scholar] [CrossRef]

| Source | # Bands | Inputs | Input Description |
|---|---|---|---|
| USDA NAIP | 9 | Red | Redness of each pixel |
| Green | Greenness of each pixel | ||
| Blue | Blueness of each pixel | ||
| Red Neighborhood * | Avg. redness of surrounding pixels | ||
| Green Neighborhood * | Avg. greenness of surrounding pixels | ||
| Blue Neighborhood * | Avg. blueness of surrounding pixels | ||
| Near-Infrared | Value of near-infrared wavelength | ||
| Near-Infrared Neighborhood * | Avg. values of near-infrared of surrounding pixels | ||
| NDVI | Calculated from NIR and red bands, indicates live green vegetation density | ||
| NSF NEON | 8 | Enhanced Vegetation Index (EVI) | Similar to NDVI, EVI estimates vegetation greenness and biomass |
| Normalized Difference Nitrogen Index (NDNI) | Relative nitrogen concentration in canopy | ||
| Normalized Difference Lignin Index (NDLI) | Uses shortwave IR to estimate lignin content in the canopy | ||
| Soil-Adjusted Vegetation Index (SAVI) | Reduces soil brightness in areas where vegetation cover is low | ||
| Atmospherically Resistant Vegetation Index (ARVI) | Reduces atmospheric noise from dust, smoke, rain, etc. | ||
| NDVI | Calculated from NIR and red bands, indicates live green vegetation density | ||
| NDVI Neighborhood * | Avg. NDVI values of surrounding pixels | ||
| Canopy Height (LiDAR) | Height of canopy above bare earth | ||
| NEON + NAIP | 17 | All of the above |
| Class | # Ground-Truthed Polygons | # Computer-Drawn Polygons | Total Polygons | Total Pixels | Total Area (m2) | % of Total Training |
|---|---|---|---|---|---|---|
| Deciduous Trees | 68 | 620 | 688 | 37,215 | 150,533.5 | 12.7% |
| Grass | 246 | 160 | 406 | 179,799 | 719,013.9 | 60.3% |
| Easter Red Cedar | 51 | 506 | 557 | 5537 | 22,751.6 | 1.9% |
| Shrubs | 341 | 1578 | 1919 | 71,101 | 285,690.1 | 24% |
| Other * | 0 | 65 | 65 | 6676 | 13,484.3 | 1.1% |
| Total | 706 | 2929 | 3635 | 300,328 | 1,191,473 | 100% |
| Source | Machine Learning Method | OA * | Kappa | Accuracy | Deciduous Trees | Grassland | ERC | Shrubs | Other ** |
|---|---|---|---|---|---|---|---|---|---|
| NAIP | SVM | 0.903 | 0.831 | PA | 0.72 | 0.98 | 0.52 | 0.85 | 0.94 |
| UA | 0.89 | 0.94 | 0.78 | 0.82 | 0.97 | ||||
| RF | 0.929 | 0.877 | PA | 0.77 | 0.98 | 0.56 | 0.93 | 0.96 | |
| UA | 0.92 | 0.97 | 0.75 | 0.85 | 0.97 | ||||
| NEON | SVM | 0.968 | 0.945 | PA | 0.96 | 0.99 | 0.84 | 0.98 | 0.97 |
| UA | 0.98 | 0.98 | 0.86 | 0.94 | 0.97 | ||||
| RF | 0.972 | 0.951 | PA | 0.93 | 0.99 | 0.83 | 0.98 | 0.96 | |
| UA | 0.98 | 0.99 | 0.85 | 0.94 | 0.97 | ||||
| NAIP + NEON | SVM | 0.977 | 0.964 | PA | 0.95 | 0.99 | 0.87 | 0.97 | 0.97 |
| UA | 0.97 | 0.98 | 0.90 | 0.96 | 0.99 | ||||
| RF | 0.977 | 0.962 | PA | 0.94 | 0.99 | 0.85 | 0.98 | 0.96 | |
| UA | 0.98 | 0.99 | 0.87 | 0.95 | 0.98 |
| NAIP SVM (Support Vector Machine) Model | |||||
|---|---|---|---|---|---|
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 9303 (72.4%) | 19 (0%) | 457 (28.1%) | 665 (3.2%) | 6 (0.4%) |
| Grassland | 325 (2.5%) | 48,825 (98.3%) | 163 (10%) | 2437 (11.8%) | 72 (4.8%) |
| ERC | 198 (1.5%) | 22 (0%) | 839 (51.6%) | 18 (0.1%) | 3 (0.2%) |
| Shrub | 2999 (23.4%) | 789 (1.6%) | 168 (10.3%) | 17,521 (84.9%) | 1 (0.1%) |
| Other | 18 (0.1%) | 19 (0%) | 0 (0%) | 0 (0%) | 1405 (94.5%) |
| NAIP RF (Random Forest) Model | |||||
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 9852 (76.7%) | 26 (0.1%) | 439 (27%) | 370 (1.8%) | 10 (0.7%) |
| Grassland | 305 (2.4%) | 48667 (98%) | 127 (7.8%) | 1010 (4.9%) | 62 (4.2%) |
| ERC | 276 (2.1%) | 28 (0.1%) | 914 (56.2%) | 3 (0%) | 3 (0.2%) |
