1. Introduction
Forest is a complex functional aggregate in the ecosystem [
1,
2]. Chlorophyll, as a key pigment in forest vegetation, has functions such as maintaining photosynthesis and indicating environmental stress. It is an important indicator for forest ecosystem evaluation [
3], and its distribution shows significant differences in space [
4]. Therefore, accurately estimating the spatial distribution of forest chlorophyll content is crucial for forest ecosystem studies [
5]. However, obtaining samples in the complex vertical and horizontal structures of forests is time-consuming, expensive, and destructive [
6]. Remote sensing technology for chlorophyll content estimation using hyperspectral data and LiDAR overcomes the limitations of traditional field measurement [
7,
8,
9]. The hyperspectral sensor can acquire hundreds of narrow bands with abundant spectral information. Continuous high-resolution spectral images of the study area can be obtained through the UAV platform with simple operation, flexible application, and low cost [
10], which avoids the disadvantages of few satellites and low resolution in the existing hyperspectral data acquisition. This provides an important data basis for the estimation of chlorophyll content in horizontal structure. Accurate estimation of chlorophyll content in the vertical direction requires LiDAR technology, which provides forest vertical structure information. LiDAR can effectively compensate for the lack of vertical information in hyperspectral data and provide more details [
11]. The fusion of hyperspectral data and LiDAR has been widely applied in forestry, including tree-species identification [
12,
13,
14], forest type classification, forest biochemical parameter inversion [
15,
16,
17,
18], and forest pest–diseases monitoring [
19,
20,
21].
In terms of chlorophyll content estimation, there are relatively few studies on the joint estimation of unmanned aerial vehicle hyperspectral and LiDAR data, which mainly rely on hyperspectral data. Models for chlorophyll content estimation include empirical models and physical models [
22]. Liao [
23] proposed a new fusion framework through deep learning. This framework realizes the complementary information of hyperspectral and radar data for mapping, thereby improving the accuracy of tree species classification. Dong [
24] obtained the Digital Surface Model (DSM) and Digital Terrain Model (DTM) from airborne lidar point cloud data. Further, by taking the difference, the Normalized Digital Surface Model (nDSM) was acquired. The normalized index was calculated using high-resolution imagery, and Principal Component Analysis (PCA) was employed for denoising and dimensionality reduction. Through the fusion of PCA images and nDSM images, the Maximum Likelihood Classification (MLC) method was used for supervised classification. The overall accuracy of extracting buildings and trees was 84.00%. For example, a backward stepwise regression model was used in Shen’s study to find a significant difference in the vertical distribution of biochemical traits on the surface of
