Estimating Urban Forests Biomass with LiDAR by Using Deep Learning Foundation Models
Abstract
1. Introduction
2. Materials and Method
2.1. Datasets
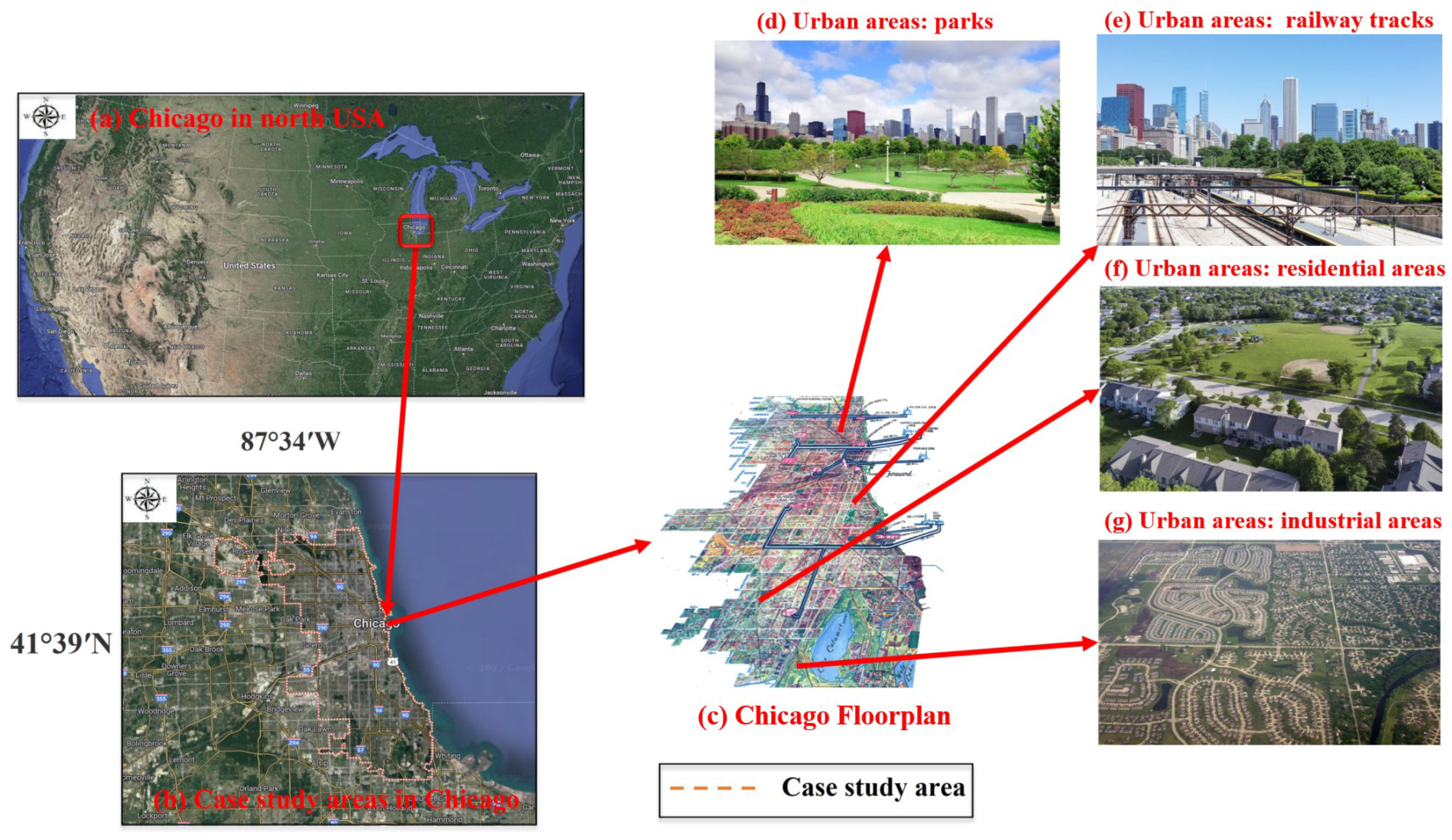
2.1.1. Data Collection
2.1.2. Data Preparation
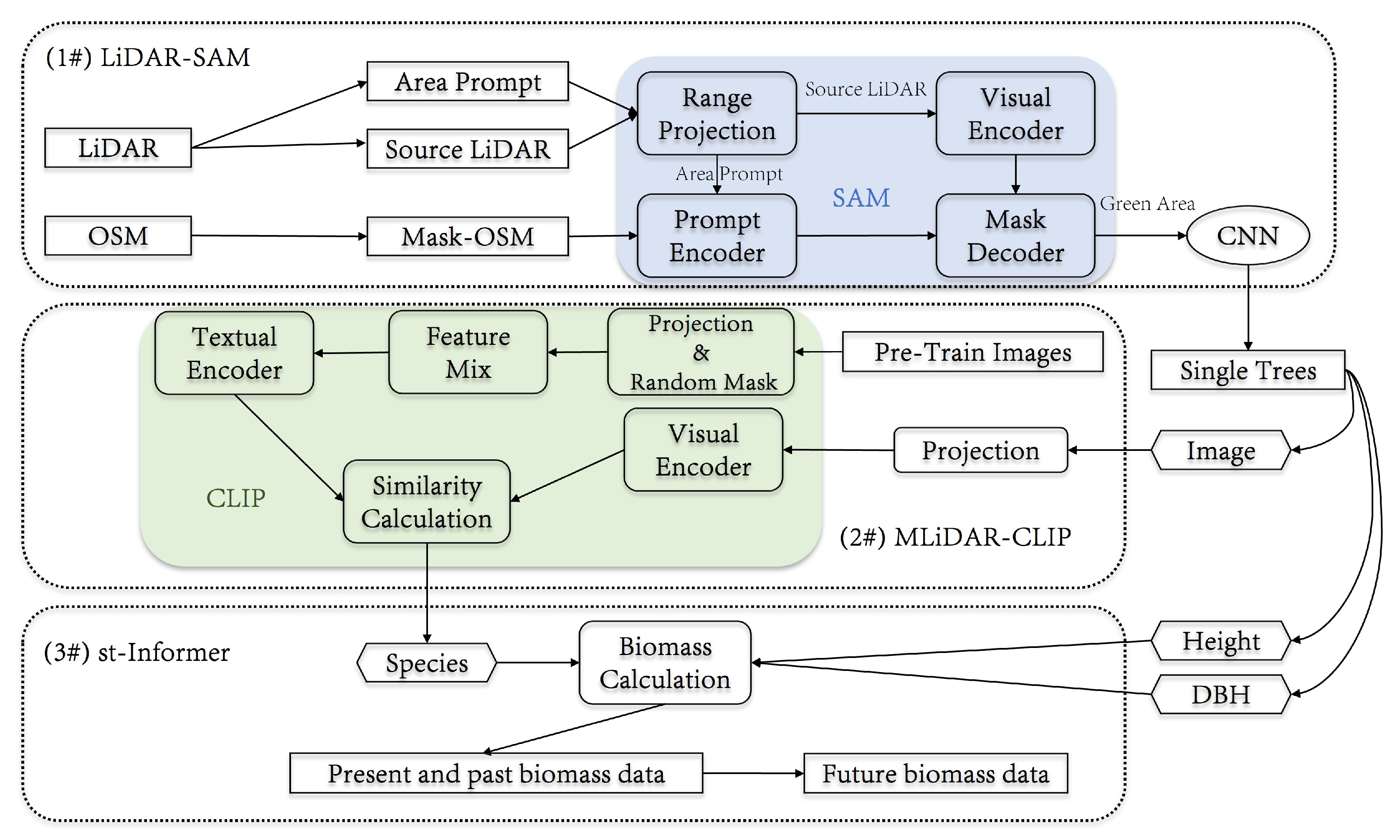
2.2. 3D-CiLBE
2.2.1. The Framework of 3D-CiLBE
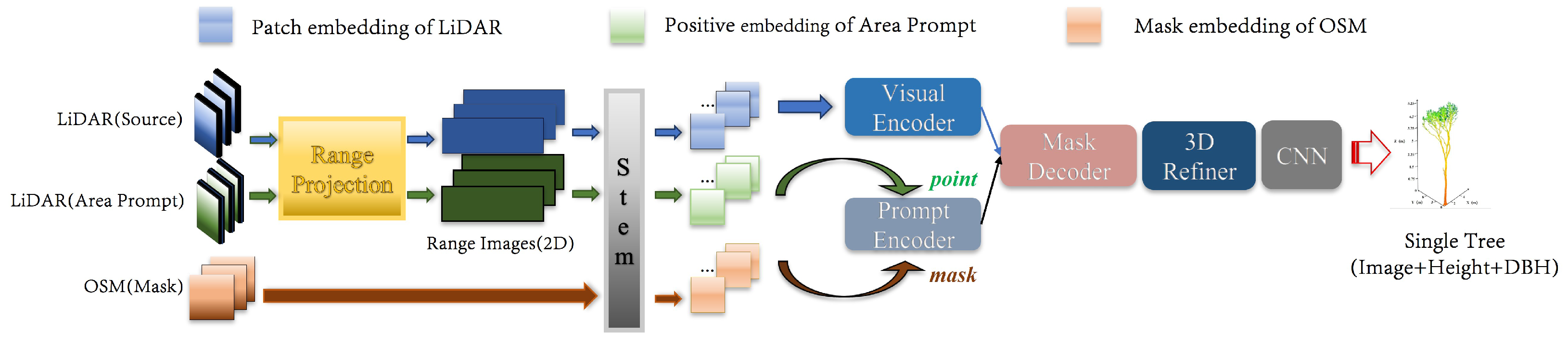
2.2.2. LiDAR-SAM
2.2.3. MLiDAR-CLIP
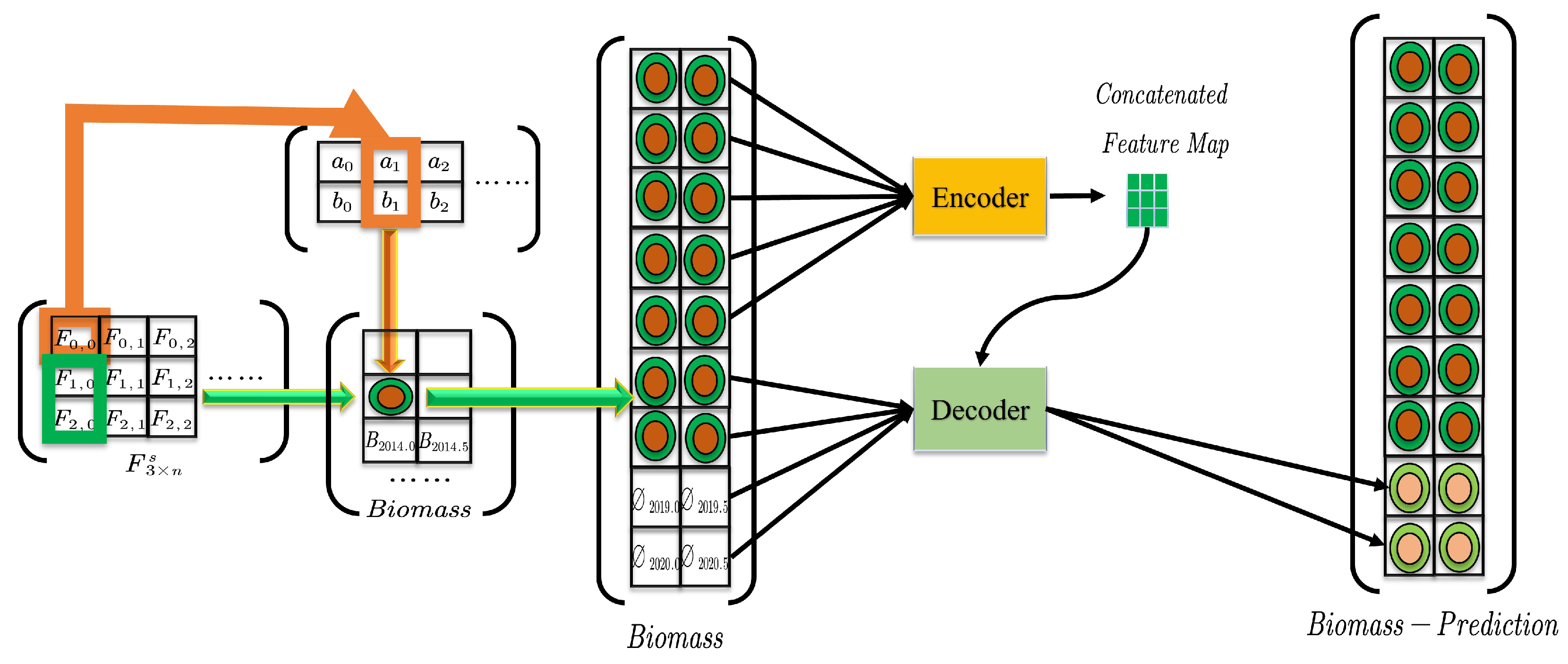
2.2.4. St-Informer
3. Results and Discussions
3.1. Experiment Setup
3.1.1. Experimental Configurations
3.1.2. Metrics
3.2. Comparative Experiments
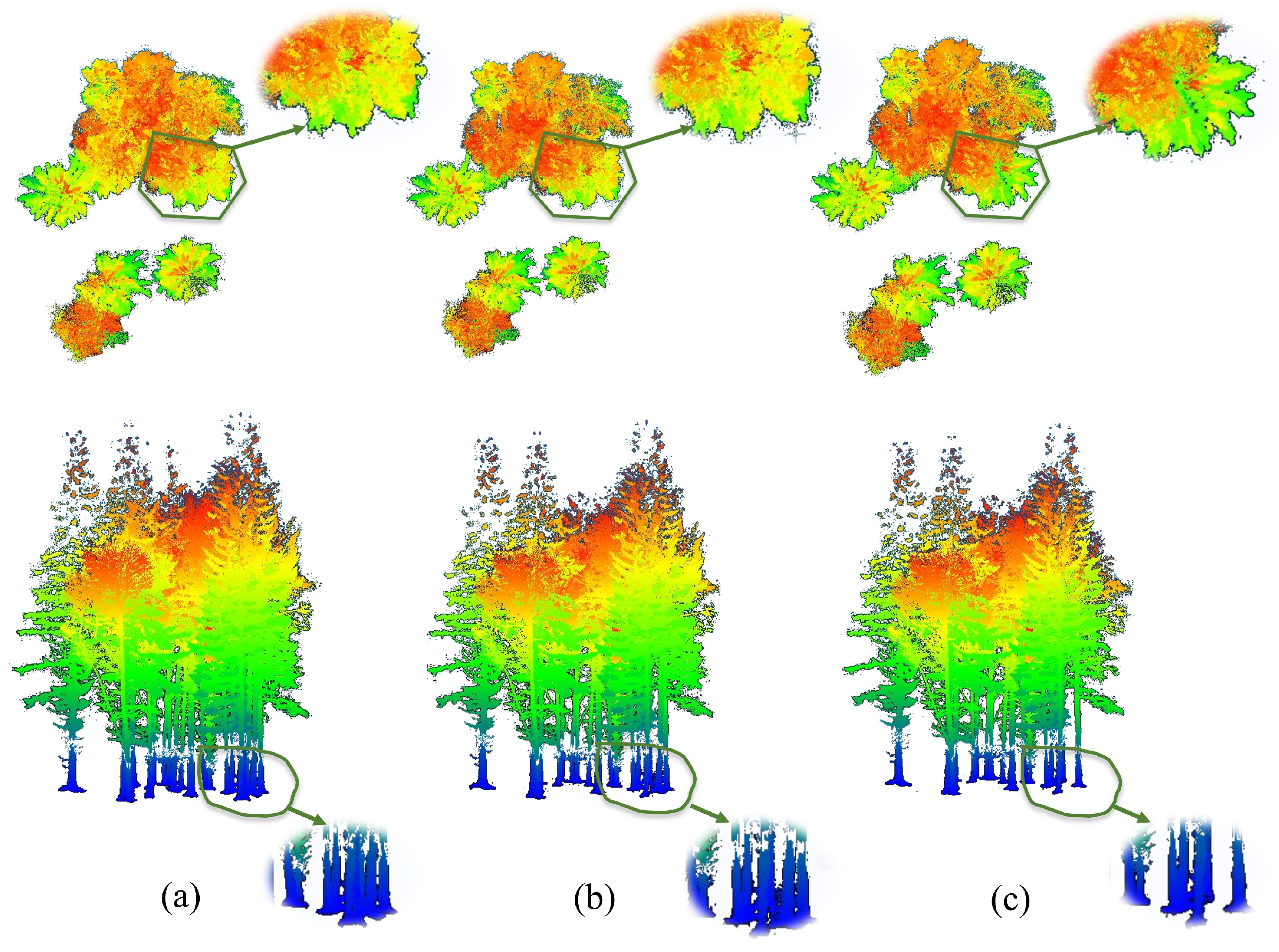
3.2.1. LiDAR-SAM
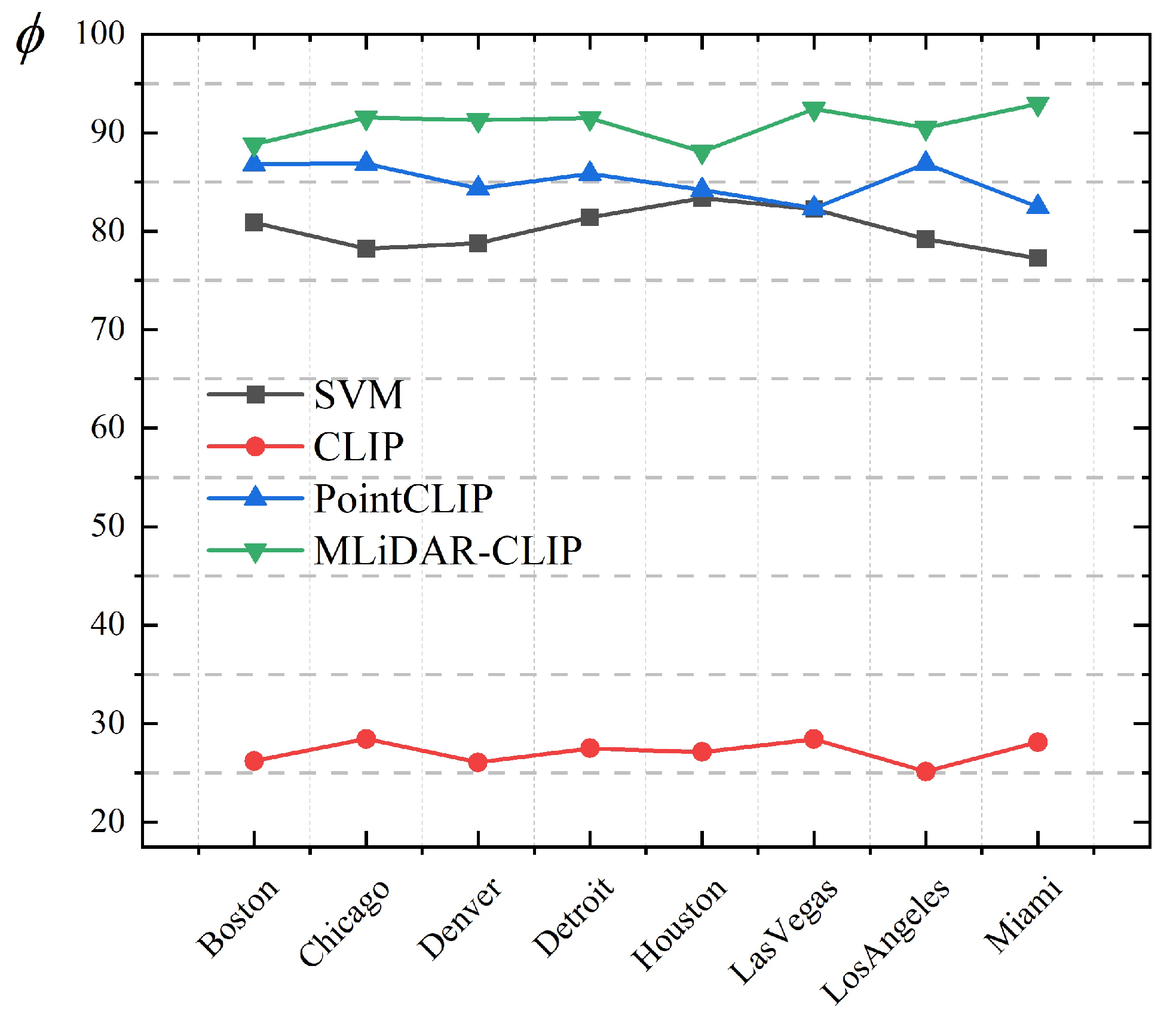
3.2.2. MLiDAR-CLIP
3.2.3. Data Dimensions
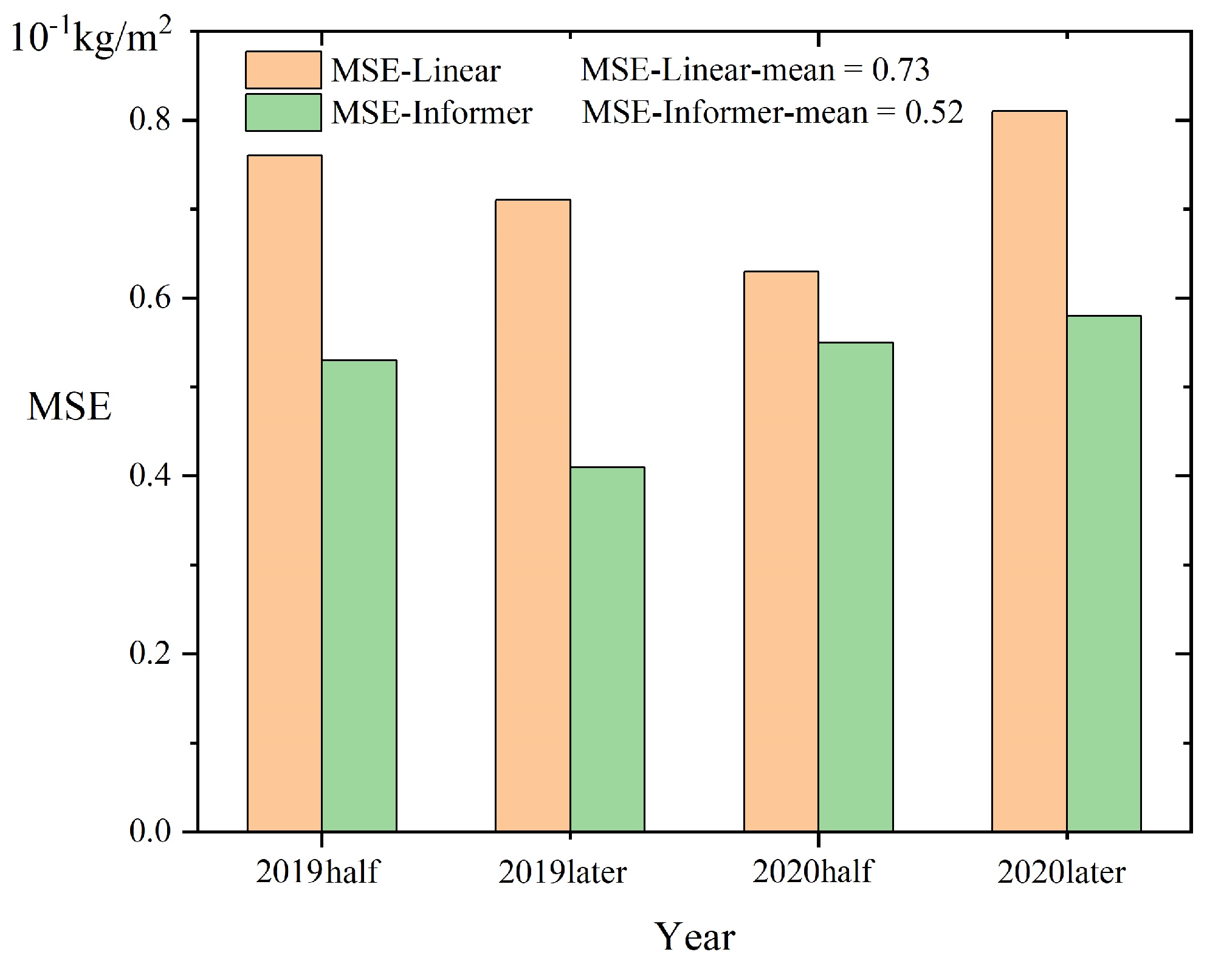
3.2.4. St-Informer
3.3. Ablation Experiments
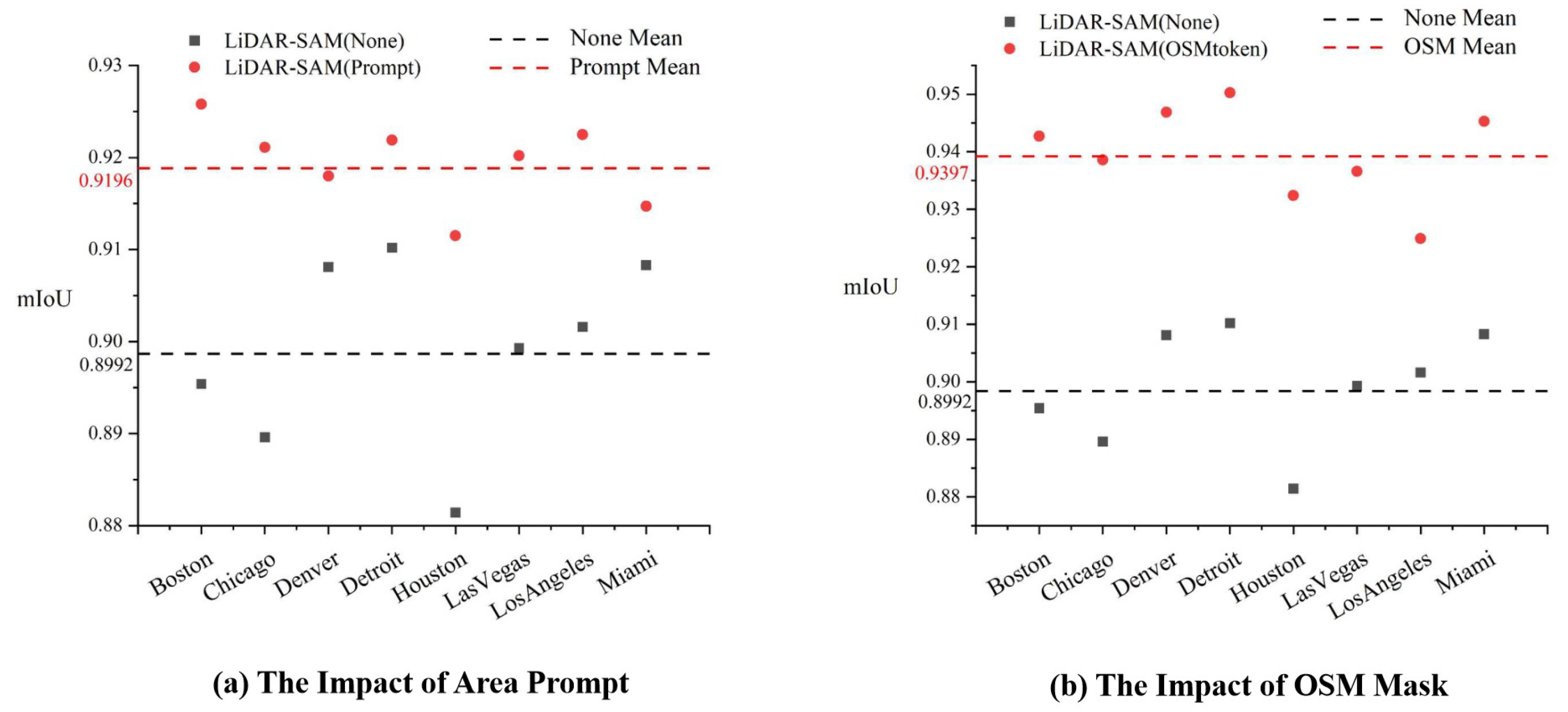
3.3.1. Area Prompt and OSM Mask
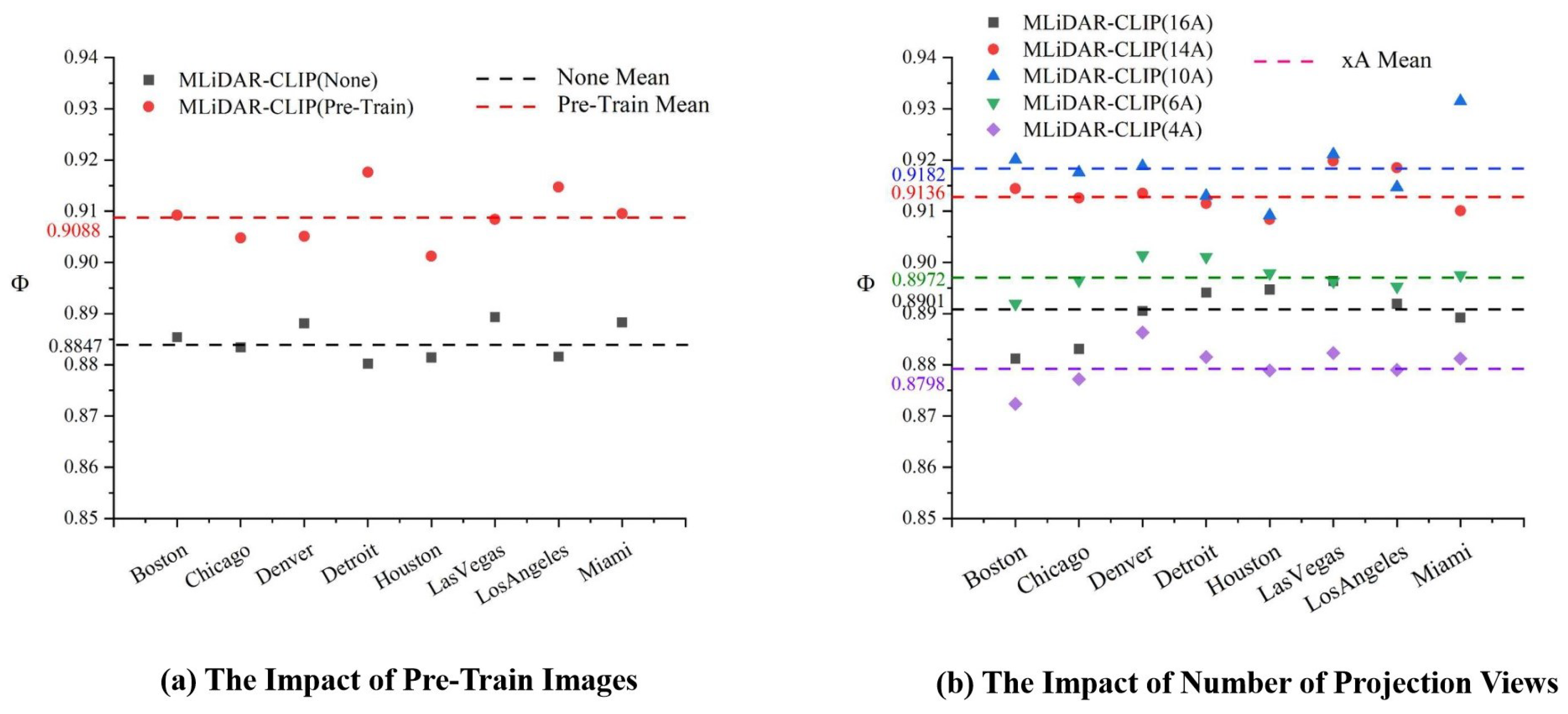
3.3.2. Pre-Training Images Feature Analysis and Number of 3D Projection Viewpoints
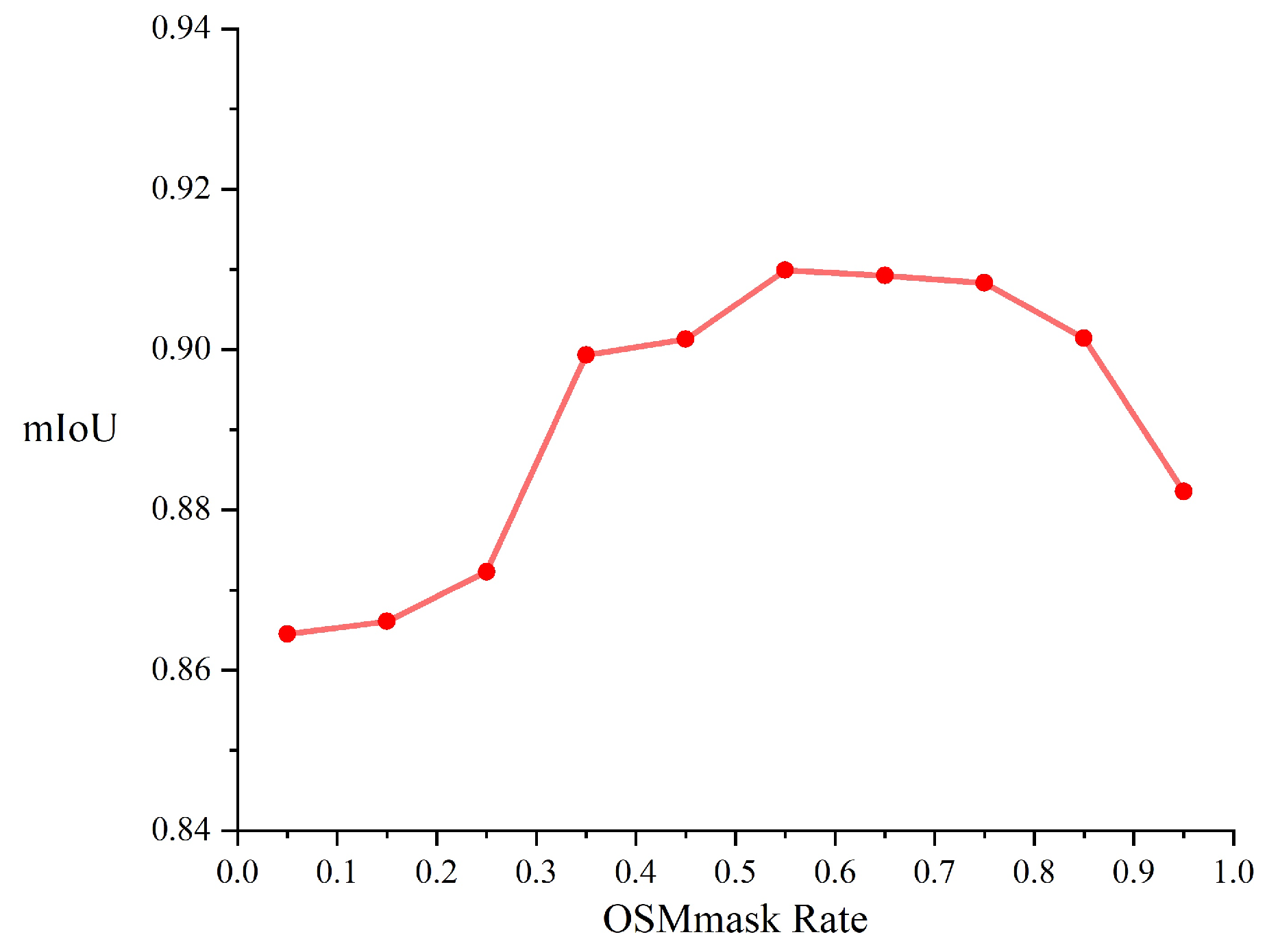
3.3.3. Value of OSMmask Rate
3.4. Case Study
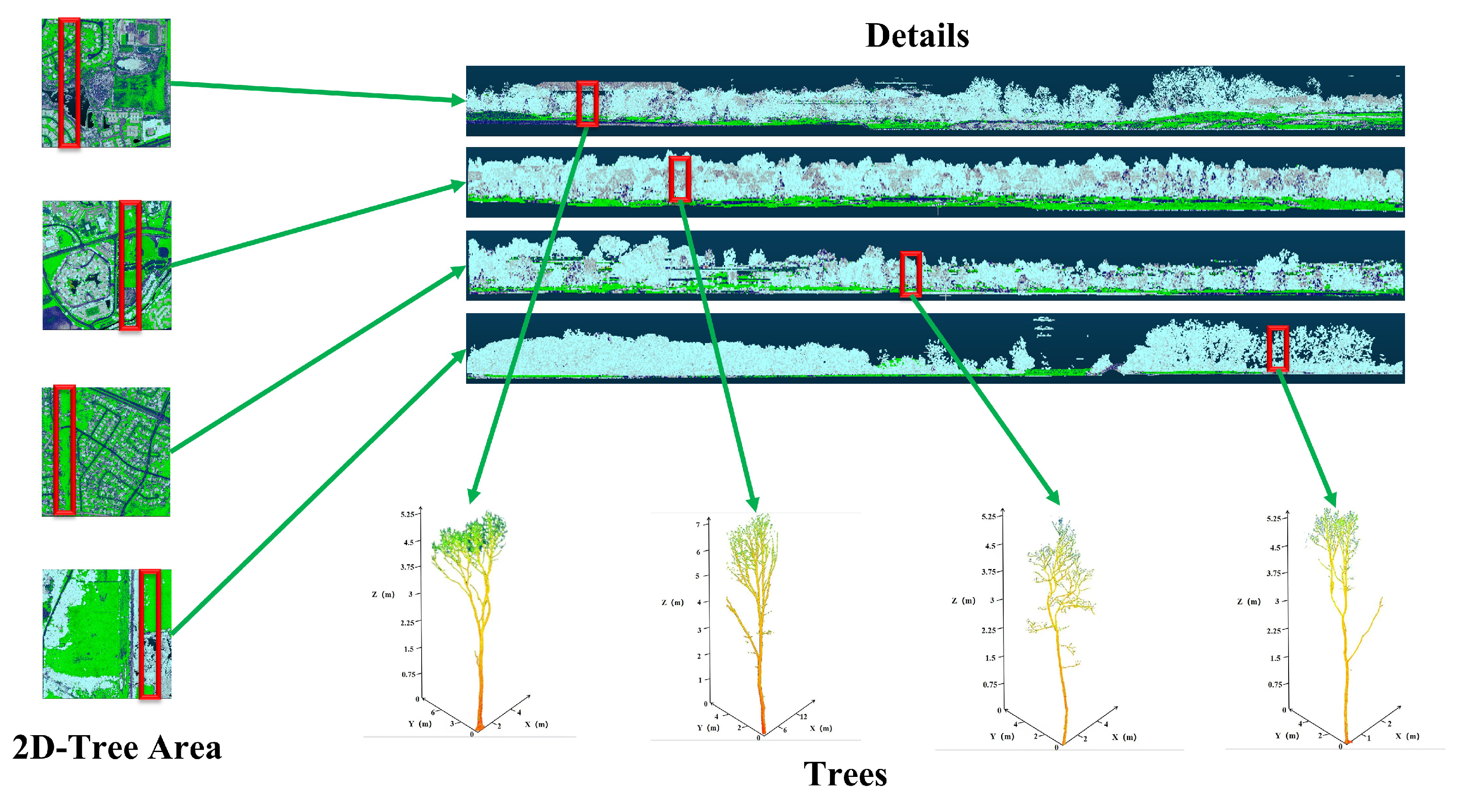
3.4.1. Data Visualization
3.4.2. Biomass Calculation and Error Analysis