| Shrub | 2389 (18.6%) | 927 (1.9%) | 147 (9%) | 19,249 (93.3%) | 0 (0%) |
| Other | 21 (0.2%) | 26 (0.1%) | 0 (0%) | 0 (0%) | 1412 (95%) |
| NEON SVM (Support Vector Machine) Model | |||||
|---|---|---|---|---|---|
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 11,877 (92.5%) | 15 (0%) | 137 (8.4%) | 114 (0.6%) | 11 (0.7%) |
| Grassland | 81 (0.6%) | 49,072 (98.8%) | 32 (2%) | 718 (3.5%) | 43 (2.9%) |
| ERC | 193 (1.5%) | 5 (0%) | 1333 (81.9%) | 12 (0.1%) | 1 (0.1%) |
| Shrub | 679 (5.3%) | 551 (1.1%) | 125 (7.7%) | 19,788 (95.9%) | 1 (0.1%) |
| Other | 13 (0.1%) | 31 (0.1%) | 0 (0%) | 0 (0%) | 1431 (96.2%) |
| NEON RF (Random Forest) Model | |||||
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 11,891 (92.6%) | 13 (0%) | 161 (9.9%) | 30 (0.1%) | 9 (0.6%) |
| Grassland | 83 (0.6%) | 48,973 (98.6%) | 15 (0.9%) | 420 (2%) | 46 (3.1%) |
| ERC | 210 (1.6%) | 8 (0%) | 1346 (82.7%) | 17 (0.1%) | 2 (0.1%) |
| Shrub | 651 (5.1%) | 642 (1.3%) | 105 (6.5%) | 20,165 (97.7%) | 2 (0.1%) |
| Other | 8 (0.1%) | 38 (0.1%) | 0 (0%) | 0 (0%) | 1428 (96%) |
| NAIP + NEON SVM (Support Vector Machine) Model | |||||
|---|---|---|---|---|---|
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 12,157 (94.7%) | 34 (0.1%) | 112 (6.9%) | 85 (0.4%) | 12 (0.8%) |
| Grassland | 55 (0.4%) | 49,388 (99.4%) | 23 (1.4%) | 511 (2.5%) | 27 (1.8%) |
| ERC | 127 (1%) | 3 (0%) | 1413 (86.8%) | 7 (0%) | 7 (0.5%) |
| Shrub | 487 (3.8%) | 232 (0.5%) | 79 (4.9%) | 20,029 (97.1%) | 0 (0%) |
| Other | 17 (0.1%) | 17 (0%) | 0 (0%) | 0 (0%) | 1465 (97%) |
| NAIP + NEON RF (Random Forest) Model | |||||
| Known Class → Predicted Class ↓ | Decid. Tree | Grassland | ERC | Shrub | Other |
| Decid. Tree | 12,043 (93.8%) | 21 (0%) | 149 (9.2%) | 22 (0.1%) | 4 (0.3%) |
| Grassland | 47 (0.4%) | 49,220 (99.1%) | 13 (0.8%) | 364 (1.8%) | 14 (0.9%) |
| ERC | 180 (1.4%) | 14 (0%) | 1388 (85.3%) | 7 (0%) | 4 (0.3%) |
| Shrub | 569 (4.4%) | 387 (0.8%) | 77 (4.7%) | 20,239 (98.1%) | 0 (0%) |
| Other | 4 (0%) | 32 (0.1%) | 0 (0%) | 0 (0%) | 1465 (98.5%) |
| Model Training Time (300,328 Pixels) | |||
|---|---|---|---|
| NAIP | NEON | NAIP + NEON | |
| SVM | 4:49:00 | 1:43:00 | 1:37:00 |
| RF | 0:30:00 | 0:23:00 | 1:05:00 |
| Model Prediction Time (8,781,520 Pixels Pixels) | |||
| Model Run Time | NAIP | NEON | NAIP + NEON |
| SVM | 6:15:25 | 3:05:08 | 2:04:59 |
| RF | 0:09:59 | 0:10:43 | 0:07:20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noble, B.; Ratajczak, Z. Combining Open-Source Machine Learning and Publicly Available Aerial Data (NAIP and NEON) to Achieve High-Resolution High-Accuracy Remote Sensing of Grass–Shrub–Tree Mosaics. Remote Sens. 2025, 17, 2224. https://doi.org/10.3390/rs17132224
Noble B, Ratajczak Z. Combining Open-Source Machine Learning and Publicly Available Aerial Data (NAIP and NEON) to Achieve High-Resolution High-Accuracy Remote Sensing of Grass–Shrub–Tree Mosaics. Remote Sensing. 2025; 17(13):2224. https://doi.org/10.3390/rs17132224
Chicago/Turabian StyleNoble, Brynn, and Zak Ratajczak. 2025. "Combining Open-Source Machine Learning and Publicly Available Aerial Data (NAIP and NEON) to Achieve High-Resolution High-Accuracy Remote Sensing of Grass–Shrub–Tree Mosaics" Remote Sensing 17, no. 13: 2224. https://doi.org/10.3390/rs17132224
APA StyleNoble, B., & Ratajczak, Z. (2025). Combining Open-Source Machine Learning and Publicly Available Aerial Data (NAIP and NEON) to Achieve High-Resolution High-Accuracy Remote Sensing of Grass–Shrub–Tree Mosaics. Remote Sensing, 17(13), 2224. https://doi.org/10.3390/rs17132224