Metasequoia glyptostroboides and
Populus tomentosa Carr [
16]. Yin used the radiation transfer model (PROSAIL) to plot the pigment distribution of
Ginkgo biloba at different ages, showing significant differences [
17]. Blackburn developed a method for estimating the chlorophyll concentration in artificial broadleaf and coniferous forests using an airborne imaging spectrometer and LiDAR [
25]. Koetz obtained comprehensive forest canopy characteristics from the combined remote sensing signals of imaging spectroscopy and large-footprint LiDAR, enabling reliable estimation of forest chlorophyll content [
26]. The empirical method is simple and easy to implement, especially in recent years, the machine learning algorithm has been well applied in the inversion of biochemical parameters. Compared with the linear model, it considers the nonlinear relationship between variables and improves the robustness of prediction. Although the physical method needs to input complex parameters, the model is not affected by time and region, and the model has good mobility, stability, and high accuracy [
22,
27]. There are more studies on the spatial distribution of chlorophyll in agriculture [
28], while in forestry, most studies focus on the horizontal direction, and some scholars are dedicated to the analysis of the vertical spatial distribution of chlorophyll [
16,
17,
18,
29]. However, studies exploring the vertical spatial distribution of chlorophyll in different forest stands are still relatively few. Meanwhile, chlorophyll content estimation is mainly focused on the single-wood level, so there is still room for exploration in the accurate estimation of chlorophyll content at the stand level. Remote sensing monitoring of vegetation chlorophyll is mainly based on hyperspectral images, and the methods adopted mainly include statistical model methods (multiple stepwise regression, neural networks, random forests) and physical model methods (radiative transfer model method) [
30,
31].
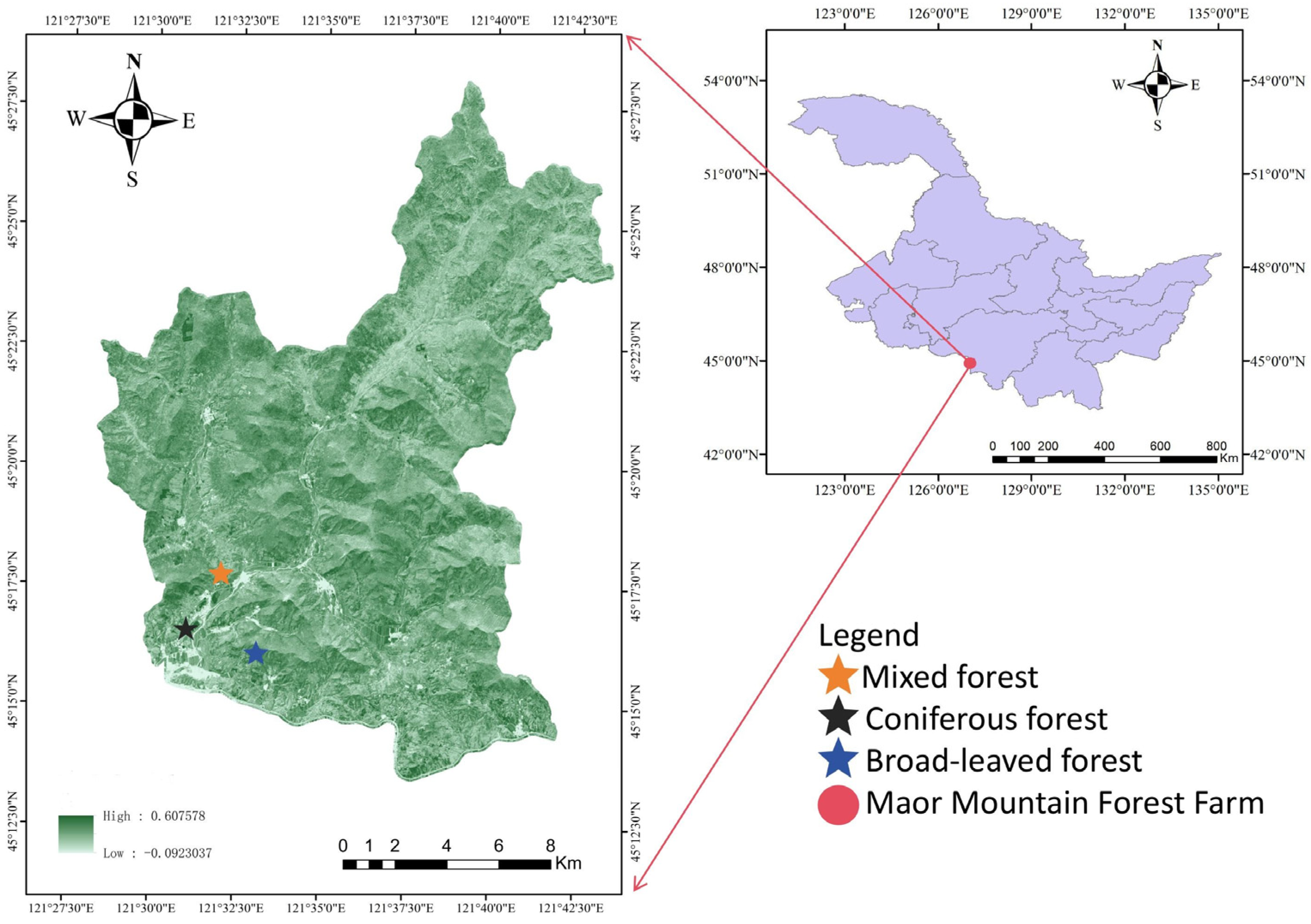
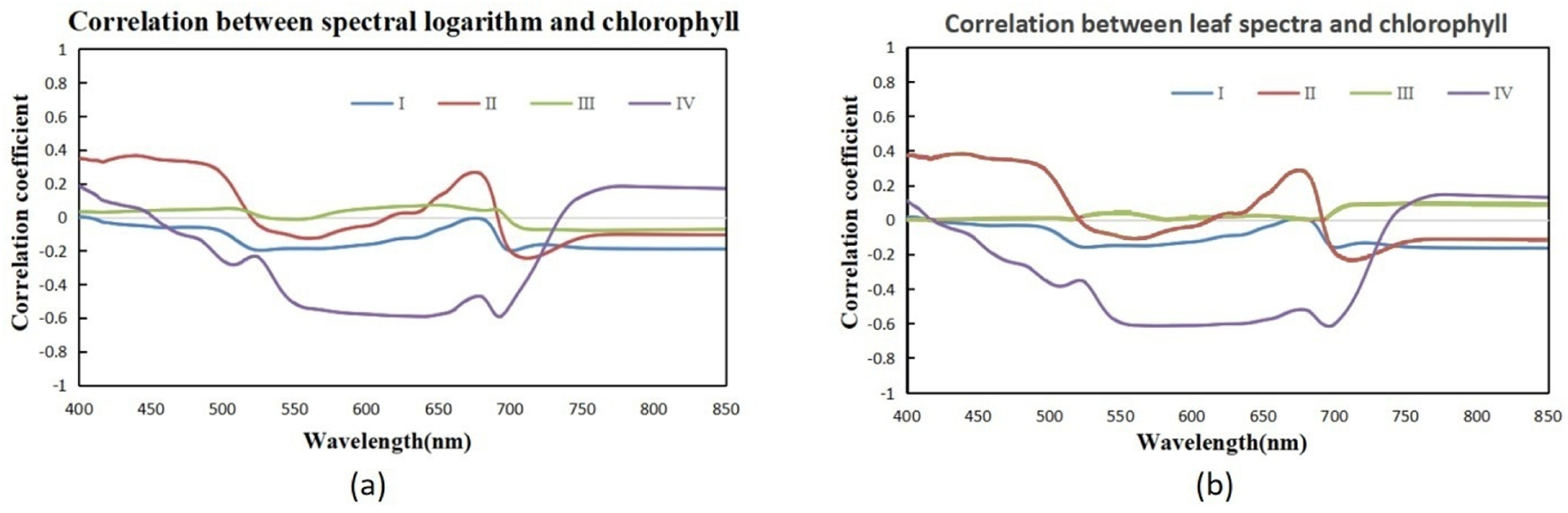
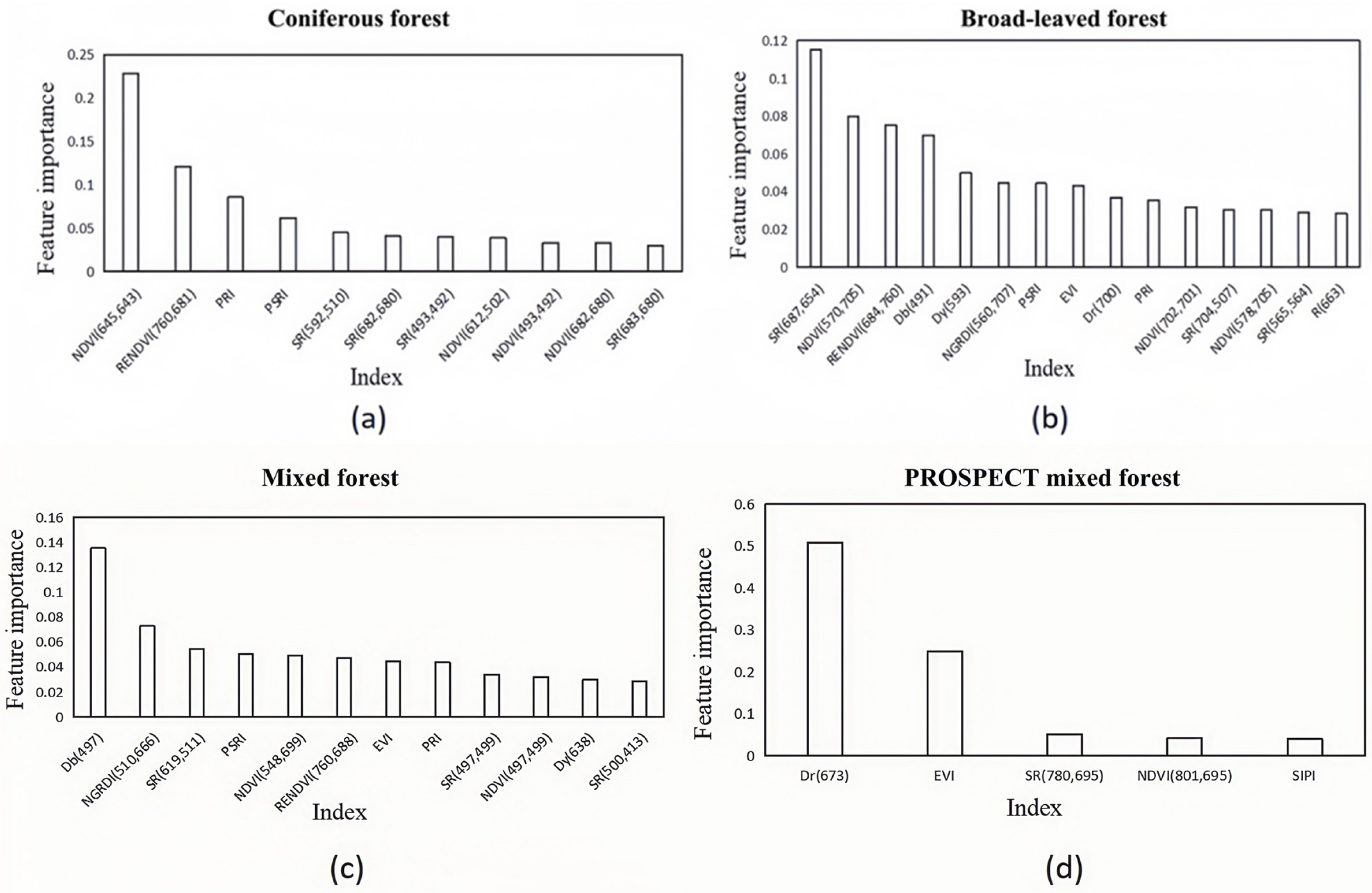
In this study, hyperspectral and LiDAR data were used to construct the three-dimensional spatial distribution of chlorophyll content among different forest types. This study explored the accuracy and differences in chlorophyll content estimation using multiple stepwise regression, random forest, BP neural network, firefly algorithm optimized BP neural network, and mechanistic modeling methods driven by a mixture of measured and simulated data. The optimal model for chlorophyll estimation was determined, and the spatial distribution pattern of chlorophyll content at the single tree and stand scale was clarified. By timely and accurately estimating the spatial distribution of chlorophyll in different forest stands and individual trees, a scientific basis is provided for effective forest management and regional carbon-neutral evaluation.