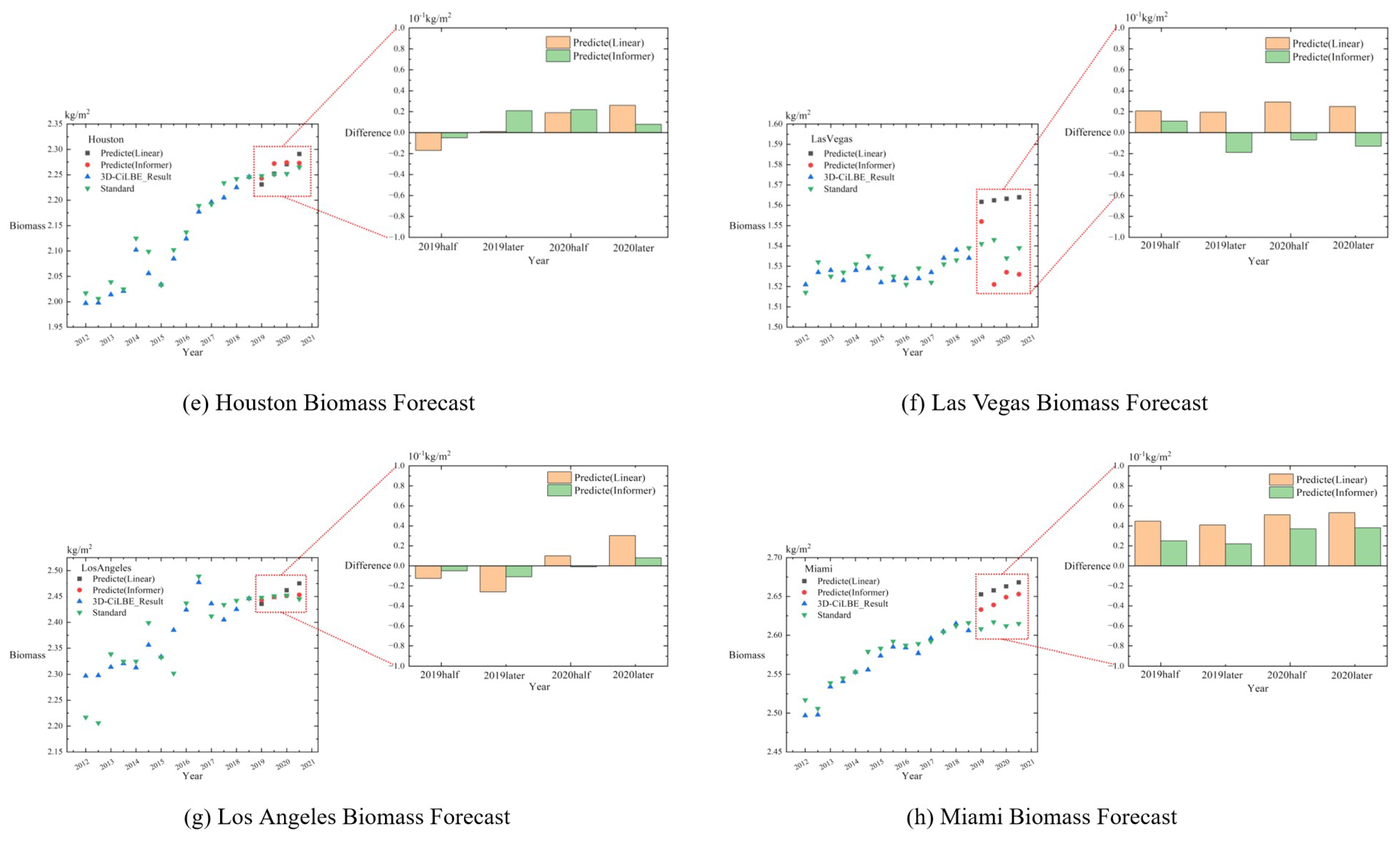
3.4.3. Predictive Analysis
3.5. Computational Resource Cost Analysis
3.6. Limitation and Scalability Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tozer, L.; Klenk, N. Urban configurations of carbon neutrality: Insights from the Carbon Neutral Cities Alliance. Environ. Plan. C Politics Space 2019, 37, 539–557. [Google Scholar] [CrossRef]
- Cao, J.; Situ, S.; Hao, Y.; Xie, S.; Li, L. Enhanced summertime ozone and SOA from biogenic volatile organic compound (BVOC) emissions due to vegetation biomass variability during 1981–2018 in China. Atmos. Chem. Phys. 2022, 22, 2351–2364. [Google Scholar] [CrossRef]
- Zhang, Y.; Shao, Z. Assessing of urban vegetation biomass in combination with LiDAR and high-resolution remote sensing images. Int. J. Remote Sens. 2021, 42, 964–985. [Google Scholar] [CrossRef]
- Chao, M.; Maimai, W.; Hanzhang, L.; Zhibo, C.; Xiaohui, C. A Spatio-Temporal Neural Network Learning System for City-Scale Carbon Storage Capacity Estimating. IEEE Access 2023, 11, 31304–31322. [Google Scholar] [CrossRef]
- Lawrence, A.; De Vreese, R.; Johnston, M.; Van Den Bosch, C.C.K.; Sanesi, G. Urban forest governance: Towards a framework for comparing approaches. Urban For. Urban Green. 2013, 12, 464–473. [Google Scholar] [CrossRef]
- Chandra, L.; Gupta, S.; Pande, V.; Singh, N. Impact of forest vegetation on soil characteristics: A correlation between soil biological and physico-chemical properties. 3 Biotech 2016, 6, 188. [Google Scholar] [CrossRef] [PubMed]
- Haq, S.M.; Calixto, E.S.; Rashid, I.; Khuroo, A.A. Human-driven disturbances change the vegetation characteristics of temperate forest stands: A case study from Pir Panchal mountain range in Kashmir Himalaya. Trees For. People 2021, 6, 100134. [Google Scholar] [CrossRef]
- Su, Y.; Wu, J.; Zhang, C.; Wu, X.; Li, Q.; Liu, L.; Bi, C.; Zhang, H.; Lafortezza, R.; Chen, X. Estimating the cooling effect magnitude of urban vegetation in different climate zones using multi-source remote sensing. Urban Clim. 2022, 43, 101155. [Google Scholar] [CrossRef]
- Ertel, W. Introduction to Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Chazdon, R.L.; Guariguata, M.R. Natural regeneration as a tool for arge-scale forest restoration in the tropics:Prospects and challenges. Biotropica 2016, 48, 716–730. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y. A big-data-based recurrent neural network method for forest energy estimation. Sustain. Energy Technol. Assessments 2023, 55, 102910. [Google Scholar] [CrossRef]