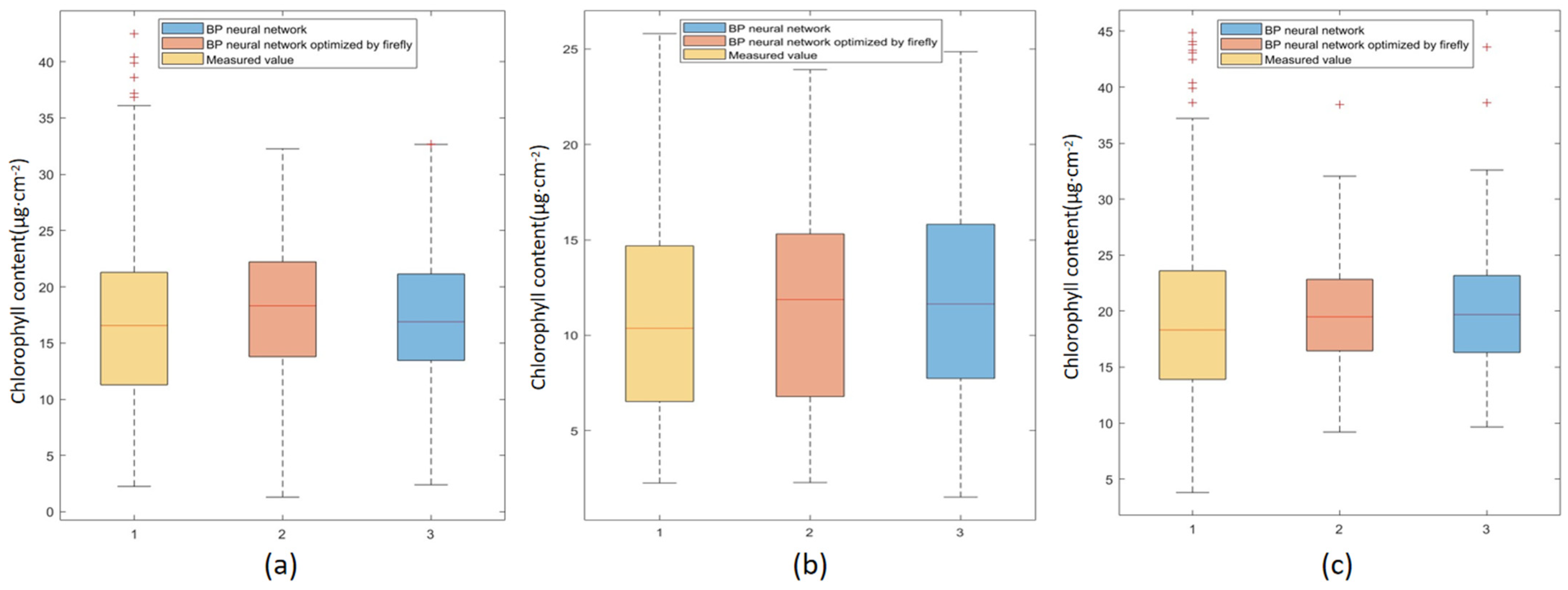
4. Discussion
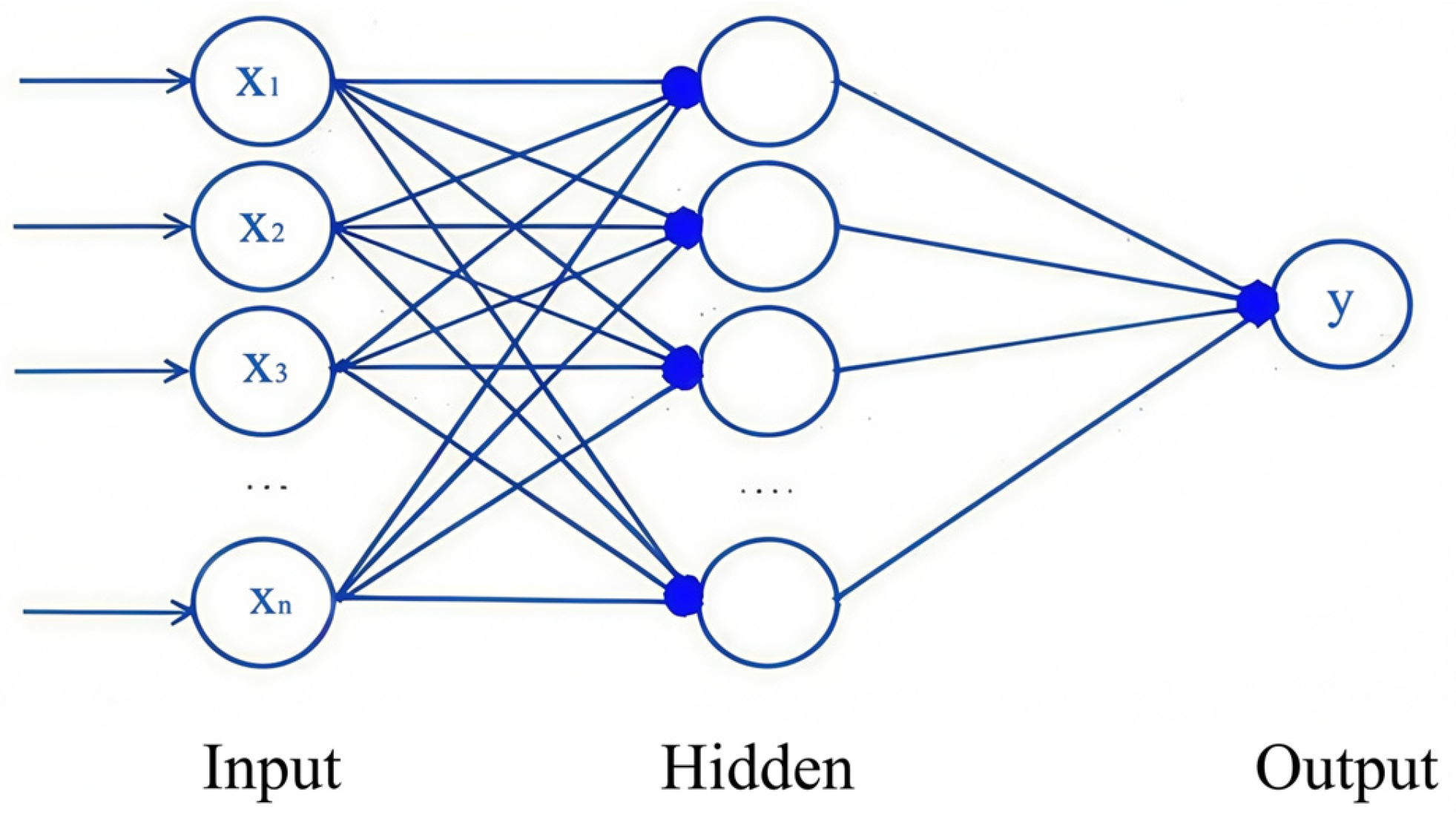
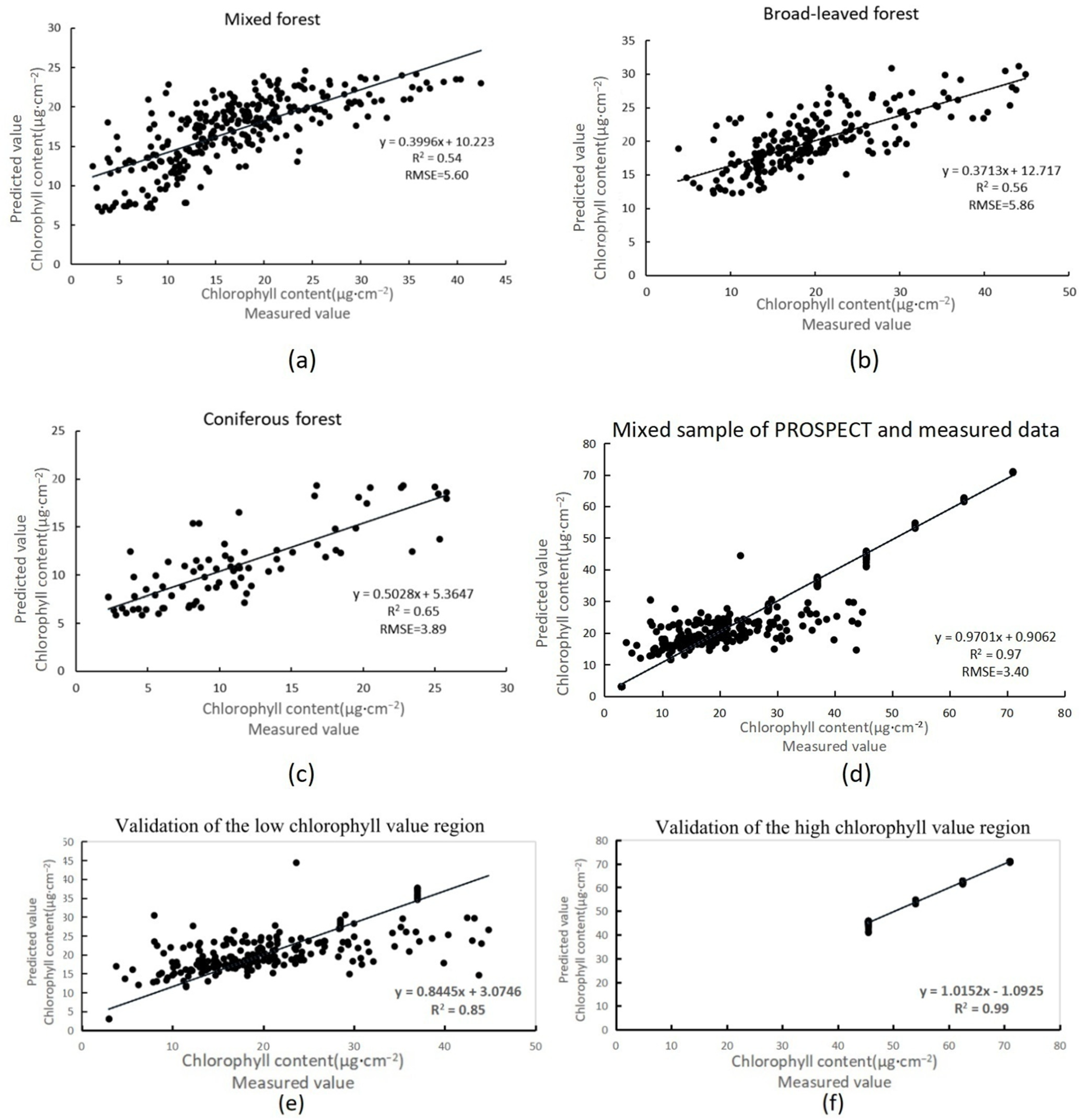
Compared with the traditional time-consuming, labor-intensive, and destructive methods for obtaining forest chlorophyll content, the remote sensing technology of UAVs equipped with hyperspectral sensors and LiDAR shortens the period of data acquisition. In this study, four statistical models were used to estimate the canopy chlorophyll content for three different stand types: coniferous forest, broad-leaved forest, and mixed coniferous broad-leaved forest. The accuracy of the nonlinear random forest model was generally higher than that of the BP neural network, multiple stepwise regression model, and BP neural network optimized by the firefly algorithm. This is because random forests can provide different interpretations of decision trees, resulting in better performance of the model. And the accuracy of the BP neural network model optimized by the firefly algorithm was better than that of the BP neural network. Neural networks require more data to be truly effective, as they can disrupt the interpretability of features. However, compared with the BP neural network, the optimized algorithm can seek the optimal solution of model parameters [
50] and pass the optimal parameters to the BP neural network for model construction. It was also found that the accuracy of the mechanism model was excellent, because the physical model could simulate leaf chlorophyll content data under different conditions in nature by inputting complex parameters [
22], and the accuracy of the model was less affected by spatial region and time factors. Compared with the statistical model, the mechanism model has higher robustness.
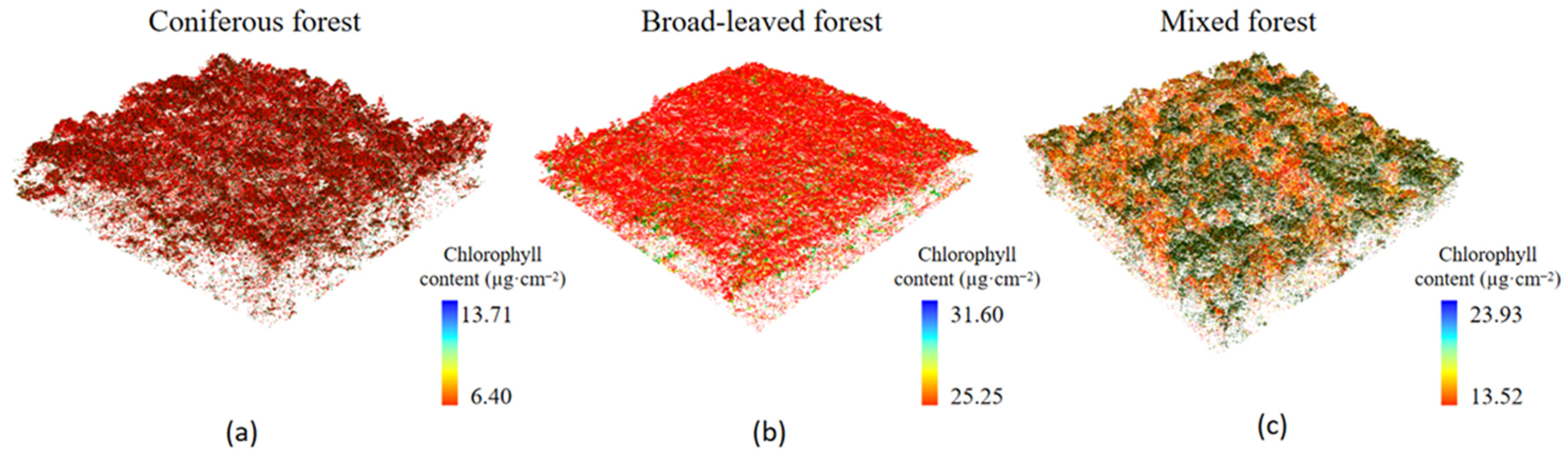
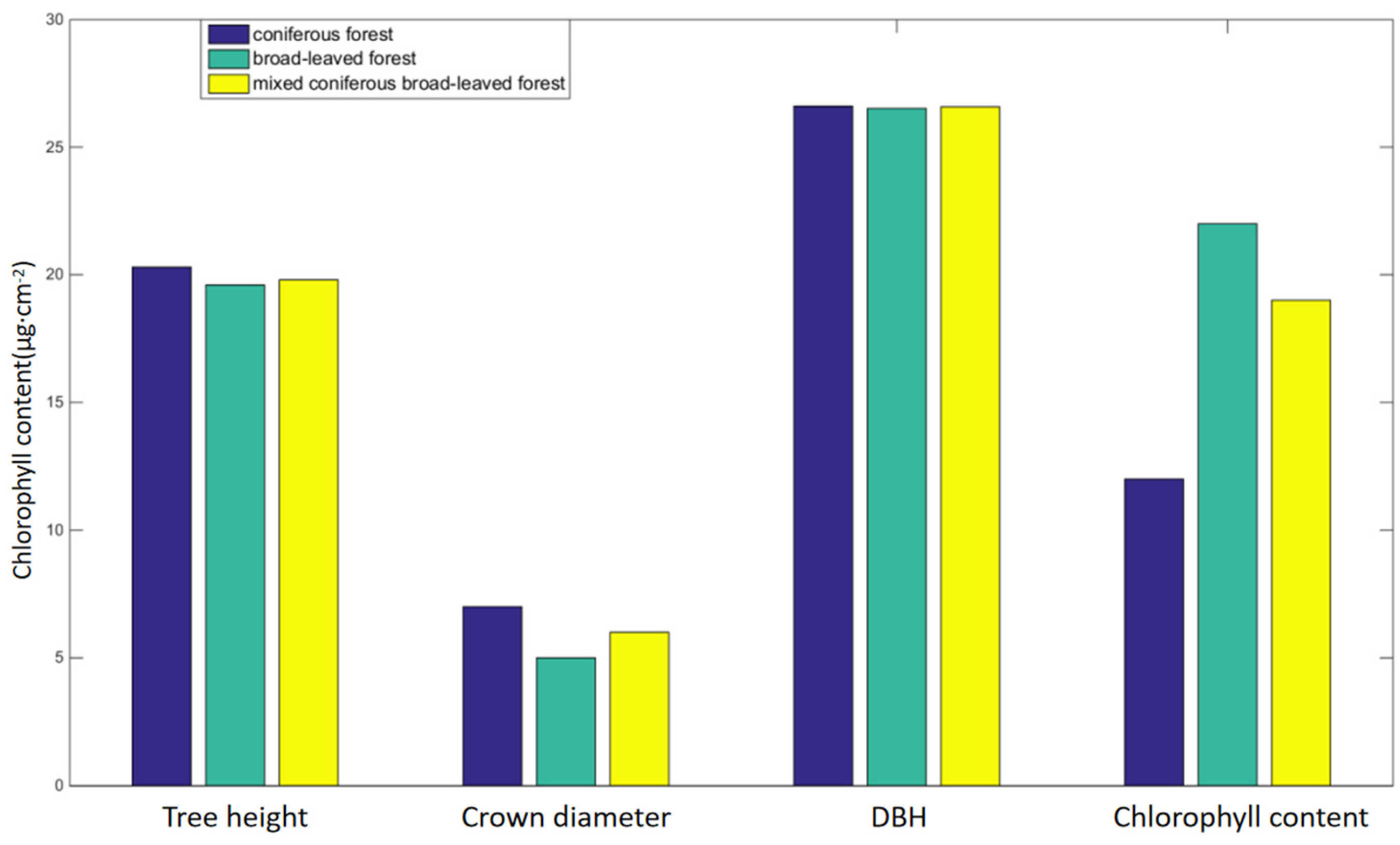
The results of chlorophyll content estimation are significantly different among different stand types. The chlorophyll content is influenced by factors such as light, temperature, moisture, and mineral elements, among which light is a necessary condition for chlorophyll. Under the condition of shading, the relative content of light-collecting pigment protein in the photosynthetic unit increased, which leads to an increase in binding chlorophyll. At the same time, it reduces the degradation and photo-oxidation of chlorophyll; therefore, the content of chlorophyll will increase after shading [
54]. When studying the shading rate of trees, Wang concluded that the diameter at breast height and crown width of most tree species were significantly correlated with the shading rate, and most of them were positively correlated [
55]. It can be seen from
Figure 14 that the broad-leaved forest has the highest chlorophyll content, and the coniferous forest has the lowest. The reason is that the largest crown width is in broad-leaved forests, and the smallest is in coniferous forests. Meanwhile, the largest chest height is in broad-leaved forests, and the smallest is in coniferous forests. Therefore, the forest with the highest shading rate is the broad-leaved forest, and the one with the lowest is the coniferous forest. Studies show that tree height mainly determines the shading range of trees with small solar altitude angles in the morning and evening and has little influence during the main periods of photosynthesis [
55]. Therefore, even if the tallest trees are in coniferous forests, and the smallest are in broad-leaved forests, it will not have a significant impact on chlorophyll content. In this study, there is little difference in DBH size among the three tree species, so the main influencing factor is crown diameter.