- Linden, J.; Gustafsson, M.; Uddling, J.; Pleijel, H. Air pollution removal through deposition on urban vegetation: The importance of vegetation characteristics. Urban For. Urban Green. 2023, 81, 127843. [Google Scholar] [CrossRef]
- Lin, J.; Kroll, C.N.; Nowak, D.J.; Greenfield, E.J. A review of urban forest modeling: Implications for management and future research. Urban For. Urban Green. 2019, 43, 126366. [Google Scholar] [CrossRef]
- Pearlmutter, D.; Calfapietra, C.; Samson, R.; O’Brien, L.; Ostoić, S.K.; Sanesi, G.; del Amo, R.A. The urban forest. In Cultivating Green Infrastructure for People and the Environment; Springer: Berlin/Heidelberg, Germany, 2017; Volume 7. [Google Scholar]
- Ordóñez, C.; Duinker, P.N. An analysis of urban forest management plans in Canada: Implications for urban forest management. Landsc. Urban Plan. 2013, 116, 36–47. [Google Scholar] [CrossRef]
- Dahar, D.; Handayani, B.; Mardikaningsih, R. Urban Forest: The role of improving the quality of the urban environment. Bull. Sci. Technol. Soc. 2022, 1, 25–29. [Google Scholar]
- Fitzky, A.C.; Sandén, H.; Karl, T.; Fares, S.; Calfapietra, C.; Grote, R.; Saunier, A.; Rewald, B. The interplay between ozone and urban vegetation—BVOC emissions, ozone deposition, and tree ecophysiology. Front. For. Glob. Chang. 2019, 2, 50. [Google Scholar] [CrossRef]
- Shi, L.; Liu, S. Methods of estimating forest biomass: A review. Biomass Vol. Estim. Valorization Energy 2017, 10, 65733. [Google Scholar]
- Chave, J.; Réjou-Méchain, M.; Búrquez, A.; Chidumayo, E.; Colgan, M.S.; Delitti, W.B.; Duque, A.; Eid, T.; Fearnside, P.M.; Goodman, R.C.; et al. Improved allometric models to estimate the aboveground biomass of tropical trees. Glob. Chang. Biol. 2014, 20, 3177–3190. [Google Scholar] [CrossRef] [PubMed]
- Monzingo, D.S.; Shipley, L.A.; Cook, R.C.; Cook, J.G. Factors influencing predictions of understory vegetation biomass from visual cover estimates. Wildl. Soc. Bull. 2022, 46, e1300. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L. Artificial intelligence for remote sensing data analysis: A review of challenges and opportunities. IEEE Geosci. Remote Sens. Mag. 2022, 10, 270–294. [Google Scholar] [CrossRef]
- Janga, B.; Asamani, G.P.; Sun, Z.; Cristea, N. A Review of Practical AI for Remote Sensing in Earth Sciences. Remote Sens. 2023, 15, 4112. [Google Scholar] [CrossRef]