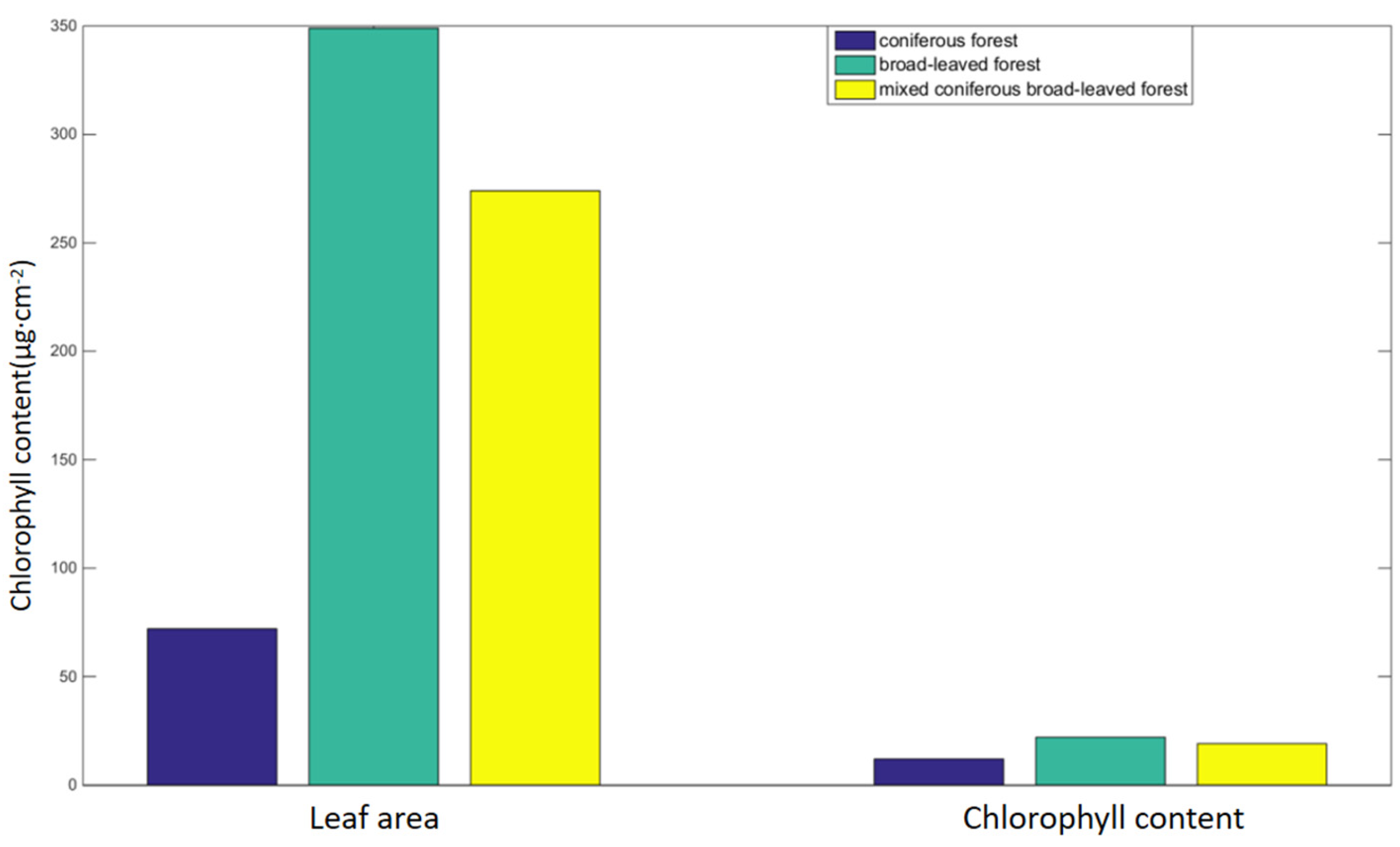
In addition, the leaf shape of trees can also affect chlorophyll content. The common leaf types of coniferous forests are narrow, mostly strip-shaped, and needle-shaped, while those of broad-leaved forests are wide, mostly round, and oval. The leaves of broad and flat broad-leaved trees contain many mesophyll tissues composed of chloroplast parenchyma cells, and the mesophyll tissues are large in quantity and volume, while the long and narrow coniferous mesophyll tissues are folded from green parenchyma tissues, and the number and volume of mesophyll tissues are smaller than those of broad-leaved leaves. According to the research, the larger the leaf area, the more light sources can be obtained by the leaves of plants, which can improve their photosynthetic capacity and correspondingly increase the chlorophyll content [
56]. As shown in
Figure 15, the size of leaf area in order is broad-leaved forest, mixed coniferous and broad-leaved forest, and coniferous forest. Therefore, the size of photosynthetic area (number and volume of mesophyll tissue) in order is broad-leaved forest, mixed coniferous and broad-leaved forest, and coniferous forest, and the corresponding chlorophyll content in order is broad-leaved forest, mixed coniferous and broad-leaved forest, and coniferous forest.