- Demagistri, L.; Mitja, D.; Delaître, E.; Yazdanparast, E.; Shahbazkia, H.; Petit, M. Palm-trees detection with very high resolution images, comparison between Geoeye and Pléiades sensors. In Pléiades Days; HAL: Lyon, France, 2014. [Google Scholar]
- Lacerda, T.H.S.; Cabacinha, C.D.; Araújo, C.A.; Maia, R.D.; Lacerda, K.W.d.S. Artificial neural networks for estimating tree volume in the Brazilian savanna. Cerne 2017, 23, 483–491. [Google Scholar] [CrossRef]
- Kakogeorgiou, I.; Karantzalos, K. Evaluating explainable artificial intelligence methods for multi-label deep earning classification tasks in remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102520. [Google Scholar]
- Corina, D.; Singleton, J. Developmental social cognitive neuroscience: Insights from deafness. Child Dev. 2009, 80, 952–967. [Google Scholar] [CrossRef] [PubMed]
- Minghini, M.; Frassinelli, F. OpenStreetMap history for intrinsic quality assessment: Is OSM up-to-date? Open Geospat. Data Softw. Stand. 2019, 4, 9. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- Huo, Y.; Zhang, M.; Liu, G.; Lu, H.; Gao, Y.; Yang, G.; Wen, J.; Zhang, H.; Xu, B.; Zheng, W.; et al. WenLan: Bridging vision and anguage by arge-scale multi-modal pre-training. arXiv 2021, arXiv:2103.06561. [Google Scholar]