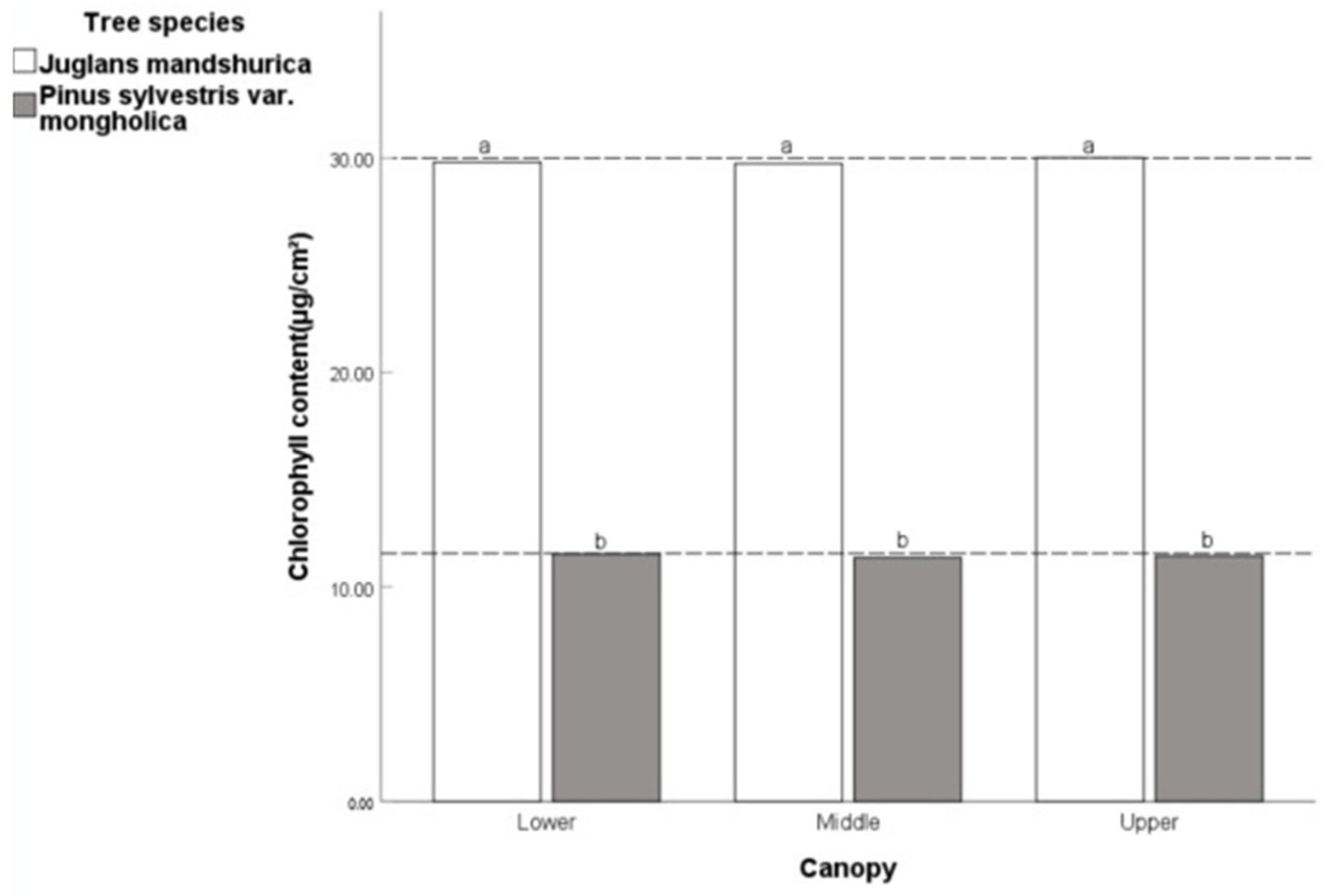
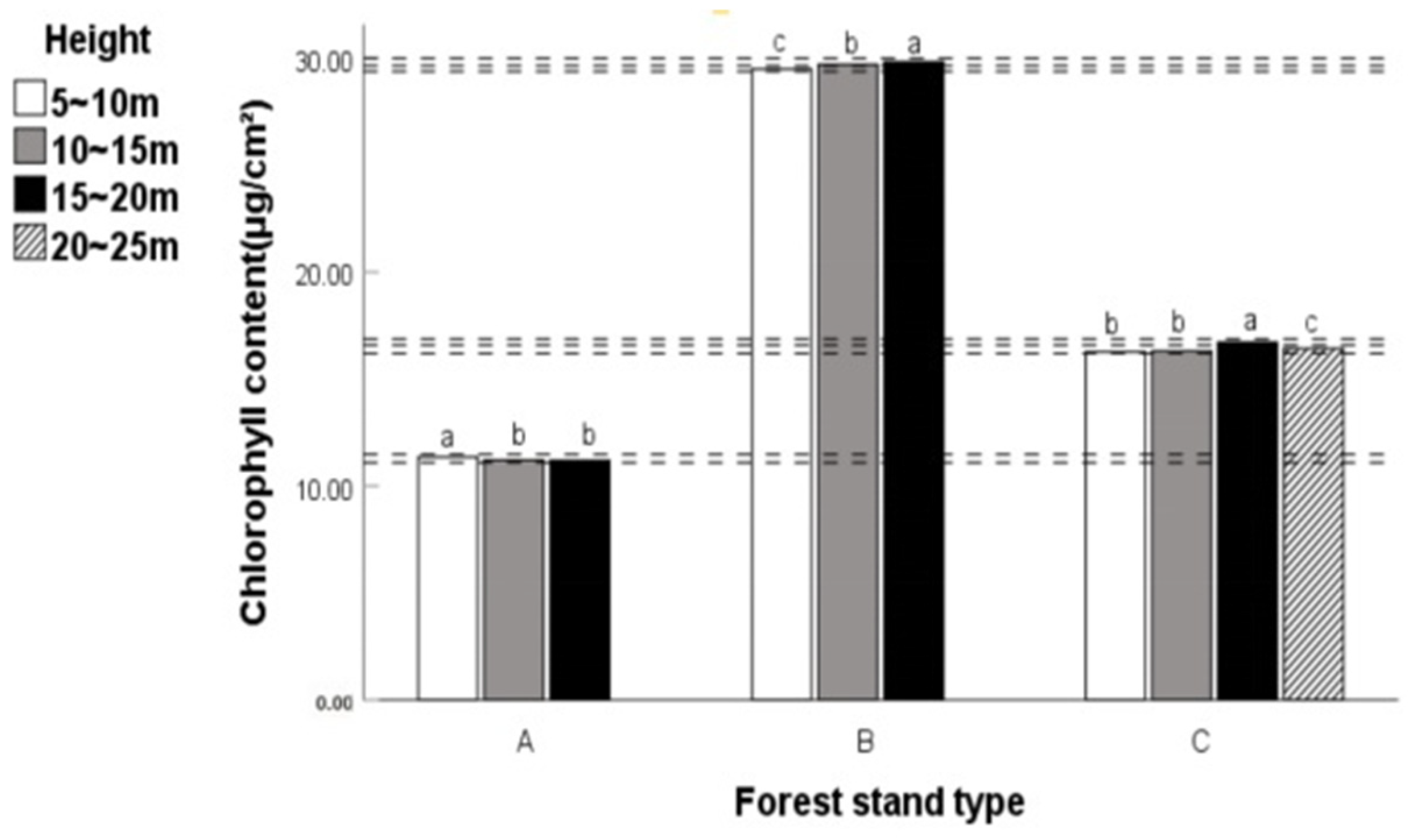
Some researchers found that plants can maximize their light utilization efficiency by using an open crown when studying the impact of plant light utilization capacity on tree architecture [
57]. As the sampling height increases, the canopy opening (
p < 0.01) of coniferous forests significantly decreases, and correspondingly, the utilization efficiency of light by leaves also decreases. Therefore, the chlorophyll content of coniferous forests decreases with the increase in height. As the sampling height increases, the crown opening of broad-leaved forests (
p < 0.05) significantly increases, and the crown opening of coniferous and broad-leaved mixed forests increases. Correspondingly, the utilization efficiency of light by leaves also increases. Therefore, the chlorophyll content of broad-leaved forests and coniferous and broad-leaved mixed forests increases with the increase in height. On the other hand, due to the sparse light that can directly illuminate the lower leaves in coniferous forests, the environmental temperature in which the leaves are located decreases with height. Leaves living in low temperatures increase their cold resistance by reducing the water content in their cells. The reduction in water inhibits the synthesis of chlorophyll, so the chlorophyll content of coniferous forests decreases with the increase in height. On the contrary, broad-leaved forests are often dense, and the light cannot reach the lower leaves, which leads to lower ambient temperatures in the lower leaves, less water in the leaf cells, and correspondingly less chlorophyll content than the upper leaves. The leaves of the broad-leaved tree species in the top layer of the coniferous and broad-leaved mixed forest are exposed to high levels of light radiation, resulting in more water evaporation and being in a water-deficient environment. Therefore, the leaves of broad-leaved tree species can avoid water loss by reducing their light-receiving area. On the other hand, the cuticle layer of the leaves at each level of coniferous tree species is thicker, which can effectively reduce water evaporation. Therefore, there will not be significant differences in the light-receiving area of the leaves at each level [
56]. This also explains why the chlorophyll content of leaves in coniferous and broad-leaved mixed forests at a height of 20~25 m is lower than that at a height of 15~20 m.