- Zhou, H.; Zhang, S.; Peng, J.; Zhang, S.; Li, J.; Xiong, H.; Zhang, W. Informer: Beyond efficient transformer for ong sequence time-series forecasting. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtually, 2–9 February 2021; Volume 35, pp. 11106–11115.
- Zou, R.; Duan, Y.; Wang, Y.; Pang, J.; Liu, F.; Sheikh, S.R. A novel convolutional informer network for deterministic and probabilistic state-of-charge estimation of ithium-ion batteries. J. Energy Storage 2023, 57, 106298. [Google Scholar] [CrossRef]
- Li, W.; Fu, H.; Han, Z.; Zhang, X.; Jin, H. Intelligent tool wear prediction based on Informer encoder and stacked bidirectional gated recurrent unit. Robot. Comput.-Integr. Manuf. 2022, 77, 102368. [Google Scholar] [CrossRef]
- Lee, J.S.; Hsiang, J. Patent claim generation by fine-tuning OpenAI GPT-2. World Pat. Inf. 2020, 62, 101983. [Google Scholar] [CrossRef]
- Seidel, D. Single Tree Point Clouds from Terrestrial aser Scanning. 2020. Available online: https://data.goettingen-research-online.de/dataset.xhtml?persistentId=doi:10.25625/FOHUJM (accessed on 4 May 2024).
- Zeybek, M.; Şanlıoğlu, İ. Point cloud filtering on UAV based point cloud. Measurement 2019, 133, 99–111. [Google Scholar] [CrossRef]
- Milioto, A.; Vizzo, I.; Behley, J.; Stachniss, C. Rangenet++: Fast and accurate idar semantic segmentation. In Proceedings of the 2019 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Macau, China, 3–8 November 2019; pp. 4213–4220. [Google Scholar]
- Shi, L.; Wang, G.; Mo, L.; Yi, X.; Wu, X.; Wu, P. Automatic Segmentation of Standing Trees from Forest Images Based on Deep Learning. Sensors 2022, 22, 6663. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Zheng, X.; Fang, L. The Semantic Segmentation of Standing Tree Images Based on the Yolo V7 Deep Learning Algorithm. Electronics 2023, 12, 929. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Ma, W.; Deng, Y.; Pan, J.; Li, J. Vegetation Extraction from Airborne Laser Scanning Data of Urban Plots Based on Point Cloud Neighborhood Features. Forests 2023, 14, 691. [Google Scholar] [CrossRef]
- Griffiths, M.J.; Garcin, C.; van Hille, R.P.; Harrison, S.T. Interference by pigment in the estimation of microalgal biomass concentration by optical density. J. Microbiol. Methods 2011, 85, 119–123. [Google Scholar] [CrossRef]
- Poley, L.G.; McDermid, G. A systematic review of the factors influencing the estimation of vegetation aboveground biomass using unmanned aerial systems. Remote Sens. 2020, 12, 1052. [Google Scholar] [CrossRef]
- Yan, X.; Li, J.; Smith, A.R.; Yang, D.; Ma, T.; Su, Y.; Shao, J. Evaluation of machine earning methods and multi-source remote sensing data combinations to construct forest above-ground biomass models. Int. J. Digit. Earth 2023, 16, 4471–4491. [Google Scholar] [CrossRef]
- Huang, T.; Ou, G.; Wu, Y.; Zhang, X.; Liu, Z.; Xu, H.; Xu, X.; Wang, Z.; Xu, C. Estimating the Aboveground Biomass of Various Forest Types with High Heterogeneity at the Provincial Scale Based on Multi-Source Data. Remote Sens. 2023, 15, 3550. [Google Scholar] [CrossRef]
- Fararoda, R.; Reddy, R.S.; Rajashekar, G.; Chand, T.K.; Jha, C.S.; Dadhwal, V. Improving forest above ground biomass estimates over Indian forests using multi source data sets with machine earning algorithm. Ecol. Inform. 2021, 65, 101392. [Google Scholar] [CrossRef]
- Meng, B.; Ge, J.; Liang, T.; Yang, S.; Gao, J.; Feng, Q.; Cui, X.; Huang, X.; Xie, H. Evaluation of remote sensing inversion error for the above-ground biomass of alpine meadow grassland based on multi-source satellite data. Remote Sens. 2017, 9, 372. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Beier, C.M.; Johnson, L.; Phoenix, D.B.; Mahoney, M. Decision tree-based machine earning models for above-ground biomass estimation using multi-source remote sensing data and object-based image analysis. Geocarto Int. 2022, 37, 12763–12791. [Google Scholar] [CrossRef]
- Tang, Z.; Xia, X.; Huang, Y.; Lu, Y.; Guo, Z. Estimation of National Forest Aboveground Biomass from Multi-Source Remotely Sensed Dataset with Machine Learning Algorithms in China. Remote Sens. 2022, 14, 5487. [Google Scholar] [CrossRef]
- Yang, Q.; Niu, C.; Liu, X.; Feng, Y.; Ma, Q.; Wang, X.; Tang, H.; Guo, Q. Mapping high-resolution forest aboveground biomass of China using multisource remote sensing data. GISci. Remote Sens. 2023, 60, 2203303. [Google Scholar] [CrossRef]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature earning on point sets in a metric space. Adv. Neural Inf. Process. Syst. 2017, 30, 1–14. [Google Scholar]
- Yang, Y.; Wu, X.; He, T.; Zhao, H.; Liu, X. SAM3D: Segment Anything in 3D Scenes. arXiv 2023, arXiv:2306.03908. [Google Scholar]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narrandes, S.; Wang, Y.; Xu, W. Applications of support vector machine (SVM) earning in cancer genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar]


| Data Types | Data Source | Data Examples | ||
|---|---|---|---|---|
| LiDAR-Urban | USGS |  |  |  |
| OSM | OpenStreetMap |  |  |  |
| Single-Trees | GRO |  |  |  |
| Abbreviation | Meaning | Formula |
|---|---|---|
| OA | Overall accuracy | |
| Recall | Recall of goal areas | |
| Precision | Precision of goal areas | |
| IoU_P | Intersection over Union of goal areas | |
| IoU_N | Intersection over Union of other areas | |
| mIoU | mean Intersection over Union | |
| Kp | Kappa coefficient | |
| Species Recognition Accuracy | ||
| MSE | Mean Square Error | |
| RMSE | Root Mean Square Error | |
| R-Square |
| Function | IoU-P | mIoU | OA | Re | Kp |
|---|---|---|---|---|---|
| PointNet++ | 0.83 | 0.85 | 0.89 | 0.91 | 0.88 |
| SAM | 0.87 | 0.91 | 0.92 | 0.97 | 0.89 |
| LiDAR-SAM | 0.93 | 0.94 | 0.98 | 0.97 | 0.92 |
| City | 2D Method | 3D-CiLBE | Ground Truth | Difference-2D | Difference-3D | DR |
|---|---|---|---|---|---|---|
| Boston | 2.149 | 2.155 | 2.164 | 0.015 | 0.009 | 1.6 |
| Chicago | 1.809 | 1.826 | 1.821 | 0.012 | 0.005 | 2.5 |
| Denver | 1.942 | 1.947 | 1.955 | 0.013 | 0.008 | 1.7 |
| Detroit | 1.614 | 1.629 | 1.625 | 0.011 | 0.004 | 2.7 |
| Houston | 2.219 | 2.225 | 2.234 | 0.015 | 0.009 | 1.7 |
| Las Vegas | 1.521 | 1.534 | 1.531 | 0.010 | 0.003 | 3.4 |
| Los Angeles | 2.418 | 2.422 | 2.434 | 0.016 | 0.012 | 1.4 |
| Miami | 2.517 | 2.546 | 2.534 | 0.017 | 0.012 | 1.4 |
| Year | 3D-CiLBE (kg/m2) | Ground Truth (kg/m2) | Difference Value (kg/m2) |
|---|---|---|---|
| 2012half | 1.79345 | 1.83254 | 0.03909 |
| 2012later | 1.74563 | 1.80435 | 0.05872 |
| 2013half | 1.77353 | 1.82995 | 0.05642 |
| 2013later | 1.76094 | 1.79392 | 0.03298 |
| 2014half | 1.81359 | 1.80823 | −0.00536 |
| 2014later | 1.73976 | 1.78137 | 0.04161 |
| 2015half | 1.70273 | 1.77316 | 0.07043 |
| 2015later | 1.67354 | 1.74782 | 0.07428 |
| 2016half | 1.68739 | 1.74964 | 0.06225 |
| 2016later | 1.64993 | 1.71875 | 0.06882 |
| 2017half | 1.67947 | 1.72359 | 0.04412 |
| 2017later | 1.68358 | 1.70461 | 0.02103 |
| 2018half | 1.71935 | 1.71874 | −0.00061 |
| 2018later | 1.69357 | 1.70125 | 0.00768 |
| Method | Params (M) | GFLOPs |
|---|---|---|
| SAM(ViT-H) | 636 | 81.34 |
| SAM(ViT-L) | 308 | 39.39 |
| SAM(ViT-B) | 91 | 11.64 |
| CLIP(RN101) | 500 | 9.9 |
| CLIP(RN50 × 46) | 1600 | 265.9 |
| LiDAR-SAM | 14 | 2.51 |
| MLiDAR-CLIP | 12 | 1.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Mou, C.; Yuan, J.; Chen, Z.; Zhong, L.; Cui, X. Estimating Urban Forests Biomass with LiDAR by Using Deep Learning Foundation Models. Remote Sens. 2024, 16, 1643. https://doi.org/10.3390/rs16091643
Liu H, Mou C, Yuan J, Chen Z, Zhong L, Cui X. Estimating Urban Forests Biomass with LiDAR by Using Deep Learning Foundation Models. Remote Sensing. 2024; 16(9):1643. https://doi.org/10.3390/rs16091643
Chicago/Turabian StyleLiu, Hanzhang, Chao Mou, Jiateng Yuan, Zhibo Chen, Liheng Zhong, and Xiaohui Cui. 2024. "Estimating Urban Forests Biomass with LiDAR by Using Deep Learning Foundation Models" Remote Sensing 16, no. 9: 1643. https://doi.org/10.3390/rs16091643
APA StyleLiu, H., Mou, C., Yuan, J., Chen, Z., Zhong, L., & Cui, X. (2024). Estimating Urban Forests Biomass with LiDAR by Using Deep Learning Foundation Models. Remote Sensing, 16(9), 1643. https://doi.org/10.3390/rs16091643