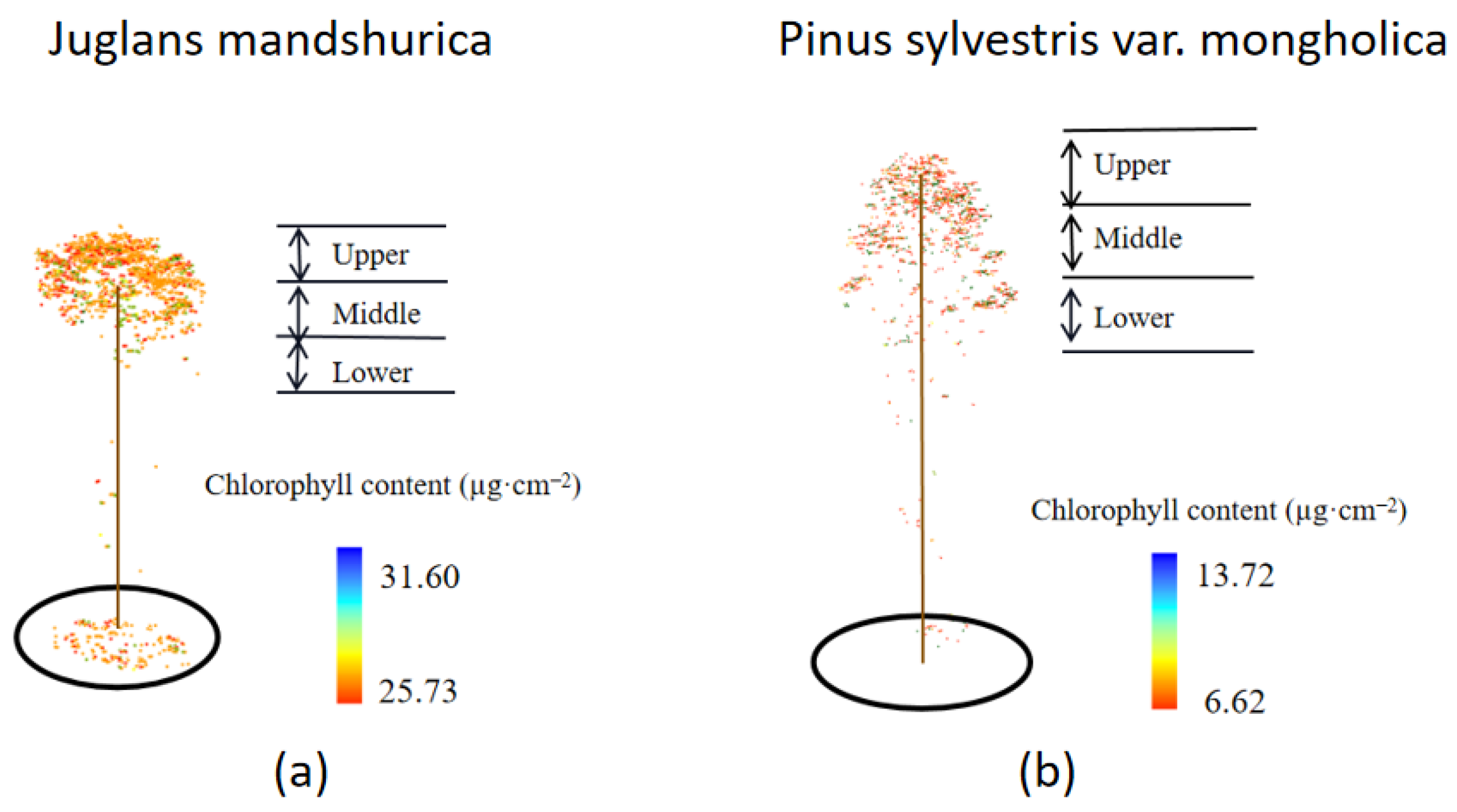
There are differences in the distribution of chlorophyll content in the canopy of different tree species at different levels and heights. The chlorophyll content in the upper layer of
Juglans mandshurica is higher than that in the middle and lower layers, while the chlorophyll content in the upper layer of
Pinus sylvestris var.
mongolica is lower than that in the lower layer. The main reason is that the complex spatial structure of the forest and the difference in the crown shape of the trees themselves lead to uneven distribution of light resources [
15]. The limited spatial position makes the trees compete fiercely for light resources [
1,
2]. The pinnacle crown of
Pinus sylvestris var.
mongolica makes the light be mainly distributed in the middle and upper layers, and the lower layer can only get a little light. In order to capture more light energy, the leaves of the lower layer can only increase their chlorophyll content for more photosynthesis. The umbrella-shaped crown of
Juglans mandshurica makes the leaves concentrate on the upper part of the canopy. As the canopy is divided from the first live branch of the trunk upwards, the number of upper leaves is higher than that of the middle and lower layers when dividing the canopy equally. Coupled with the spatial competition among trees, the number of leaves in the lower layer decreases sharply during the self-thinning process of trees. Secondly, it is difficult for light to penetrate the leaves and branches to reach the middle and lower layers [
29] because of the cover of the upper broad-leaved leaves, which reduces the resource allocation of middle and lower layer leaves and leads to a decrease in chlorophyll content in the middle and lower layers. The chlorophyll content of
Juglans mandshurica and
Pinus sylvestris var.
mongolica in the horizontal direction of a single tree is significantly different in the shady and sunny directions, which further shows that the uneven distribution of light energy in the shady and sunny directions affects the chlorophyll distribution.
However, the models studied in this paper are all limited, and all models are based on certain assumptions and simplifications, which may not be completely valid in the actual estimation. This leads to errors in the estimates. At the same time, ecological complexity will also affect the accuracy of the chlorophyll estimation model.
The chlorophyll content can indicate the status of forest productivity [
4,
5]. The fluctuation of chlorophyll content in coniferous and broad-leaved mixed forests is greater than that in coniferous and broad-leaved forests, indicating that coniferous and broad-leaved mixed forests can adapt well to changes in light and make more full use of light energy. Simultaneously increasing the solidification rate of carbon and enhancing forest productivity. The shortcoming of the study is that the effects of climate change on the chlorophyll content distribution of different stands are not considered. In the future, the effects of climate change on the spatial distribution of chlorophyll content of stands can be evaluated by measuring the chlorophyll content of sample trees by setting plots in different climatic zones.